Volume 21 Supplement 9

Selected Articles from the 20th International Conference on Bioinformatics & Computational Biology (BIOCOMP 2019)
- Introduction
- Open access
- Published: 03 December 2020
Current trend and development in bioinformatics research
- Yuanyuan Fu 1 ,
- Zhougui Ling 1 , 2 ,
- Hamid Arabnia 3 &
- Youping Deng 1
BMC Bioinformatics volume 21 , Article number: 538 ( 2020 ) Cite this article
10k Accesses
19 Citations
4 Altmetric
Metrics details
This is an editorial report of the supplements to BMC Bioinformatics that includes 6 papers selected from the BIOCOMP’19—The 2019 International Conference on Bioinformatics and Computational Biology. These articles reflect current trend and development in bioinformatics research.
The supplement to BMC Bioinformatics was proposed to launch during the BIOCOMP’19—The 2019 International Conference on Bioinformatics and Computational Biology held from July 29 to August 01, 2019 in Las Vegas, Nevada. In this congress, a variety of research areas was discussed, including bioinformatics which was one of the major focuses due to the rapid development and requirement of using bioinformatics approaches in biological data analysis, especially for omics large datasets. Here, six manuscripts were selected after strict peer review, providing an overview of the bioinformatics research trend and its application for interdisciplinary collaboration.
Cancer is one of the leading causes of morbidity and mortality worldwide. There exists an urgent need to identify new biomarkers or signatures for early detection and prognosis. Mona et al. identified biomarker genes from functional network based on the 407 differential expressed genes between lung cancer and healthy populations from a public Gene Expression Omnibus dataset. The lower expression of sixteen gene signature is associated with favorable lung cancer survival, DNA repair, and cell regulation [ 1 ]. A new class of biomarkers such as alternative splicing variants (ASV) have been studied in recent years. Various platforms and methods, for example, Affymetrix Exon-Exon Junction Array, RNA-seq, and liquid chromatography tandem mass spectrometry (LC–MS/MS), have been developed to explore the role of ASV in human disease. Zhang et al. have developed a bioinformatics workflow to combine LC–MS/MS with RNA-seq which provide new opportunities in biomarker discovery. In their study, they identified twenty-six alternative splicing biomarker peptides with one single intron event and one exon skipping event; further pathways indicated the 26 peptides may be involved in cancer, signaling, metabolism, regulation, immune system and hemostasis pathways which validated by the RNA-seq analysis [ 2 ].
Proteins serve crucial functions in essentially all biological processes and the function directly depends on their three-dimensional structures. Traditional approaches to elucidation of protein structures by NMR spectroscopy are time consuming and expensive, however, the faster and more cost-effective methods are critical in the development of personalized medicine. Cole et al. improved the REDRAFT software package in the important areas of usability, accessibility, and the core methodology which resulted in the ability to fold proteins [ 3 ].
The human microbiome is the aggregation of microorganisms that reside on or within human bodies. Rebecca et al. discussed the tissue-associated microbial detection in cancer using next generation sequencing (NGS). Various computational frameworks could shed light on the role of microbiota in cancer pathogenesis [ 4 ]. How to analyze the human microbiome data efficiently is a huge challenge. Zhang et al. developed a nonparametric test based on inter-point distance to evaluate statistical significance from a Bayesian point of view. The proposed test is more efficient and sensitive to the compositional difference compared with the traditional mean-based method [ 5 ].
Human disease is also considered as the cause of the interaction between genetic and environmental factors. In the last decades, there was a growing interest in the effect of metal toxicity on human health. Evaluating the toxicity of chemical mixture and their possible mechanism of action is still a challenge for humans and other organisms, as traditional methods are very time consuming, inefficient, and expensive, so a limited number of chemicals can be tested. In order to develop efficient and accurate predictive models, Yu et al. compared the results among a classification algorithm and identified 15 gene biomarkers with 100% accuracy for metal toxicant using a microarray classifier analysis [ 6 ].
Currently, there is a growing need to convert biological data into knowledge through a bioinformatics approach. We hope these articles can provide up-to-date information of research development and trend in bioinformatics field.
Availability of data and materials
Not applicable.
Abbreviations
The 2019 International Conference on Bioinformatics and Computational Biology
Liquid chromatography tandem mass spectrometry
Alternative splicing variants
Nuclear Magnetic Resonance
Residual Dipolar Coupling based Residue Assembly and Filter Tool
Next generation sequencing
Mona Maharjan RBT, Chowdhury K, Duan W, Mondal AM. Computational identification of biomarker genes for lung cancer considering treatment and non-treatment studies. 2020. https://doi.org/10.1186/s12859-020-3524-8 .
Zhang F, Deng CK, Wang M, Deng B, Barber R, Huang G. Identification of novel alternative splicing biomarkers for breast cancer with LC/MS/MS and RNA-Seq. Mol Cell Proteomics. 2020;16:1850–63. https://doi.org/10.1186/s12859-020-03824-8 .
Article Google Scholar
Casey Cole CP, Rachele J, Valafar H. Increased usability, algorithmic improvements and incorporation of data mining for structure calculation of proteins with REDCRAFT software package. 2020. https://doi.org/10.1186/s12859-020-3522-x .
Rebecca M, Rodriguez VSK, Menor M, Hernandez BY, Deng Y. Tissue-associated microbial detection in cancer using human sequencing data. 2020. https://doi.org/10.1186/s12859-020-03831-9 .
Qingyang Zhang TD. A distance based multisample test for high-dimensional compositional data with applications to the human microbiome . 2020. https://doi.org/10.1186/s12859-020-3530-x .
Yu Z, Fu Y, Ai J, Zhang J, Huang G, Deng Y. Development of predicitve models to distinguish metals from non-metal toxicants, and individual metal from one another. 2020. https://doi.org/10.1186/s12859-020-3525-7 .
Download references
Acknowledgements
This supplement will not be possible without the support of the International Society of Intelligent Biological Medicine (ISIBM).
About this supplement
This article has been published as part of BMC Bioinformatics Volume 21 Supplement 9, 2020: Selected Articles from the 20th International Conference on Bioinformatics & Computational Biology (BIOCOMP 2019). The full contents of the supplement are available online at https://bmcbioinformatics.biomedcentral.com/articles/supplements/volume-21-supplement-9 .
Publication of this supplement has been supported by NIH grants R01CA223490 and R01 CA230514 to Youping Deng and 5P30GM114737, P20GM103466, 5U54MD007601 and 5P30CA071789.
Author information
Authors and affiliations.
Department of Quantitative Health Sciences, John A. Burns School of Medicine, University of Hawaii at Manoa, Honolulu, HI, 96813, USA
Yuanyuan Fu, Zhougui Ling & Youping Deng
Department of Pulmonary and Critical Care Medicine, The Fourth Affiliated Hospital of Guangxi Medical University, Liuzhou, 545005, China
Zhougui Ling
Department of Computer Science, University of Georgia, Athens, GA, 30602, USA
Hamid Arabnia
You can also search for this author in PubMed Google Scholar
Contributions
YF drafted the manuscript, ZL, HA, and YD revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Correspondence to Youping Deng .
Ethics declarations
Ethics approval and consent to participate, consent for publication, competing interests.
The authors declare that they have no competing interests.
Additional information
Publisher's note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ . The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
Reprints and permissions
About this article
Cite this article.
Fu, Y., Ling, Z., Arabnia, H. et al. Current trend and development in bioinformatics research. BMC Bioinformatics 21 (Suppl 9), 538 (2020). https://doi.org/10.1186/s12859-020-03874-y
Download citation
Published : 03 December 2020
DOI : https://doi.org/10.1186/s12859-020-03874-y
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Bioinformatics
- Human disease
BMC Bioinformatics
ISSN: 1471-2105
- General enquiries: [email protected]
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
Computational biology and bioinformatics articles from across Nature Portfolio
Computational biology and bioinformatics is an interdisciplinary field that develops and applies computational methods to analyse large collections of biological data, such as genetic sequences, cell populations or protein samples, to make new predictions or discover new biology. The computational methods used include analytical methods, mathematical modelling and simulation.
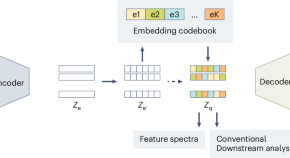
Discrete latent embeddings illuminate cellular diversity in single-cell epigenomics
CASTLE, a deep learning approach, extracts interpretable discrete representations from single-cell chromatin accessibility data, enabling accurate cell type identification, effective data integration, and quantitative insights into gene regulatory mechanisms.
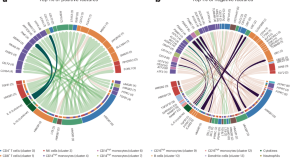
Immune signatures of disease stages and outcomes in myocardial infarction
A longitudinal multiomic dataset was assembled to characterize the immune landscape in myocardial infarction and chronic coronary syndromes. Multiomics factor analysis (MOFA) revealed immune signatures that associate with disease stage or treatment outcomes. This work opens new directions for future mechanistic and clinical studies on coronary artery disease and myocardial infarction.
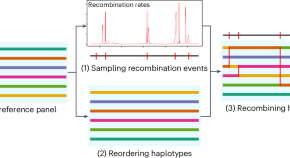
Shuffling haplotypes to share reference panels for imputation
We present a method to alleviate re-identification risks behind sharing haplotype reference panels for imputation. In an anonymized reference panel, one might try to infer the genomes’ phenotypes to re-identify their owner. Our method protects against such attack by shuffling the reference panels genomes while maintaining imputation accuracy.
Related Subjects
- Biochemical reaction networks
- Cellular signalling networks
- Classification and taxonomy
- Communication and replication
- Computational models
- Computational neuroscience
- Computational platforms and environments
- Data acquisition
- Data integration
- Data mining
- Data processing
- Data publication and archiving
- Functional clustering
- Gene ontology
- Gene regulatory networks
- Genome informatics
- Hardware and infrastructure
- High-throughput screening
- Image processing
- Literature mining
- Machine learning
- Microarrays
- Network topology
- Predictive medicine
- Probabilistic data networks
- Programming language
- Protein analysis
- Protein design
- Protein folding
- Protein function predictions
- Protein structure predictions
- Proteome informatics
- Quality control
- Scale invariance
- Sequence annotation
- Statistical methods
- Virtual drug screening
Latest Research and Reviews

Chromosome-level genome assembly of ridgetail white shrimp Exopalaemon carinicauda
- Jiajia Wang
- Jianjian Lv
Chromosome-level genome assembly of Solanum pimpinellifolium
- Chuanyou Li
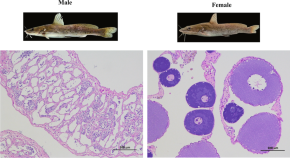
Gap-free chromosome-level genomes of male and female spotted longbarbel catfish Hemibagrus guttatus

Evolutionary and phylogenetic insights from the mitochondrial genomic analysis of Diceraeus melacanthus and D. furcatus (Hemiptera: Pentatomidae)
- Lilian Cris Dallagnol
- Fernando Luís Cônsoli

A novel framework based on explainable AI and genetic algorithms for designing neurological medicines
- Vishakha Singh
- Sanjay Kumar Singh
- Ritesh Sharma
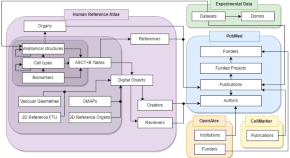
Publication, funding, and experimental data in support of Human Reference Atlas construction and usage
- Yongxin Kong
- Katy Börner
News and Comment

Superfast Microsoft AI is first to predict air pollution for the whole world
The model, called Aurora, also forecasts global weather for ten days — all in less than a minute.
- Carissa Wong

Accelerating AI: the cutting-edge chips powering the computing revolution
Engineers are harnessing the powers of graphics processing units (GPUs) and more, with a bevy of tricks to meet the computational demands of artificial intelligence.
- Dan Garisto

Integrating computational and experimental worlds
Dr Kelly Ruggles, associate professor at New York University Langone Health, discusses with Nature Computational Science how she uses computational approaches to gain insights into cancer, inflammation and cardiovascular disease, as well as the importance of mentorship.
- Ananya Rastogi
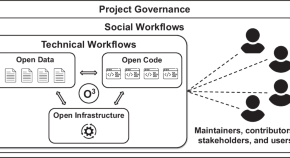
The O3 guidelines: open data, open code, and open infrastructure for sustainable curated scientific resources
Curated resources that support scientific research often go out of date or become inaccessible. This can happen for several reasons including lack of continuing funding, the departure of key personnel, or changes in institutional priorities. We introduce the Open Data, Open Code, Open Infrastructure (O3) Guidelines as an actionable road map to creating and maintaining resources that are less susceptible to such external factors and can continue to be used and maintained by the community that they serve.
- Charles Tapley Hoyt
- Benjamin M. Gyori
A surprising abundance of pancreatic pre-cancers
AI-based three-dimensional genomic mapping reveals a large abundance of cancer precursors in normal pancreatic tissue — prompting new insights and research directions.
- Karen O’Leary

Affordable and simplified whole-body MRI
A whole-body scanner developed using a permanent 0.05-tesla magnet and deep learning has demonstrated its versatility in imaging various anatomical structures, showcasing its potential to address unmet clinical needs.
- Sonia Muliyil
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Navigation Menu
Search code, repositories, users, issues, pull requests..., provide feedback.
We read every piece of feedback, and take your input very seriously.
Saved searches
Use saved searches to filter your results more quickly.
To see all available qualifiers, see our documentation .
Bioinformatics
Bioinformatics is an interdisciplinary field that intersects with biology, computer science, mathematics and statistics. It concerns itself with the development and use of methods and software tools for collecting and analyzing biological data.
Here are 9,251 public repositories matching this topic...
Developer-y / cs-video-courses.
List of Computer Science courses with video lectures.
- Updated May 30, 2024
plotly / dash
Data Apps & Dashboards for Python. No JavaScript Required.
- Updated Jun 3, 2024
biopython / biopython
Official git repository for Biopython (originally converted from CVS)
- Updated Jun 4, 2024
google / deepvariant
DeepVariant is an analysis pipeline that uses a deep neural network to call genetic variants from next-generation DNA sequencing data.
- Updated Mar 19, 2024
danielecook / Awesome-Bioinformatics
A curated list of awesome Bioinformatics libraries and software.
- Updated Apr 2, 2024
seandavi / awesome-single-cell
Community-curated list of software packages and data resources for single-cell, including RNA-seq, ATAC-seq, etc.
- Updated Mar 27, 2024
nextflow-io / nextflow
A DSL for data-driven computational pipelines
OpenGene / fastp
An ultra-fast all-in-one FASTQ preprocessor (QC/adapters/trimming/filtering/splitting/merging...)
- Updated Apr 7, 2024
scverse / scanpy
Single-cell analysis in Python. Scales to >1M cells.
lh3 / minimap2
A versatile pairwise aligner for genomic and spliced nucleotide sequences
- Updated May 22, 2024
allenai / scispacy
A full spaCy pipeline and models for scientific/biomedical documents.
- Updated Mar 30, 2024
broadinstitute / gatk
Official code repository for GATK versions 4 and up
bioconda / bioconda-recipes
Conda recipes for the bioconda channel.
Burrow-Wheeler Aligner for short-read alignment (see minimap2 for long-read alignment)
- Updated Apr 15, 2024
lh3 / seqtk
Toolkit for processing sequences in FASTA/Q formats
- Updated Oct 24, 2023
galaxyproject / galaxy
Data intensive science for everyone.
soedinglab / MMseqs2
MMseqs2: ultra fast and sensitive search and clustering suite
- Updated May 23, 2024
shenwei356 / seqkit
A cross-platform and ultrafast toolkit for FASTA/Q file manipulation
- Updated May 17, 2024
MultiQC / MultiQC
Aggregate results from bioinformatics analyses across many samples into a single report.
lightaime / deep_gcns_torch
Pytorch Repo for DeepGCNs (ICCV'2019 Oral, TPAMI'2021), DeeperGCN (arXiv'2020) and GNN1000(ICML'2021): https://www.deepgcns.org
- Updated Jul 31, 2022
Related Topics
- Director’s Welcome
- Participating Departments
- Frontiers in Computational Biosciences Seminar Series
- Current Ph.D. Students
- Current M.S. Students
- Bioinformatics Department Handbook
- B.I.G. Summer Institute
- The Collaboratory
- Diversity and Inclusiveness
- Helpful Information for Current Students
- Joint UCLA-USC Meeting
- Student Blog and Twitter Feed
- Social Gatherings
- Introduction to the Program
- Admissions Information
- Admissions FAQs
- Student Funding
- Curriculum and Graduate Courses
- Research Rotations
- Qualifying Exams
- Doctoral Dissertation
- Student Publications
- Capstone Project
- Undergraduate Courses
- Undergraduate and Masters Research
- Bioinformatics Minor Course Requirements
- Bioinformatics Minor FAQs
- Bioinformatics Minor End-of-Year Celebration
- For Engineering Students

General Information There are plenty of opportunities for Bioinformatics research projects at UCLA. This program is designed to help interested students find research projects related to Bioinformatics across campus. Typically, these projects are for credit; in exceptional circumstances they may offer funding. Participation in research projects can both significantly improve your chances of admittance into top graduate programs and make you a much more competitive employment candidate. Even better, it gives you something to talk about during an interview. Feel free to contact us even if you do not know exactly whether or not you want to work on a research project or know the field you wish to research in. Please remember that every undergraduate and masters student is welcome to participate in research, regardless of your background or year in the program. Undergraduates are STRONGLY encouraged to participate in research as early as possible in their careers. Ideally, you should start a research project during your sophomore year, but it is never too late or to early to start! Undergraduate students may receive up to 8 units credit toward the minor with enrollment in Computer Science 194/199 or Bioinformatics 194/199.
General Procedure If you are reasonably sure which project you would like to work on, use the contact information listed under the project to contact the person responsible for the project directly to set up a meeting. If you are not sure, but you are even slightly interested in research, feel free to email us or drop in to help chose an appropriate project. Most students take a project for course credit, although funding may be available in some cases. You can contact Eleazar Eskin (eeskin [at] cs [dot] ucla [dot] edu) if you have any questions.
Research Projects Below is a list of research projects that are accepting undergraduate researchers.
Featured News
Researchers awarded $4.7 million to study genomic variation in stem cell production, dr. nandita garud recognized for her research on gut microbiome, ucla study reveals how immune cells can be trained to fight infections, ucla scientists decode the ‘language’ of immune cells, dr. eran halperin elected as fellow of international society for computational biology, upcoming events, spring 2024 quarter instruction ends, spring 2024 quarter finals week, spring 2024 quarter ends, recent student publications.
RECENT STUDENT PUBLICATIONS LINK-PLEASE CLICK!
Updates Coming Soon!

College of Agricultural Sciences
Project Examples
In this section.
- Bioinformatics
Here are some examples of Bioinformatic analyses we have expertise in conducting.
We have experience working with many diverse data and organism types, so even if your topic is not listed in our project examples, we are likely to be able to assist you.
Deliverables for Basic/Standard Analysis
MAX Turnaround time – 2 months depending on application and sample size
1. Whole Genome Sequencing
Prokaryotes.
- RE-SEQUENCING: Raw Data QC and Report, Alignment Statistics and Report, Variation Calling Report (SNP, InDels), Gene Annotation Table with Variations.
- DENOVO: Raw Data QC and Report, Assembly Statistics and Report, Genome Finishing using Closest homolog, rRNA identification and analysis report, Phage Identification and analysis report, Plasmid Identification, and analysis report, RAST Annotation.
- RE-SEQUENCING/TARGETED/EXOME: Raw Data QC and Report, Alignment Report, Variation calling Report (SNP, InDels), Basic Variation Annotation, and Effect Analysis Report.
- DENOVO: Raw Data QC and Report, Assembly Statistics and Report, Gene Prediction and Annotation Report. Data generation depends on predicted genome size
2. Transcriptome Sequencing
- RE-SEQUENCING: Ribo Depletion (rRNA Depletion) – Raw Data QC and Report, Read Alignment to reference genome and transcript Identification, Comprehensive Transcript Annotation, Functional Classification of Annotated Transcript, Expression Profiling, Quantification & Expression Profiling of transcripts, Differential Analysis among the conditions, Biological Significance Analysis of differentials.
- RE-SEQUENCING: – Raw Data QC and Report, Read Alignment to reference genome and transcript Identification, Quantification & Expression Profiling of transcripts, Differential, Analysis among the conditions, Biological Significance Analysis of differentials (n-1). All pictorial representations of comparisons will be according to n-1
- DENOVO: Raw Data QC and Report, De novo assembly, Assembly Evaluation & Filtering, Sequence homology-based Transcript Annotation using Blast2Go – REFSEQ, Expression Profiling, Differential Analysis among the conditions, Biological Significance Analysis of differentials (n-1). ALL pictorial representation LL of comparisons will be according to n-1.
3. Chip Sequencing
Raw Data QC and Report, Alignment Report, Peak Identification, and Enrichment Report, Peak Annotation Report
4. Metagenome Sequencing
Sample Grouping or individual as per experimental design, Group-wise OTU Clustering and abundance Report, OTU identification and taxonomic annotation Report (Sample Wise – Genius Level) and OTU Fasta file will be provided, Pie chart representation TOP 10 taxonomic classification; phylum to species-level.
5. SmallRNA Sequencing
Sample wise Raw Data QC, Unique tags and abundance Report, Known Small RNA analysis report, Identification and Quantitation of Known miRNAs, Expression Profiling and Differential Expression Analysis of Known miRNAs.
6. Microbiome Sequencing
Pre-processing of reads including Quality Filtering, trimming low-quality reads, De-Replication, Sequence reconstruction and grouping, Gene prediction, Functional Annotation.
Deliverables for Advanced Analysis
MAX TAT – 3 months depending on the project requirement and sample size
- RE-SEQUENCING: Raw Data QC and Report, Alignment Statistics and Report, Variation Calling Report (SNP, InDels), Gene Annotation Table with Variations, Structural Variations (Inversion, Deletion, Insertion, Translocation, Transversion) analysis report, Comparative Genome analysis – Across selected genomes, High SNP and Low SNP Region, Generic and NonGeneic SNPs, SNP Density Analysis, Synonymous and Non-synonymous SNPs, Effect of Frameshift Indels on Gene Prediction, Submitting Data to NCBI -SRA, Support in providing write up on methods for the manuscript purpose (Time Limit: 3-6 month)
- DENOVO: Raw Data QC and Report, Assembly Statistics and Report, Genome Finishing using Closest homolog, rRNA identification and analysis report, Phage Identification and analysis report, Plasmid Identification and analysis report, Phylogeny 16s RNA based, COG Analysis, Interproscan Analysis, AAI and ANI analysis with the selected reference genome, Antibiotic resistance gene analysis with reference to transposable elements, PAN and Core genome analysis, Synteny Analysis, Chromosome Mapping, Plasmid Re-construction from whole-genome, Submitting Data to NCBI- SRA, Support in providing write up on methods for the manuscript purpose (Time Limit: 3-6 month)
- RE-SEQUENCING/TARGETED/EXOME: Raw Data QC and Report, Alignment Report, Variation calling Report (SNP, InDels), Basic Variation Annotation and Effect Analysis Report, All the deliverables from Standard Analysis, Structural Variation Analysis Report, Variation Effect Analysis Report, Pathway and GO analysis of variations, Copy Number Variation Analysis, Data Submission to NCBI, Comparative Exome Analysis, Submitting Data to NCBI- SRA, Support in providing write up on methods for the manuscript purpose.
- DENOVO: Raw Data QC and Report, Assembly Statistics and Report, Gene Prediction and Annotation Report, Prediction of rRNAs, tRNAs, Repeat Analysis, Identification of Transposons, Domain Identification, Analysis of Virulence genes, Analysis of CaZymes, Synteny Analysis, Comparative Exome Analysis, Submitting Data to NCBI- SRA, Support in providing write up on methods for the manuscript purpose.
- RE-SEQUENCING: Ribo Depletion (rRNA Depletion) – Raw Data QC and Report, Read Alignment to reference genome and transcript Identification, Comprehensive Transcript Annotation, Functional Classification of Annotated Transcript, Expression Profiling, Quantification & Expression Profiling of transcripts, Differential Analysis among the conditions, Biological Significance Analysis of differentials, Inter and Intra Gene List Comparisons, Gene and Pathway enrichment analysis, GO and Pathways based Gene Regulatory Network Modelling, Submitting Data to NCBI- SRA, Support in providing write up on methods for the manuscript purpose.
- RE-SEQUENCING: Raw Data QC and Report, Read Alignment to reference genome and transcript Identification, Expression Profiling, Quantification & Expression Profiling of transcripts, Differential Analysis among the conditions, Biological Significance Analysis of differentials, Inter and Intra Gene List Comparisons, Gene and Pathway enrichment analysis, GO and Pathways based Gene Regulatory Network Modeling, Functional classification of expressed transcripts Submitting, Data to NCBI-SRA, Support in providing write up on methods for the manuscript purpose.
- DENOVO: Raw Data QC and Report, De novo assembly, Assembly Evaluation & Filtering, Sequence homology-based Transcript Annotation using Blast2Go – NRDB, Expression Profiling, Differential Analysis among the conditions, Biological Significance Analysis of differentials, Sequence homology-based Transcript Annotation against the customized database, Inter and Intra Gene List Comparisons, Gene and Pathway enrichment analysis, Functional Classification of Annotated Transcript, GO and Pathways based Gene Regulatory Network Modeling, Data to NCBI-SRA, Support in providing write up on methods for the manuscript purpose.
Raw Data QC and Report, Alignment Report, Peak Identification, and Enrichment Report, Peak Annotation Report, Motif Identification, Statistical analysis of Peak Reproducibility (If replicates are provided), Significant GO and Pathway Analysis, Data to NCBI-SRA, Support in providing write up on methods for the manuscript purpose.
Sample Grouping/Individual (either one) as per experimental design, Group-wise OTU Clustering and abundance Report, OTU identification and taxonomic annotation Report (Sample Wise – Genius Level) and OTU Fasta file will be provided, Pie chart representation TOP 10 taxonomic classification (Phylum to Species-level), Differential Metagenome based on sample conditions, Diversity Analysis (Alpha and Beta), Rarefaction Curves, PCoA Plot (required minimum six samples), Krona Plot at the genus level, Heat-Maps for comparisons, Species-level annotation (If V3 & V4 is covered), Data to NCBI-SRA, Support in providing write up on methods for the manuscript purpose.
Raw Data QC and Report, Known Small RNA analysis report, Identification and Quantitation of Known miRNAs, Expression Profiling and Differential Expression Analysis of Known miRNAs, Novel miRNA Identification (In case of reference genome availability) and analysis report, Characterization of other small RNAs like siRNA, piRNA, snoRNA, miRNA Target Prediction / Identification, Significant GO and Pathway Analysis of targets of differentially expressed miRNAs, DData to NCBI-SRA, Support in providing write up on methods for the manuscript purpose.
Pre-processing of reads including Quality Filtering, Trimming low quality reads, De-Replication, Sequence reconstruction and grouping, Gene and regulatory element prediction, Functional Annotation, Differential Microbiome based on sample parameters, Statistical analysis of Microbiome based on OTUs, Diversity Analysis (Alpha and Beta), Rarefaction Curves, Species-level annotation, Seed Subsystem classification, COG, KEGG Analysis, Gene Ontology and Pathway Analysis (Functional Microbiome Analysis), Data to NCBI-SRA, Support in providing write up on methods for the manuscript purpose.
5 Machine Learning Projects in Bioinformatics For Practice
Explore Top Machine Learning Projects Ideas to Understand the Applications of Machine Learning in Bioinformatics| ProjectPro

The term "bioinformatics" represents the use of computation and analysis methods to collect and analyze biological data. It's a multidisciplinary field that combines genetics, biology, statistics, mathematics, and computer science. Various branches of bioinformatics, including genomics, proteomics, and microarrays, extensively use machine learning for better outcomes.

Personalized Medicine: Redefining Cancer Treatment
Downloadable solution code | Explanatory videos | Tech Support

Top 5 Machine Learning Projects in Bioinformatics
Here are five exciting machine learning projects for bioinformatics to help you understand the application of machine learning in healthcare , mainly bioinformatics.

1. Anti-Cancer Drug Efficacy Prediction
Predicting which patients are likely to benefit or not from a specific therapy is a significant concern in cancer treatment because, generally speaking, not all patients will benefit from a particular medication. This enhances the efficacy of treatment and reduces the suffering and misery experienced by non-responders. Thus, there is an immediate need to find reliable biomarkers (i.e., genes or proteins) that can precisely predict which patients respond best to which medications. For this project, you will use fundamental data science techniques , such as data processing, integration, analysis, and visualization, to determine the most effective biomarkers for various cancer types.

2. Autism Mutation Detection
In this machine learning project for bioinformatics, you will develop a deep-learning-based system that predicts the accurate regulatory effects and the harmful impacts of genetic variants to address the issue of detecting the impact of noncoding mutations on disease. This predictive genomics framework is likely relevant to complex human diseases, illustrates the significance of noncoding mutations in ASD [autism spectrum disorder], and identifies mutations with higher effects for further analysis. If you want to add some unique project to your machine learning portfolio , you must try working on this project.
Here's what valued users are saying about ProjectPro

Director Data Analytics at EY / EY Tech

Gautam Vermani
Data Consultant at Confidential
Not sure what you are looking for?
3. Personalized Cancer Medication
This deep learning project can predict how different genetic variations affect a patient's health. You can use the MSKCC (Memorial Sloan Kettering Cancer Center) database, including thousands of mutations that top-notch scientists and physicians have thoroughly classified. For this machine learning project, you will create a machine learning algorithm using the Keras deep learning library and LSTM that automatically categorizes genetic variants utilizing this data set as a starting point. Additionally, this project entails using various NLP text processing techniques such as Lemmatization, Stemming, Tokenization, etc.
You don't have to remember all the machine learning algorithms by heart because of amazing libraries in Python. Work on these Machine Learning Projects in Python with code to know more!
4. Human Disease Genetic Basis Identification
Human genomes vary between individuals by.1%. Our genetic inclination to specific disorders, such as hypertension, is encoded within this small degree of variation. We can accurately define which gene variants belong to each disease by comparing populations of healthy and diseased people and their variations in the genes responsible for the diseases. In this bioinformatics, AI and machine learning project, strategies for finding the variation corresponding to disease are developed, along with statistics to support the predictions. Furthermore, this project develops methods for predicting how a gene mutation can alter the structure of the protein or the regulatory structure. You can also estimate the disease risk factor's history and evolution by recreating the genes' phylogeny.
5. Build a DNA Sequence Classifier
You will use a classification model in this project that can predict a gene's function just from the DNA sequence of the coding sequence. You will create a function that will extract from any sequence string all overlapping k-mers of a given length, count the k-mers and convert the k-mers list for each gene into string sequences using scikit-learn NLP tools.

About the Author

Daivi is a highly skilled Technical Content Analyst with over a year of experience at ProjectPro. She is passionate about exploring various technology domains and enjoys staying up-to-date with industry trends and developments. Daivi is known for her excellent research skills and ability to distill
© 2024
© 2024 Iconiq Inc.
Privacy policy
User policy
Write for ProjectPro
Loading metrics
Open Access
Bioinformatics Projects Supporting Life-Sciences Learning in High Schools
Affiliation Instituto Gulbenkian de Ciência, Oeiras, Portugal
Affiliation Escola Secundária Stuart de Carvalhais, Queluz, Portugal
* E-mail: [email protected]
- Isabel Marques,
- Paulo Almeida,
- Renato Alves,
- Maria João Dias,
- Ana Godinho,
- José B. Pereira-Leal

Published: January 23, 2014
- https://doi.org/10.1371/journal.pcbi.1003404
- Reader Comments
The interdisciplinary nature of bioinformatics makes it an ideal framework to develop activities enabling enquiry-based learning. We describe here the development and implementation of a pilot project to use bioinformatics-based research activities in high schools, called “Bioinformatics@school.” It includes web-based research projects that students can pursue alone or under teacher supervision and a teacher training program. The project is organized so as to enable discussion of key results between students and teachers. After successful trials in two high schools, as measured by questionnaires, interviews, and assessment of knowledge acquisition, the project is expanding by the action of the teachers involved, who are helping us develop more content and are recruiting more teachers and schools.
Citation: Marques I, Almeida P, Alves R, Dias MJ, Godinho A, Pereira-Leal JB (2014) Bioinformatics Projects Supporting Life-Sciences Learning in High Schools. PLoS Comput Biol 10(1): e1003404. https://doi.org/10.1371/journal.pcbi.1003404
Editor: Fran Lewitter, Whitehead Institute, United States of America
Copyright: © 2014 Marques et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Funding: This work was funded by the Instituto Gulbenkian de Ciência. The funders had no role in the preparation of the manuscript.
Competing interests: The authors have declared that no competing interests exist.
Background and Motivation
Our lives are increasingly touched by science and technology, from the everyday activities of browsing the internet, taking a prescription drug, etc., to major societal discussions involving, for example, genetically modified foods, cloning, or stem cells. It is therefore imperative that we engage young people in science. We witnessed in the past shrinking numbers of students choosing science degrees for their university education [1] . This trend seems, however, to have been inverted both in Europe and in the United States [2] , [3] . A recent study points to the development of new and more attractive curricula and teaching methods as the driver for this increased interest [3] . In light of the growing evidence of a direct link between attitudes towards science and the way science is taught [1] , there is increasing recognition of the need to couple the traditional teacher-centred “deductive approach” to the learner-centred “inductive approach,” relying on observation, experimentation, and teacher guidance in constructing students' knowledge. This “bottom-up” approach, called enquiry-based learning (also known as problem-based learning or case-based learning) [4] recapitulates the scientific process (raising questions, collecting data, reasoning, reviewing evidence, drawing conclusions, and discussing results), thus promoting both ideas of science (scientific concepts) and ideas about science (process, practices, and critical thinking), i.e., about the Nature of Science (NOS).
Bioinformatics is a discipline at the intersection of biology, computer science, information science, mathematics, and to some extent also of chemistry and physics. It developed in response to the increasingly complex data types and relationships in biological research, addressing the need to manage and interpret biological information. This interdisciplinary nature makes bioinformatics an ideal framework to engage high school students, as it illustrates the interplay between different scientific areas, while touching on many aspects that are relevant to the younger generations—health, environment, etc. This has been recognized by many others who have implemented bioinformatics-training programs. Examples are a web-based, problem-oriented approach aimed at introducing students to bioinformatics [5] and the use of bioinformatics activities as a way to teach evolution [6] or notions of polymorphisms in the context of human genetic variation and disease [7] . Bioinformatics has also integrated with wet-lab activities in initiatives like the student-aimed “Cus-Mi-Bio” project [8] , which include gene finding activities, or in projects aimed at high school and college teachers, such as the ones at the Dolan DNA learning centre of Cold Spring Harbor Laboratory involving plant genome annotation [9] . More recently, activities that aim to introduce high school students to bioinformatics itself have also been reported [10] , and, as of 2012, an exercise using Basic Local Alignment Search Tool (BLAST) has been included on the Advanced Placement, high school biology, national test in the US ( http://apcentral.collegeboard.com/apc/members/courses/teachers_corner/218954.html ). Note, however, that these are likely isolated cases rather than the norm, as a survey revealed that in 2008 bioinformatics was still absent from the classroom in the US [11] , and likely elsewhere.
The “Bioinformatics@school” Program
We run a Bioinformatics Core at the Instituto Gulbenkian de Ciência, in Portugal, that has long been engaged in outreach activities. In 2007, we decided to implement a genomics/bioinformatics activity that would enable enquiry-based learning; link to the national curricula in biology in secondary education; introduce students to bioinformatics, genomics, and molecular biology, areas that underlie many of the key debates and products in our societies; foster active learning, making use of technologies that younger generations are increasingly comfortable with; and help teachers incorporate the latest advances in science into their teaching. We developed a prototype system that we describe the following components of here: its development, implementation, and the results of nearly five years of activity.
We developed and implemented a framework for the use of bioinformatics-based research projects in high schools to support the life-sciences curricula, which we named, in Portuguese, “Bioinformática na escola,” loosely translating to “Bioinformatics@school.” It consists of research projects that may be conducted independently by high school students of different ages, either under direct teacher supervision or as homework. Each work unit in a research project is designed to be carried out in 90 minutes, which is a standard class length in Portuguese high schools. We implemented it as a web portal ( Figure 1A, 1B )— www.bioinformatica-na-escola.org . Although primarily written in Portuguese, the site makes use of external, freely accessible bioinformatics tools and databases available in English. This is not a problem for Portuguese high school students that typically start learning English at the age of nine. Because of the dependency on external sites, we have ensured that students are given alternative access to any data on which progression to the following activity depends.
- PPT PowerPoint slide
- PNG larger image
- TIFF original image
( A ) Screenshots of the home page. ( B ) Screenshots of exercises pages.
https://doi.org/10.1371/journal.pcbi.1003404.g001
The whole program is structured as a set of projects with open-ended questions. A project may have a single activity or several, each having focused questions. Answering these focused questions enables students to discuss and/or solve the project's main question. Individual activities in the multi-activity projects were designed to also be used independently (discussed below). The concept lends itself both to classroom use, individually or in pairs, or as homework. We designed individual activities to explore specific concepts that are part of the school curriculum and the projects to be coherent with the curriculum of specific age groups, with the active collaboration of teachers in choosing the topics.
Projects are organized as follows. Once a project is selected, the student has access to a page that summarizes the problem to be solved and a link to the first activity. As the student enters one activity, s/he is presented with a sequential series of pages, each giving some background information on the specific problem the student has to follow and a brief description of the bioinformatics resources/tools to be used. At the end of each activity, the student is taken to a summary page (“now you know that…”) with an overview of the basic concepts that were addressed in the activity. All pages include links to additional information on key concepts, mostly on Wikipedia ( www.wikipedia.org ), including explanations about the resources and algorithms used in the analysis. Once the activity/final activity is complete, the student is taken to a summary page that reviews the key concepts of the project as a whole and a series of questions that act as primers for discussion amongst students and with the teacher(s) (see Figure 1A, 1B for screenshots). Table 1 summarizes the questions, concepts, and software and resources that are covered in each individual activity of “Vision,” the first multi-activity project that we have implemented in the Bioinformatics@school portal (further detailed in Text S1 ). Its implementation in schools is discussed below.
https://doi.org/10.1371/journal.pcbi.1003404.t001
Implementing “Bioinformatics@school”
Iterative development of project modules.
We started to develop “Bioinformatics@school” as a pilot project in 2007, in close collaboration with high school students and teachers. The first stage of the project consisted of identifying the topics within the high school curricula that would be amenable to bioinformatics treatment, as well as the ideal school year for the pilot to be developed. We chose 12th grade biology, in the last year of high school in the Portuguese educational system, as their curricula included multiple themes that were ideal to address using bioinformatics (such as genes, genomes, genetics, evolution, mutation, etc.), and these students would all have had several years of English language schooling (discussed above). The next phase of the project consisted in the enrollment of schools. Two secondary schools located in the Lisbon area were recruited, representing two different demographics. Escola Secundária Miguel Torga (ESMT), in Queluz, is a large suburban school that covers a variety of social strata, while Escola Secundária Quinta do Marquês (ESQM), in Oeiras, is located in a high income area with high levels of graduates and post-graduates. We engaged seven 12th grade Biology teachers, two from ESMQ and five from ESMT. One hundred and fifty students were involved in this initial pilot phase, representing multiple science-related career ambitions, ranging from engineering, health, biology, psychology, sports, etc.
We conceived the general framework described above and developed the first full project consisting of five activities, aimed at understanding how animals see different colours (“Vision” project, Table 1 ). We chose this question because we believed it to be sufficiently intriguing and relevant to engage the students (natural variants cause differential colour perception between species and between different people), but also for practical reasons—the biology of light detection via opsins is well understood, as is the 3D structure of opsins. The aim of the project is to motivate a discussion about evolution, molecular mechanisms, and disease, all inferred from bioinformatics analysis, while helping teachers and students engage with specific topics of the Life Sciences curriculum via the individual bioinformatics activities.
An innovative aspect of this project was the collaboration between scientists, teachers, and students on different aspects of the development, implementation, and testing—a three-way dialogue with continual updating in response to feedback of students and teachers. The development was iterative, first within our Bioinformatics Unit, and then in discussions with teachers. Once a first prototype was in place, one of us (IM) went to the schools to guide the students in the first activities of the project, with the help of the teacher. Student feedback was then used to improve the activities, in terms of rationale, language, and presentation.
Teacher training
Keeping up to date with the rapid developments in genomics and bioinformatics represents a challenge for high school teachers, particularly when many may have completed their training decades ago. In fact, in our experience, bioinformatics is a novel subject area to most Portuguese high school teachers. This led us to implement a parallel teacher-training program, again co-developed with the first set of teachers. Teachers were trained by bioinformatics experts, with the main goal of training to guide the students in the bioinformatics-based projects and to understand the basics of the bioinformatics methods and resources underlying each activity. We developed a teacher's manual that described the activities step by step and provided additional background information for the teacher to be comfortable with all the concepts in each activity. The teacher training consisted of having the teachers follow the same activities as the students, with the help of the teacher manual and under the supervision of a bioinformatician. We have expanded the teacher training to include seminars about applications of bioinformatics to human health, biotechnology, etc. A typical teacher training program lasts about 25 hours.
Extending the Program and Sustainability
After the successful pilot stage in 2007 the project has expanded to other geographical areas of Portugal. Thirty three new schools have joined the program, some via previously engaged teachers who took the program with them when they moved to a new school, others by new teachers who contacted us, after hearing about the project, and asked us to help them implement it in their schools. In total, schools of 11 municipalities in four Portuguese districts are currently following the program ( Figure 2A ). On their own initiative, some teachers have adapted the individual activities within the “Vision” project for use with younger students. They have also picked individual or subsets of activities and re-used them with different genes/systems, combining them in novel ways, to create new projects. They have also engaged with us to develop new projects (“Tasting Bitter”) and activities (“Tree of Life”). Furthermore, teachers are recruiting and training new teachers to use our activities. Interestingly, we observed that teachers tailored the activities to their own teaching style, some engaging the students almost at every mouse click, whereas others would only focus on explaining the basic ideas at the beginning and then discussing the outcomes at the end.
( A ) Map of schools participating, coloured by year of joining the project. ( B ) Summary of responses to confidential questionnaire. ( C ) Knowledge acquisition—each dot represents one class and the average score that students in that class achieved in the test before and after finishing the “Vision” project. ( D ) Confidence—each dot represents one class and the percent of answers that students in that class answered True or False, as opposed to answering “I don't know,” before and after finishing the “Vision” project.
https://doi.org/10.1371/journal.pcbi.1003404.g002
One aspect that worried us from early on was how to motivate teachers to engage with projects like ours when they are overwhelmed with teaching and administrative work. We realized that certification of the training is important for career progression within the Portuguese public educational system. We invested in having the project certified for teachers' continuous professional development by the national educational authorities (Conselho Científico-Pedagógico da Formação Contínua), thus making engagement with Bioinformatics@school even more appealing to the teachers. Recently we established a partnership with a teacher training centre (Centro de Formação Lezíria - Oeste) to enable other teachers in another Portuguese region to receive training in Bioinformatics activities and further promote the decentralization of “Bioinformatics@school.”
We have, thus, reason to believe that the use of the Bioinformatics@school platform is spreading on its own, with a dynamic beyond the ability of the small staff at the Bioinformatics Core that developed it.
Impact Assessment
We wished to evaluate how students and teachers perceive the program and to what extent it is an effective learning tool. These are independent questions that we addressed using different approaches. Conversations with students participating in the program suggested that they were motivated to participate in “hands-on” activities We implemented a simple confidential questionnaire to capture students' views beyond anecdotal opinions, that was given to 150 students (two schools, seven classes), during the implementation phase of the project. The results are shown in Figure 2B and reveal that the majority of the students found the approach used in this project more motivating than traditional teaching methods (58%), and enjoyed participating in it (60%). About 80% considered it had not been a waste of time and 80% would recommend the project to next year's colleagues. This type of questionnaire is useful in gauging attitudes towards the program, but it has caveats, namely that the students at this stage were very involved with the development of the Bioinformatics@school project and may be overly positive because of that. In addition, it gives no information about student learning. To address this, we devised a simple test on the concepts explored in the program, with “True/False/I Don't Know” answers ( Table S1 ). We asked four classrooms to take the test before and after the activities (this test was irrelevant for their grades). Plotting the percentage of correct answers per student before and after the activities ( Figure 2C ) revealed a dramatic increase in the proportion of correct answers, indicating that students actually gain knowledge. One surprising result was that the students appeared more confident after doing the activities: they increasingly answered the test questions as false or true, rarely using “I don't know” ( Figure 2D ). Since most of the concepts in our activities are part of the school curricula and were being covered in class by their teachers, we speculate that the decrease in “I don't know” answers may indicate that students are less afraid of venturing answers to scientific questions after doing the activities. Fear of science (“too complicated”) has been pointed out as a reason for the decreasing number of students pursuing scientific degrees [1] . This is an exciting finding that we will need to specifically evaluate further in the future. Regarding the teachers, we developed the whole program in close collaboration with them and obtained continuous feedback on the content and presentation. Although we have not as yet conducted a systematic evaluation of teachers' views about the program, the continuous contact with the currently more than seventy teachers involved suggests to us that this is a useful teaching/learning tool. In particular, teachers mention that these activities allow them to overcome the lack of laboratory-based practicals associated with some of the content in the curricula, like genetics and molecular biology. The fact that the program is spreading, with new teachers and schools recruited by word of mouth by the teachers themselves, underscores its interest and usefulness to teachers.
Discussion and Future Directions
In summary, we implemented a set of bioinformatics multi-activity research projects designed to enable enquiry-based learning in high schools. Assessment of this project has shown that students find it enjoyable and teachers believe it to be useful as a teaching aid. Objective assessment of knowledge acquisition revealed a clear positive effect both in knowledge and confidence of the students. Teachers have taken the initiative to adapt the activities to their own teaching settings and are also recruiting other teachers, which gives us further confidence in the usefulness of this project.
We have focused the projects on addressing specific biological questions, to serve the Life Sciences curriculum. This means that we don't explore the algorithmic or technological side of bioinformatics. For the future, we hope to engage teachers from mathematics, information technology, physics, and chemistry to develop projects that can serve the curricula of those particular subjects.
Recently, Form and Lewitter proposed a simple set of ten rules to guide the use of bioinformatics in high schools [12] . While these were not available at the time we were developing this project, it is interesting to note that we independently “discovered” several of these principles. We implemented individual activities with clear, simple goals (rule 1) that built on each other (rule 4), enabling students to “discover” concepts on their own (rules 5 and 8). Throughout this project we were always mindful that these activities need to serve the pre-existing curricula (Rule 3). In the future we would like to have multiple projects serving the same concepts that would allow students in each class to choose an individual project (rule 6: personalization) that they could then present and contrast to other projects pursued by their colleagues (rule 10: produce a product). We would like to develop a mapping of activities to concepts in the curricula, so that it becomes even easier for teachers to mix and match the individual activities to different contexts, thus using our project as a means to empower the teachers. Based on our experience in setting up this program, we would like to suggest two additional “simple rules” that we believe to be important when developing contents to be used in high schools:
- Engage teachers and students in the development of the activities, as a means of empowering them and ensuring that the end product meets all the cognitive and pedagogical requirements (e.g., engage the teachers in choosing the specific topics of the curricula that would benefit from bioinformatics-based projects as well as to advise on time or practical constraints on their use in the school setting; engage both teachers and students to identify weak/unappealing points in the contents and formats of the activities and to suggest better solutions, etc.).
- Evaluate the impact of the activities on engagement/enthusiasm for science and, in particular, on knowledge acquisition, as demonstrated effectiveness is the best way to get bioinformatics into the classroom. In our opinion, perpetuating useless activities just for the sake of their perceived modernity is more likely to harm the use of bioinformatics as a tool for high school science education than to advance it.
Our program was developed in Portuguese as it is targeted at Portuguese students. While this gives us potential access to a universe of more than 200 million Portuguese speakers worldwide, it is hard to use by speakers of other languages. We have started translating the whole set of activities into English, thus making Bioinformatics@school accessible to a much larger target audience. Equally, besides developing novel activities, we would like to adapt those from successful experiments elsewhere, and in due time will contact their authors directly. In this regard, the existence of a central repository of bioinformatics exercises to be used in high schools, with clear explanations according to pre-defined standards and mapping to specific concepts, would facilitate the adoption of bioinformatics in high schools. Developing standards and repositories should come naturally to the bioinformatics community!
Supporting Information
Questionnaire for impact assessment.
https://doi.org/10.1371/journal.pcbi.1003404.s001
Activities in the “Vision” project.
https://doi.org/10.1371/journal.pcbi.1003404.s002
Acknowledgments
We wish to thank all the high school teachers who have engaged with the Bioinformatics@school project, in particular Lurdes Louro (ESMT, Queluz), Filomena Delgado (ESQM, Oeiras), and Teresa Palma (Escola Secundária de Camões [ESdeC], Lisboa). We wish also to thank for their generosity and enthusiasm the initial batch of students from ESQM and ESMT who helped us develop ever better activities. Finally, we thank João Garcia and Gil Neto at the IGC, who provided invaluable IT support. We also wish to thank the Instituto Gulbenkian de Ciência for hosting this program.
- View Article
- Google Scholar
- 2. Kang K (2012) Graduate Enrollment in Science and Engineering Grew Substantially in the Past Decade but Slowed in 2010. National Center for Science and Engineering Studies. Available: http://www.nsf.gov/statistics/infbrief/nsf12317/ . Accessed 20 December 2013.
- 3. Kearney C (2010) Efforts to Increase Students' Interest in Pursuing Mathematics, Science and Technology Studies and Careers. Wastiau P, Gras-Velázquez A, Grečnerová B, Baptista R, editors. Brussels: European Schoolnet. Available: http://cms.eun.org/shared/data/pdf/spice_kearney_mst_report_nov2010.pdf . Accessed 20 December 2013.
You are using an outdated browser. Please upgrade your browser .
T4Tutorials.com
Bioinformatics research topics ideas.
List of Bioinformatics Research Topics Ideas for.
1. Data access control in the cloud computing environment for bioinformatics 2. The bioinformatics toolbox for circRNA discovery and analysis 3. Want to track pandemic variants faster? Fix the bioinformatics bottleneck 4. A constructivist-based proposal for bioinformatics teaching practices during lockdown 5. Virus-CKB: an integrated bioinformatics platform and analysis resource for COVID-19 research 6. Therapeutic targets and signaling mechanisms of vitamin C activity against sepsis: a bioinformatics study 7. Bioinformatics helping to mitigate the impact of COVID-19–Editorial 8. Network bioinformatics analysis provides insight into drug repurposing for COVID-19 9. Deep learning-based clustering approaches for bioinformatics 10. User-friendly bioinformatics pipeline gDAT (graphical downstream analysis tool) for analysing rDNA sequences 11. Bioinformatics analysis of SARS-CoV-2 to approach an effective vaccine candidate against COVID-19 12. The Bio3D packages for structural bioinformatics 13. Metabolic Basis of Creatine in Health and Disease: A Bioinformatics-Assisted Review 14. The European Bioinformatics Institute: empowering cooperation in response to a global health crisis 15. Epigenetic dysregulation of immune-related pathways in cancer: bioinformatics tools and visualization 16. Implementing FAIR data management within the German Network for Bioinformatics Infrastructure (de. NBI) exemplified by selected use cases 17. Bioinformatics resources for SARS-CoV-2 discovery and surveillance 18. Bioinformatics and system biology approach to identify the influences of SARS-CoV-2 infections to idiopathic pulmonary fibrosis and chronic obstructive … 19. Insights into mineralocorticoid receptor homodimerization from a combined molecular modeling and bioinformatics study 20. Analysis and identification of novel biomarkers involved in neuroblastoma via integrated bioinformatics 21. Bioinformatics resources facilitate understanding and harnessing clinical research of SARS-CoV-2 22. BioContainers Registry: Searching Bioinformatics and Proteomics Tools, Packages, and Containers 23. A Review of Pharmacological and Toxicological Effects of Sophora tonkinensis with Bioinformatics Prediction 24. Application of Multilayer Network Models in Bioinformatics 25. Identification of potential biomarkers of polycystic ovary syndrome via integrated bioinformatics analysis 26. Bioinformatics analysis and verification of gene targets for renal clear cell carcinoma 27. Bioinformatics tools developed to support BioCompute Objects 28. Bioinformatics-based prediction of conformational epitopes for Enterovirus A71 and Coxsackievirus A16 29. Emulsifier peptides derived from seaweed, methanotrophic bacteria, and potato proteins identified by quantitative proteomics and bioinformatics 30. … of the molecular targets and mechanisms of compound mylabris capsules for hepatocellular carcinoma treatment through network pharmacology and bioinformatics … 31. Improving the Thermostability of Xylanase A from Bacillus subtilis by Combining Bioinformatics and Electrostatic Interactions Optimization 32. Physical exercise, obesity, inflammation and neutrophil extracellular traps (NETs): a review with bioinformatics analysis 33. MMP7 as a potential biomarker of colon cancer and its prognostic value by bioinformatics analysis 34. Bioinformatics: new tools and applications in life science and personalized medicine 35. OPCML Methylation and the Risk of Ovarian Cancer: A Meta and Bioinformatics Analysis 36. … mechanisms of GegenQinlian decoction on improving insulin resistance in adipose, liver, and muscle tissue by integrating system pharmacology and bioinformatics … 37. Determination of Potential Therapeutic Targets and Prognostic Markers of Ovarian Cancer by Bioinformatics Analysis 38. Structure–function engineering of novel fish gelatin-derived multifunctional peptides using high-resolution peptidomics and bioinformatics 39. Chemical composition, biological properties and bioinformatics analysis of two Caesalpina species: A new light in the road from nature to pharmacy shelf 40. … Cloud-Based Tutorials That Combine Bioinformatics Software, Interactive Coding, and Visualization Exercises for Distance Learning on Structural Bioinformatics 41. Functional characterization of ABCC8 variants of unknown significance based on bioinformatics predictions, splicing assays, and protein analyses: Benefits for the … 42. Integrative pharmacological mechanism of vitamin C combined with glycyrrhizic acid against COVID-19: findings of bioinformatics analyses 43. NGS-µsat: Bioinformatics framework supporting high throughput microsatellite genotyping from next generation sequencing platforms 44. Integrated bioinformatics analysis for the identification of key genes and signaling pathways in thyroid carcinoma 45. Integrative bioinformatics and omics data source interoperability in the next-generation sequencing era 46. Comprehensive bioinformatics analysis reveals kinase activity profiling associated with heart failure 47. Nimodipine attenuates dibutyl phthalate-induced learning and memory impairment in kun ming mice: An in vivo study based on bioinformatics analysis 48. Construction of circRNA-miRNA-mRNA network in the pathogenesis of recurrent implantation failure using integrated bioinformatics study 49. Bioinformatics analysis of differentially expressed miRNAs in non-small cell lung cancer 50. A systematic evaluation of bioinformatics tools for identification of long noncoding RNAs 51. Using Integrated Bioinformatics Analysis to Identify Abnormally Methylated Differentially Expressed Genes in Hepatocellular Carcinoma 52. Bioinformatics analysis of the microRNA-mRNA network in sebaceous gland carcinoma of the eyelid 53. A Bioinformatics Pipeline to Identify a Subset of SNPs for Genomics-Assisted Potato Breeding 54. Bioinformatics analysis of candidate genes involved in ethanol-induced microtia pathogenesis based on a human genome database: GeneCards 55. Bioinformatics and machine learning methodologies to identify the effects of central nervous system disorders on glioblastoma progression 56. Integrative analysis of miRNA–mRNA network in high altitude retinopathy by bioinformatics analysis 57. Screening and verification of hub genes involved in osteoarthritis using bioinformatics 58. Identification of a Gene Prognostic Signature for Oral Squamous Cell Carcinoma by RNA Sequencing and Bioinformatics 59. TRIB3 Promotes the Malignant Progression of Bladder Cancer: An Integrated Analysis of Bioinformatics and in vitro Experiments 60. Identified GNGT1 and NMU as Combined Diagnosis Biomarker of Non-Small-Cell Lung Cancer Utilizing Bioinformatics and Logistic Regression 61. A bioinformatics WGS workflow for clinical Mycobacterium tuberculosis complex isolate analysis, validated using a reference collection extensively characterized with … 62. A bioinformatics Approach for Identification of the core ontologies and signature genes of Pulmonary Disease and Associated Disease 63. Author Correction: Single-cell RNA sequencing technologies and bioinformatics pipelines 64. A new bioinformatics tool to recover missing gene expression in single-cell RNA sequencing data 65. Comprehensive analysis of PLOD family members in low-grade gliomas using bioinformatics methods 66. Identification of four genes and biological characteristics associated with acute spinal cord injury in rats integrated bioinformatics analysis 67. Identification of a prognostic gene signature of colon cancer using integrated bioinformatics analysis 68. Identification of Inflammatory Genes, Pathways, and Immune Cells in Necrotizing Enterocolitis of Preterm Infant by Bioinformatics Approaches 69. Bioinformatics Resources for RNA Editing 70. … /immune system-specific expressed genes are considered as the potential biomarkers for the diagnosis of early rheumatoid arthritis through bioinformatics … 71. A Critical Review on the Application of Artificial Neural Network in Bioinformatics 72. Identification of candidate biomarkers of liver hydatid disease via microarray profiling, bioinformatics analysis, and machine learning 73. Identification of Hub Genes in Different Stages of Colorectal Cancer through an Integrated Bioinformatics Approach 74. Key Genes and Molecular Mechanism Investigation in the Synthesis of Maize Quercetin Based on SNP and Bioinformatics Analysis 75. Bioinformatics analysis of common key genes and pathways of intracranial, abdominal, and thoracic aneurysms 76. A Bioinformatics Systems Biology Analysis of the Current Oral Proteomic Biomarkers and Implications for Diagnosis and Treatment of External Root Resorption 77. Bioinformatics Analyses of Potential miRNA-mRNA Regulatory Axis in HBV-related Hepatocellular Carcinoma 78. … the Mechanisms and Molecular Targets of Qishen Yiqi Formula for the Treatment of Pulmonary Arterial Hypertension using a Bioinformatics/Network Topology-based … 79. Introduction to Unsupervised Learning in Bioinformatics 80. Insight into molecular profile changes after skeletal muscle contusion using microarray and bioinformatics analyses 81. Bioinformatics analysis and biochemical characterisation of ABC transporter-associated periplasmic substrate-binding proteins ModA and MetQ from Helicobacter … 82. Identification of biomarkers and construction of a microRNA mRNA regulatory network for clear cell renal cell carcinoma using integrated bioinformatics … 83. Identifying the p65-Dependent Effect of Sulforaphene on Esophageal Squamous Cell Carcinoma Progression via Bioinformatics Analysis 84. Clinical heterogeneity of the SLC26A4 gene in UAE patients with hearing loss and bioinformatics investigation of DFNB4/Pendred syndrome missense mutations 85. Identification of differentially expressed genes, signaling pathways and immune infiltration in rheumatoid arthritis by integrated bioinformatics analysis 86. Bioinformatics in Plant Pathology 87. Identification of hub genes in triple-negative breast cancer by integrated bioinformatics analysis 88. Functional Bioinformatics Analyses of the Matrisome and Integrin Adhesome 89. Identification of potential markers for differentiating epithelial ovarian cancer from ovarian low malignant potential tumors through integrated bioinformatics … 90. Integrated bioinformatics analysis reveals novel key biomarkers and potential candidate small molecule drugs in gestational diabetes mellitus 91. … Neurotrophic Factor Functions as a Potential Candidate Gene in Obstructive Sleep Apnea Based on a Combination of Bioinformatics and Targeted Capture … 92. Bioinformatics analysis indicates that microRNA 628 5p overexpression may alleviate Alzheimer’s disease by targeting TYROBP 93. POS0851 IDENTIFICATION OF HUB GENES AND PATHWAYS IN DERMATOMYOSITIS BY BIOINFORMATICS ANALYSIS 94. Bioinformatics analysis of Myelin Transcription Factor 1 95. Development and Optimization of Clinical Informatics Infrastructure to Support Bioinformatics at an Oncology Center 96. A Systems Bioinformatics Approach to Interconnect Biological Pathways 97. Bioinformatics identification of green tea anticancer properties: a network-based approach 98. Diatom metabarcoding and microscopic analyses from sediment samples at Lake Nam Co, Tibet: The effect of sample-size and bioinformatics on the identified … 99. Bioinformatics Applied to the Development of Biomolecules of Pharmaceutical Interest 100. Bioinformatics analysis of WRKY transcription factors in grape and their potential roles prediction in sugar and abscisic acid signaling pathway 101. The clinical and prognostic significance of LGR5 in GC: A meta-analysis of IHC assay and bioinformatics analysis. 102. Bioinformatics Investigation and Contribution of Other Chromosomes Besides Chromosome 21 in the Risk of Down Syndrome Development 103. Network Pharmacological Analysis through a Bioinformatics Approach of Novel NSC765600 and NSC765691 Compounds as Potential Inhibitors of CCND1/CDK4 … 104. Cohort Identification for Translational Bioinformatics Studies 105. Advances in Omics and Bioinformatics Tools for Phyllosphere Studies 106. Harmonic Progression in Bioinformatics and Recurrent Series in Inherited Biostructures 107. OverCOVID: an integrative web portal for SARS-CoV-2 bioinformatics resources 108. Statistical and Bioinformatics Analysis of Data from Bulk and Single-Cell RNA Sequencing Experiments 109. A Bioinformatics Tutorial for Comparative Development Genomics in Diverse Meiofauna 110. Multidrug resistance protein structure of Trypanosoma evansi isolated from buffaloes in Ngawi District, Indonesia: A bioinformatics analysis 111. Integrative Bioinformatics Analysis Reveals Noninvasive miRNA Biomarkers for Lung Cancer 112. Quantifying plasmid dynamics using single-cell microfluidics and image bioinformatics 113. Identification of four differentially expressed genes associated with acute and chronic spinal cord injury based on bioinformatics data 114. Identification of key pathways and gene expression in the activation of mast cells via calcium flux using bioinformatics analysis 115. Identification of Significant Genes and Therapeutic Agents for Breast Cancer by Integrated Bioinformatics 116. Using supercomputer to finish M1 Bioinformatics Exercise from Ogata Lab 117. Transcriptomic Alterations Induced by Vemurafenib After Treatment of Melanoma: A Comprehensive Bioinformatics Analysis 118. … OF MOLECULAR PHENOTYPES AND IMMUNE CELL INFILTRATION IN PSORIATIC ARTHRITIS PATIENTS’SKIN TISSUES BY INTEGRATED BIOINFORMATICS … 119. Bioinformatics: A New Insight Tool to Deal with Environment Management 120. Identification of acute spinal cord injury and autophagy-related potential key genes, pathways, and targeting drugs through bioinformatics analysis 121. Identification of Molecular Mechanisms Underlying Sex-Associated Differences in the Chronic Obstructive Pulmonary Disease through Bioinformatics Analysis 122. Bioinformatics and In Vitro Studies Reveal the Importance of p53, PPARG and Notch Signaling Pathway in Inhibition of Breast Cancer Stem Cells by … 123. Bioinformatics Approaches for Functional Prediction of Long Noncoding RNAs 124. A Comprehensive Phylogenetic and Bioinformatics Survey of Lectins in the Fungal kingdom 125. Deep networks and network representation in bioinformatics 126. Identifying Potential Prognostic Biomarkers Associated With Clinicopathologic Characteristics of Hepatocellular Carcinoma by Bioinformatics Analysis 127. Identification of key pathways and hub genes in the myogenic differentiation of pluripotent stem cell: a bioinformatics and experimental study 128. … of intestinal microbiome in a process of faecal microbiota transplantation in a patient with Clostridioides difficile infection: NGS analysis with different bioinformatics … 129. Clinical significance of long noncoding RNA MNX1-AS1 in human cancers: a meta-analysis of cohort studies and bioinformatics analysis based on TCGA datasets 130. Anticancer property of Zika virus proteins: Lack of evidence from predictive clinical bioinformatics study 131. Identification of Differentially Expressed Genes Using Deep Learning in Bioinformatics 132. Bioinformatics Analysis Predicts hsa_circ_0026337/miR-197-3p as a Potential Oncogenic ceRNA Network for Non-small Cell Lung Cancers 133. … Molecular Mechanism of Xiao Huoluo Pills in the Treatment of Cartilage Degeneration of Knee Osteoarthritis Based on Bioinformatics Analysis and Molecular … 134. A bioinformatics analysis of differentially expressed proteins in plasma exosome of acute-on-chronic liver failure patients with different prognoses 135. Bioinformatics Analyses of Serine Acetyltransferase (SAT) Gene Family in Rice (Oryza sativa) and their Expressions under Salt Stress 136. Expression profiling and bioinformatics analysis of exosomal long noncoding RNAs in patients with myasthenia gravis by RNA sequencing 137. Bioinformatics Analysis in Different Expression Genes and Potential Pathways of CD4+ Cells in Childhood Allergic Asthma 138. … Key Genes in Anaplastic Thyroid Cancer Using Bioinformatics AnalysisIdentification of Potential Key Genes in Anaplastic Thyroid Cancer using Bioinformatics … 139. … predicts poor prognosis in patients with surgically resected Lung Adenocarcinoma: A study based on Immunohistochemical Analysis and Bioinformatics 140. High-throughput screening and bioinformatics analysis of 2,000 177Lu-PSMA and Radiotherapy+ drug combinations 141. Bioinformatics Analysis of C3 and CXCR4 act as Potential Prognostic Biomarkers in Clear Cell Renal Cell Carcinoma (ccRCC) 142. Complexity matters: Evaluating the impact of bioinformatics parameters on eukaryotic MOTU delimitation and taxonomy assignment 143. Bioinformatics analysis of the expression and role of microRNA-221-3p in head and neck squamous cell carcinoma 144. In-silico analysis of BCL2 gene using multiple bioinformatics tools to identify the most lethal mutations that are crucial for its structural and functional integrity 145. A Novel Ferroptosis-related Lncrna Prognostic Signature for Colorectal Cancer by Bioinformatics Analysis 146. Correction to “Complementary Genomic Bioinformatics and Chemical Approaches Facilitate the Absolute Structure Assignment of Ionostatin, a Linear Polyketide from … 147. Correction to: A Bioinformatics Tutorial for Comparative Genomics of Meiofauna 148. Want to track pandemic variants faster? Fix the bioinformatics bottleneck 149. Use of Bioinformatics Technologies and Databases to Teach Analysis of Genetic Sequences to Undergraduate Students in Physics, Biotechnology, and … 150. A Single-Cell Bioinformatics Analysis of the Host Transcriptional Response to Infection Consisting of Natural Combinations of Influenza A Virus Gene Segments 151. Erratum to comprehensive bioinformatics analysis of the TP53 signaling pathway in Wilms’ tumor 152. Bioinformatics Analysis of DNA Methylation Through Bisulfite Sequencing Data 153. Correction to: Development and Optimization of Clinical Informatics Infrastructure to Support Bioinformatics at an Oncology Center 154. Comparison of bioinformatics pipelines for eDNA metabarcoding data analysis of fish populations in Czech reservoirs 155. Bioinformatics analysis combined with experiments predicts CENPK as a potential prognostic factor for lung adenocarcinoma 156. Bioinformatics Analysis of the Lycopene ß-Cyclase Gene in Jujube (Ziziphus jujube Mill) 157. Gene expression collective data analysis for studying the effects of high-LET ionizing radiation: A bioinformatics approach 158. CoV-AbDab: the coronavirus antibody database 159. BERT4Bitter: a bidirectional encoder representations from transformers (BERT)-based model for improving the prediction of bitter peptides 160. Sanhuang Jiangtang tablet protects type 2 diabetes osteoporosis via AKT-GSK3ß-NFATc1 signaling pathway by integrating bioinformatics analysis and experimental … 161. … Tuning onto Recurrent Neural Network and Long Short-Term Memory (RNN-LSTM) Network for Feature Selection in Classification of High-Dimensional Bioinformatics … 162. pyGenomeTracks: reproducible plots for multivariate genomic datasets 163. IS900 RFLP Analysis of Mycobacterium avium subsp. Paratuberculosis of Iranian Isolates and Analyze Using Bioinformatics Tools 164. Screening druggable targets and predicting therapeutic drugs for COVID-19 via integrated bioinformatics analysis 165. Supplementary Material to “Integrated analysis of label-free quantitative proteomics and bioinformatics reveal insights into signaling pathways in male breast … 166. … gallate interaction in SARS-CoV-2 spike-protein central channel with reference to the hydroxychloroquine interaction: bioinformatics and molecular docking study 167. CAFE: a software suite for analysis of paired-sample transposon insertion sequencing data 168. Multidrug resistance protein structure of Trypanosoma evansi isolated from buffaloes in Ngawi District, Indonesia: A bioinformatics analysis, Veterinary World, 14 (1) … 169. A Complete Bibliography of IEEE/ACM Transactions on Computational Biology and Bioinformatics 170. Ribbon: intuitive visualization for complex genomic variation 171. Bioinformatics analysis of gene expression profile and key pathways related to fatty infiltration after rotator cuff injury 172. Significance and Mechanisms Analyses of RB1 Mutation in Bladder Cancer Disease Progression and Drug Selection by Bioinformatics Analysis 173. Towards Investigating the Role of Proprotein Convertase Subtilisin/Kexin Family (PCSK/7/9) in Cancer by Using Bioinformatics Motif Detection Technique 174. CNVfilteR: an R/bioconductor package to identify false positives produced by germline NGS CNV detection tools 175. Potential prediction of phenolic compounds in red ginger (Zingiber officinale var. rubrum) as an AT1R antagonist by bioinformatics approach for antihypertensive oral … 176. [PS][PS] Interval Versions of Statistical Techniques, with Applications to Environmental Analysis, Bioinformatics, and Privacy in Statistical Databases 177. Using Interpretable Deep Learning to Model Cancer Dependencies 178. iCarPS: a computational tool for identifying protein carbonylation sites by novel encoded features 179. iEnhancer-XG: interpretable sequence-based enhancers and their strength predictor 180. Outlier detection in Bioinformatics with Mixtures of Gaussian and heavy-tailed distributions 181. A Modelling Framework for Embedding-based Predictions for Compound-Viral Protein Activity 182. SimText: A text mining framework for interactive analysis and visualization of similarities among biomedical entities 183. UniBioDicts: unified access to biological dictionaries 184. GraphDTA: Predicting drug–target binding affinity with graph neural networks 185. COVID-KOP: integrating emerging COVID-19 data with the ROBOKOP database 186. Discovering footprints of evolutionary chromatin response to transposons activity: merging biophysics with bioinformatics 187. mzRAPP: a tool for reliability assessment of data pre-processing in non-targeted metabolomics 188. In-silico prediction of in-vitro protein liquid-liquid phase separation experiments outcomes with multi-head neural attention 189. Pentraxin 3 is a diagnostic and prognostic marker for ovarian epithelial cancer patients based on comprehensive bioinformatics and experiments 190. ViralMSA: Massively scalable reference-guided multiple sequence alignment of viral genomes 191. PBSIM2: a simulator for long-read sequencers with a novel generative model of quality scores 192. MELODI Presto: a fast and agile tool to explore semantic triples derived from biomedical literature 193. UglyTrees: a browser-based multispecies coalescent tree visualizer 194. Mutation-Simulator: fine-grained simulation of random mutations in any genome 195. SAIGEgds—an efficient statistical tool for large-scale PheWAS with mixed models 196. Unsupervised protein embeddings outperform hand-crafted sequence and structure features at predicting molecular function 197. Detecting Let-7 isoforms of Salmonids by bioinformatics data 198. PICS2: next-generation fine mapping via probabilistic identification of causal SNPs 199. mixtureS: a novel tool for bacterial strain genome reconstruction from reads 200. Genozip: a universal extensible genomic data compressor 201. A systems-biology model of the tumor necrosis factor (TNF) interactions with TNF receptor 1 and 2 202. Early cancer detection from genome-wide cell-free DNA fragmentation via shuffled frog leaping algorithm and support vector machine 203. MAT2: Manifold alignment of single-cell transcriptomes with cell triplets 204. MDeePred: novel multi-channel protein featurization for deep learning-based binding affinity prediction in drug discovery 205. Narrative Scientific Data Visualization in an Immersive Environment 206. Assessing the fit of the multi-species network coalescent to multi-locus data 207. Predicting candidate genes from phenotypes, functions and anatomical site of expression 208. BamSnap: a lightweight viewer for sequencing reads in BAM files 209. ILoReg: a tool for high-resolution cell population identification from single-cell RNA-seq data 210. Characterizing protein conformers by cross-linking mass spectrometry and pattern recognition 211. GraphQA: protein model quality assessment using graph convolutional networks 212. CaNDis: a web server for investigation of causal relationships between diseases, drugs and drug targets 213. ProkSeq for complete analysis of RNA-seq data from prokaryotes 214. HATK: HLA analysis toolkit 215. A combined recall and rank framework with online negative sampling for chinese procedure terminology normalization 216. Systematic determination of the mitochondrial proportion in human and mice tissues for single-cell RNA-sequencing data quality control 217. DELPHI: accurate deep ensemble model for protein interaction sites prediction 218. SOLQC: Synthetic oligo library quality control tool 219. shinyÉPICo: A graphical pipeline to analyze Illumina DNA methylation arrays 220. dream: Powerful differential expression analysis for repeated measures designs 221. ASimulatoR: splice-aware RNA-Seq data simulation 222. SARS-CoV-2 Through the Lens of Computational Biology: How bioinformatics is playing a key role in the study of the virus and its origins 223. Interactive gene networks with KNIT 224. A database of flavivirus RNA structures with a search algorithm for pseudoknots and triple base interactions 225. IGD: high-performance search for large-scale genomic interval datasets 226. NetSets. js: a JavaScript framework for compositional assessment and comparison of biological networks through Venn-integrated network diagrams 227. Proteo-chemometrics interaction fingerprints of protein–ligand complexes predict binding affinity 228. GWASinspector: comprehensive quality control of genome-wide association study results 229. FASTRAL: Improving scalability of phylogenomic analysis 230. A network-based deep learning methodology for stratification of tumor mutations 231. Deuteros 2.0: peptide-level significance testing of data from hydrogen deuterium exchange mass spectrometry 232. The Russian Drug Reaction Corpus and neural models for drug reactions and effectiveness detection in user reviews 233. eHSCPr discriminating the cell identity involved in endothelial to hematopoietic transition 234. EM-stellar: benchmarking deep learning for electron microscopy image segmentation 235. mzRecal: universal MS1 recalibration in mzML using identified peptides in mzIdentML as internal calibrants 236. MR-Clust: clustering of genetic variants in Mendelian randomization with similar causal estimates 237. PyRice: a Python package for querying Oryza sativa databases 238. Bipartite graph-based approach for clustering of cell lines by gene expression-drug response associations 239. The iPPI-DB initiative: A Community-centered database of Protein-Protein Interaction modulators 240. MotifGenie: A Python Application for Searching Transcription Factor Binding Sequences Using ChIP-Seq Datasets 241. Network-guided search for genetic heterogeneity between gene pairs 242. Annotating high-impact 5′ untranslated region variants with the UTRannotator 243. Few shot domain adaptation for in situ macromolecule structural classification in cryoelectron tomograms 244. Recognition of small molecule-RNA binding sites using RNA sequence and structure 245. Machine Boss: rapid prototyping of bioinformatic automata 246. Automated download and clean-up of family-specific databases for kmer-based virus identification 247. PC2P: Parameter-free network-based prediction of protein complexes 248. CRAFT: Compact genome Representation toward large-scale Alignment-Free daTabase 249. CABEAN: a software for the control of asynchronous Boolean networks 250. Analysis of Collagen type X alpha 1 (COL10A1) expression and prognostic significance in gastric cancer based on bioinformatics 251. Large-scale entity representation learning for biomedical relationship extraction 252. Higher infectivity of the SARS-CoV-2 new variants is associated with K417N/T, E484K, and N501Y mutants: An insight from structural data 253. EARRINGS: an efficient and accurate adapter trimmer entails no a priori adapter sequences 254. ProteomeExpert: a docker image based web-server for exploring, modeling, visualizing, and mining quantitative proteomic data sets 255. PhyloCorrelate: inferring bacterial gene-gene functional associations through large-scale phylogenetic profiling 256. VIDHOP, viral host prediction with Deep Learning 257. CCmed: cross-condition mediation analysis for identifying replicable trans-associations mediated by cis-gene expression 258. Network-adjusted Kendall’s Tau Measure for Feature Screening with Application to High-dimensional Survival Genomic Data 259. Comparison of observation-based and model-based identification of alert concentrations from concentration–expression data 260. xGAP: A python based efficient, modular, extensible and fault tolerant genomic analysis pipeline for variant discovery 261. P69. 02 Identification of Potential Core Gene in Immune Infiltrates of EGFR Mutant Lung Adenocarcinoma using Bioinformatics Analysis 262. CellTracker: An Automated Toolbox for Single-Cell Segmentation and Tracking of Time-lapse Microscopy Images 263. Inferring cancer progression from single-cell sequencing while allowing mutation losses 264. ResiRole: residue-level functional site predictions to gauge the accuracies of protein structure prediction techniques 265. MendelVar: gene prioritization at GWAS loci using phenotypic enrichment of Mendelian disease genes 266. Robust and ultrafast fiducial marker correspondence in electron tomography by a two-stage algorithm considering local constraints 267. KORP-PL: a coarse-grained knowledge-based scoring function for protein–ligand interactions 268. A Method for Subtype Analysis with Somatic Mutations 269. PoSeiDon: a Nextflow pipeline for the detection of evolutionary recombination events and positive selection 270. FuSe: a tool to move RNA-Seq analyses from chromosomal/gene loci to functional grouping of mRNA transcripts 271. A statistical approach for tracking clonal dynamics in cancer using longitudinal next-generation sequencing data 272. Diamond: A Multi-Modal DIA Mass Spectrometry Data Processing Pipeline 273. TreeMap: a structured approach to fine mapping of eQTL variants 274. Co-phosphorylation networks reveal subtype-specific signaling modules in breast cancer 275. TSPTFBS: a docker image for Trans-Species Prediction of Transcription Factor Binding Sites in Plants 276. Lnc2Cancer 3.0: an updated resource for experimentally supported lncRNA/circRNA cancer associations and web tools based on RNA-seq and scRNA-seq data 277. Network approach to mutagenesis sheds insight on phage resistance in mycobacteria 278. QAlign: Aligning nanopore reads accurately using current-level modeling 279. Machine-OlF-Action: A unified framework for developing and interpreting machine-learning models for chemosensory research 280. Backward Pattern Matching on Elastic Degenerate Strings. 281. Coordinate Systems for Pangenome Graphs based on the Level Function and Minimum Path Covers. 282. CoRC: the COPASI R connector 283. MSL-ST: Development of Mass Spectral Library Search Tool to Enhance Compound Identification. 284. Genome-wide identification and bioinformatics characterization of superoxide dismutases in the desiccation-tolerant cyanobacterium Chroococcidiopsis … 285. Unpaired data empowers association tests 286. DataRemix: a universal data transformation for optimal inference from gene expression datasets 287. … -regulated Differentially Expressed Genes and Related Pathways in Hepatocellular Carcinoma: A Study Based on TCGA Database and Bioinformatics … 288. Discovering a sparse set of pairwise discriminating features in high-dimensional data 289. Gastric cancer-associated microRNA expression signatures: integrated bioinformatics analysis, validation, and clinical significance 290. Cataloguing experimentally confirmed 80.7 kb-long ACKR1 haplotypes from the 1000 Genomes Project database 291. BoardION: real-time monitoring of Oxford Nanopore sequencing instruments 292. Augur: a bioinformatics toolkit for phylogenetic analyses 293. TANTIGEN 2.0: a knowledge base of tumor T cell antigens and epitopes 294. Exploring the potential of Galangin in Cholangiocarcinoma cells using a bioinformatics approach 295. Efflux proteins at the blood-brain barrier: review and bioinformatics analysis (vol 48, pg 506, 2018) 296. GLEANER: a web server for GermLine cycle Expression ANalysis and Epigenetic Roadmap visualization 297. Open Targets Genetics: systematic identification of trait-associated genes using large-scale genetics and functional genomics 298. Advances in the prediction of mouse liver microsomal studies: From machine learning to deep learning 299. Advantages of using graph databases to explore chromatin conformation capture experiments 300. P* R* O* P: a web application to perform phylogenetic analysis considering the effect of gaps 301. Introduction to the JBCB Special Issue on Selected Papers from BICOB-2020 302. ViR: a tool to solve intrasample variability in the prediction of viral integration sites using whole genome sequencing data 303. Improving high-resolution copy number variation analysis from next generation sequencing using unique molecular identifiers 304. Genome-resolved metagenomics using environmental and clinical samples. 305. Set-theory based benchmarking of three different variant callers for targeted sequencing 306. Targeting a cytokine checkpoint enhances the fitness of armored cord blood CAR-NK cells 307. Construction and Analysis of mRNA and lncRNA Regulatory Networks Reveal the Key Genes Associated with Prostate Cancer Related Fatigue During Localized … 308. Current RNA-seq methodology reporting limits reproducibility 309. Search for SINE repeats in the rice genome using correlation-based position weight matrices 310. AutoDTI++: deep unsupervised learning for DTI prediction by autoencoders 311. Mining Biomedical Texts for Pediatric Information. 312. Identification of Key mRNAs, miRNAs, and mRNA-miRNA Network Involved in Papillary Thyroid Carcinoma 313. RaacLogo: a new sequence logo generator by using reduced amino acid clusters 314. NGlyAlign: an automated library building tool to align highly divergent HIV envelope sequences 315. CeNet Omnibus: an R/Shiny application to the construction and analysis of competing endogenous RNA network 316. Measurements of venous oxygen saturation in the superior sagittal sinus using conventional 3D multiple gradient-echo MRI: Effects of flow velocity and acceleration 317. SARS-CoV-2 hot-spot mutations are significantly enriched within inverted repeats and CpG island loci 318. Six genes involved in prognosis of hepatocellular carcinoma identified by Cox hazard regression 319. Deep Learning-Based Experimentation for Predicting Secondary Structure of Amino Acid Sequence 320. A novel end-to-end method to predict RNA secondary structure profile based on bidirectional LSTM and residual neural network 321. MMFGRN: a multi-source multi-model fusion method for gene regulatory network reconstruction 322. Identification of genetic variations associated with drug resistance in non-small cell lung cancer patients undergoing systemic treatment 323. BugSeq: a highly accurate cloud platform for long-read metagenomic analyses 324. The complete genome sequence of Hafnia alvei A23BA; a potential antibiotic-producing rhizobacterium 325. Web tools to fight pandemics: the COVID-19 experience 326. Single-cell transcriptomic profiling of satellite glial cells in stellate ganglia reveals developmental and functional axial dynamics 327. Computational strategies to combat COVID-19: useful tools to accelerate SARS-CoV-2 and coronavirus research 328. Toll-like Receptor 4 Gene Polymorphisms in Chinese Population After Allogeneic Hematopoietic Stem Cell Transplantation 329. ReCGBM: a gradient boosting-based method for predicting human dicer cleavage sites 330. NIDM: network impulsive dynamics on multiplex biological network for disease-gene prediction 331. Comparison study of differential abundance testing methods using two large Parkinson disease gut microbiome datasets derived from 16S amplicon … 332. Interaction of Nucleic Acids: Hidden Order of Interaction 333. Identification of Glioma Specific Genes as Diagnostic and Prognostic Markers for Glioma 334. Visual4DTracker: a tool to interact with 3D+ t image stacks 335. Establishing a consensus for the hallmarks of cancer based on gene ontology and pathway annotations 336. Temperature and latitude correlate with SARS-CoV-2 epidemiological variables but not with genomic change worldwide 337. Comparing de novo transcriptome assembly tools in di-and autotetraploid non-model plant species 338. SPServer: split-statistical potentials for the analysis of protein structures and protein–protein interactions 339. Identifying the sequence specificities of circRNA-binding proteins based on a capsule network architecture 340. Network-based identification genetic effect of SARS-CoV-2 infections to Idiopathic pulmonary fibrosis (IPF) patients 341. MATHLA: a robust framework for HLA-peptide binding prediction integrating bidirectional LSTM and multiple head attention mechanism 342. DeepDist: real-value inter-residue distance prediction with deep residual convolutional network 343. CHTKC: a robust and efficient k-mer counting algorithm based on a lock-free chaining hash table 344. DeepLPI: a multimodal deep learning method for predicting the interactions between lncRNAs and protein isoforms 345. COVID-19: disease pathways and gene expression changes predict methylprednisolone can improve outcome in severe cases. 346. Modeling drug mechanism of action with large scale gene-expression profiles using GPAR, an artificial intelligence platform 347. Brain Interface: Nano-Scaled Device as an Improvement in the Process of Learning 348. Profile hidden Markov model sequence analysis can help remove putative pseudogenes from DNA barcoding and metabarcoding datasets 349. Grafting Methionine on 1F1 Ab Increases the Broad-Activity on HA Structural-Conserved Residues of H1, H2, and H3 Influenza a Viruses 350. Accurate prediction of multi-label protein subcellular localization through multi-view feature learning with RBRL classifier 351. Pathway Tools version 23.0 update: software for pathway/genome informatics and systems biology 352. A decade of de novo transcriptome assembly: Are we there yet? 353. Feature selection based on fuzzy joint mutual information maximization [J] 354. tidyMicro: a pipeline for microbiome data analysis and visualization using the tidyverse in R 355. A survey of gene expression meta-analysis: methods and applications 356. Drug perturbation gene set enrichment analysis (dpGSEA): a new transcriptomic drug screening approach 357. PCirc: random forest-based plant circRNA identification software 358. Alvis: a tool for contig and read ALignment VISualisation and chimera detection 359. Small noncoding RNA discovery and profiling with sRNAtools based on high-throughput sequencing 360. DeepDRK: a deep learning framework for drug repurposing through kernel-based multi-omics integration 361. CoronaPep: An Anti-coronavirus Peptide Generation Tool 362. Identification of deregulation mechanisms specific to cancer subtypes 363. Computational resources for identifying and describing proteins driving liquid–liquid phase separation 364. Successful identification of predictive profiles for infection utilising systems-level immune analysis: a pilot study in patients with relapsed and refractory multiple … 365. SCC: an accurate imputation method for scRNA-seq dropouts based on a mixture model 366. MicrobeAnnotator: a user-friendly, comprehensive functional annotation pipeline for microbial genomes 367. Propedia: a database for protein–peptide identification based on a hybrid clustering algorithm 368. Epidemiological data analysis of viral quasispecies in the next-generation sequencing era 369. Prediction of tumor purity from gene expression data using machine learning 370. Practical Workflow from High-Throughput Genotyping to Genomic Estimated Breeding Values (GEBVs) 371. Design powerful predictor for mRNA subcellular location prediction in Homo sapiens 372. First Complete Genome of the Thermophilic Polyhydroxyalkanoates Producing Bacterium Schlegelella thermodepolymerans DSM 15344 373. recoup: flexible and versatile signal visualization from next generation sequencing 374. Predicting chemosensitivity using drug perturbed gene dynamics 375. Learning curves for drug response prediction in cancer cell lines 376. Anticancer peptides prediction with deep representation learning features 377. Structured sparsity regularization for analyzing high-dimensional omics data 378. MADGAN: unsupervised medical anomaly detection GAN using multiple adjacent brain MRI slice reconstruction 379. Novel perspectives for SARS-CoV-2 genome browsing 380. DISTEVAL: a web server for evaluating predicted protein distances 381. Fast and Accurate Multiple Sequence Alignment with MSAProbs-MPI 382. Prediction of RNA-binding protein and alternative splicing event associations during epithelial–mesenchymal transition based on inductive matrix completion 383. BioMedR: an R/CRAN package for integrated data analysis pipeline in biomedical study 384. Murine induced pluripotent stem cell-derived neuroimmune cell culture models emphasize opposite immune-effector functions of interleukin 13-primed microglia and … 385. A novel essential protein identification method based on PPI networks and gene expression data 386. Repeat DNA expands our understanding of autism spectrum disorder 387. Error-corrected estimation of a diagnostic accuracy index of a biomarker against a continuous gold standard 388. Using deep neural networks and biological subwords to detect protein S-sulfenylation sites 389. ProtFold-DFG: protein fold recognition by combining Directed Fusion Graph and PageRank algorithm 390. G-Tric: generating three-way synthetic datasets with triclustering solutions 391. Twelve years of SAMtools and BCFtools 392. mixIndependR: a R package for statistical independence testing of loci in database of multi-locus genotypes 393. PredCID: prediction of driver frameshift indels in human cancer 394. Dualmarker: a flexible toolset for exploratory analysis of combinatorial dual biomarkers for clinical efficacy 395. A novel computational framework for genome-scale alternative transcription units prediction 396. H2V: a database of human genes and proteins that respond to SARS-CoV-2, SARS-CoV, and MERS-CoV infection 397. Isolating SARS-CoV-2 strains from countries in the same meridian: genome evolutionary analysis 398. Federated sharing and processing of genomic datasets for tertiary data analysis 399. SARS-CoV-2 3D database: understanding the coronavirus proteome and evaluating possible drug targets 400. Postoperative radiotherapy is associated with improved overall survival for alveolar ridge squamous cell carcinoma with adverse pathologic features 401. Mass spectrometry–based protein identification in proteomics—a review 402. CodAn: predictive models for precise identification of coding regions in eukaryotic transcripts 403. Genome-wide discovery of pre-miRNAs: comparison of recent approaches based on machine learning 404. Meta-i6mA: an interspecies predictor for identifying DNA N6-methyladenine sites of plant genomes by exploiting informative features in an integrative machine … 405. Graph and Convolution Recurrent Neural Networks for Protein-Compound Interaction Prediction 406. FoldRec-C2C: protein fold recognition by combining cluster-to-cluster model and protein similarity network 407. Next generation sequencing of SARS-CoV-2 genomes: challenges, applications and opportunities 408. A computational platform to identify origins of replication sites in eukaryotes 409. Toli c N, Jaitly N, Shaw JL, Adkins JN, Smith RD 410. Adverse events associated with potential drugs for COVID-19: a case study from real-world data 411. KFGRNI: A robust method to inference gene regulatory network from time-course gene data based on ensemble Kalman filter. 412. Updates to HCOP: the HGNC comparison of orthology predictions tool 413. Bicuspid aortic valve sparing root replacement 414. Unsupervised and self-supervised deep learning approaches for biomedical text mining 415. HapSolo: An optimization approach for removing secondary haplotigs during diploid genome assembly and scaffolding 416. Computational Antigen Discovery for Eukaryotic Pathogens Using Vacceed 417. Wireless Wi-Fi module Testing Procedure in Gigabyte Passive Optical Network to Optical Network Terminal of Equipment 418. Receiver for m-ary Radio Communication System Between Motile Objects in the Microwave Range 419. Impact of perioperative factors on nadir serum prostate-specific antigen levels after holmium laser enucleation of prostate 420. gutMEGA: a database of the human gut MEtaGenome Atlas 421. Computer-aided prediction and design of IL-6 inducing peptides: IL-6 plays a crucial role in COVID-19 422. Toxicological assessment of newly expressed proteins (NEPs) in genetically modified (GM) plants 423. SubLocEP: a novel ensemble predictor of subcellular localization of eukaryotic mRNA based on machine learning 424. Determining Cell Death Pathway and Regulation by Enrichment Analysis 425. The European Nucleotide Archive in 2020 426. iCysMod: an integrative database for protein cysteine modifications in eukaryotes 427. A review on viral data sources and search systems for perspective mitigation of COVID-19 428. Searching for universal model of amyloid signaling motifs using probabilistic context-free grammars 429. Current challenges and best-practice protocols for microbiome analysis 430. The Coronavirus Network Explorer: Mining a large-scale knowledge graph for effects of SARS-CoV-2 on host cell function 431. Deep-belief network for predicting potential miRNA-disease associations 432. DeepATT: a hybrid category attention neural network for identifying functional effects of DNA sequences 433. A molecular modelling approach for identifying antiviral selenium-containing heterocyclic compounds that inhibit the main protease of SARS-CoV-2: an in silico … 434. SG-LSTM-FRAME: A computational frame using sequence and geometrical information via LSTM to predict miRNA–gene associations 435. Common low complexity regions for SARS-CoV-2 and human proteomes as potential multidirectional risk factor in vaccine development 436. OrthoDB in 2020: evolutionary and functional annotations of orthologs 437. PDB-tools web: A user-friendly interface for the manipulation of PDB files 438. Comparative evaluation of full-length isoform quantification from RNA-Seq 439. SurvivalMeth: a web server to investigate the effect of DNA methylation-related functional elements on prognosis 440. The peripheral and core regions of virus-host network of COVID-19. 441. Accucopy: accurate and fast inference of allele-specific copy number alterations from low-coverage low-purity tumor sequencing data 442. Unveiling COVID-19-associated organ-specific cell types and cell-specific pathway cascade 443. Drug-induced cell viability prediction from LINCS-L1000 through WRFEN-XGBoost algorithm 444. MeSHHeading2vec: a new method for representing MeSH headings as vectors based on graph embedding algorithm 445. M6A2Target: a comprehensive database for targets of m6A writers, erasers and readers 446. Computational recognition of lncRNA signature of tumor-infiltrating B lymphocytes with potential implications in prognosis and immunotherapy of bladder cancer 447. MNDR v3. 0: mammal ncRNA–disease repository with increased coverage and annotation 448. CNA2Subpathway: identification of dysregulated subpathway driven by copy number alterations in cancer 449. DeepTorrent: a deep learning-based approach for predicting DNA N4-methylcytosine sites 450. Severe acute respiratory syndrome coronavirus (SARS-CoV)-2 infection induces dysregulation of immunity: in silico gene expression analysis 451. A stack LSTM structure for decoding continuous force from local field potential signal of primary motor cortex (M1) 452. The Distinction of Omics in Amelioration of Food Crops Nutritional Value 453. DeepCNV: a deep learning approach for authenticating copy number variations 454. Key residues influencing binding affinities of 2019-nCoV with ACE2 in different species 455. A survey on computational models for predicting protein–protein interactions 456. ADeditome provides the genomic landscape of A-to-I RNA editing in Alzheimer’s disease 457. Integrated hybrid de novo assembly technologies to obtain high-quality pig genome using short and long reads 458. WIND (Workflow for pIRNAs aNd beyonD): a strategy for in-depth analysis of small RNA-seq data 459. Current Situation and Prospect of EMDB/EMPIAR-China 460. isomiRs–Hidden Soldiers in the miRNA Regulatory Army, and How to Find Them? 461. Identifying the natural polyphenol catechin as a multi-targeted agent against SARS-CoV-2 for the plausible therapy of COVID-19: an integrated computational … 462. rmvp: A memory-efficient, visualization-enhanced, and parallel-accelerated tool for genome-wide association study 463. Roles of host small RNAs in the evolution and host tropism of coronaviruses 464. CpG-island-based annotation and analysis of human housekeeping genes 465. Classification and gene selection of triple-negative breast cancer subtype embedding gene connectivity matrix in deep neural network 466. A novel privacy-preserving federated genome-wide association study framework and its application in identifying potential risk variants in ankylosing spondylitis 467. QAUST: Protein Function Prediction Using Structure Similarity, Protein Interaction, and Functional Motifs 468. Comparative host–pathogen protein–protein interaction analysis of recent coronavirus outbreaks and important host targets identification 469. Adult polyglucosan body disease—an atypical compound heterozygous with a novel GBE1 mutation 470. Differential expression of Triggering Receptor Expressed on Myeloid cells 2 (Trem2) in tissue eosinophils 471. Breast Tumor Microenvironment in Black Women: A Distinct Signature of CD8+ T Cell Exhaustion 472. MiBiOmics: An interactive web application for multi-omics data exploration and integration 473. Computational prediction and interpretation of both general and specific types of promoters in Escherichia coli by exploiting a stacked ensemble-learning framework 474. Development of a novel immune-related lncRNA signature as a prognostic classifier for endometrial carcinoma 475. SCDC: bulk gene expression deconvolution by multiple single-cell RNA sequencing references 476. MolAICal: a soft tool for 3D drug design of protein targets by artificial intelligence and classical algorithm 477. Homoeologous gene expression and co-expression network analyses and evolutionary inference in allopolyploids 478. Exosomal circRNA as a novel potential therapeutic target for multiple myeloma-related peripheral neuropathy 479. Predicting protein subchloroplast locations: the 10th anniversary 480. Tensor decomposition with relational constraints for predicting multiple types of microRNA-disease associations 481. GPS-Palm: a deep learning-based graphic presentation system for the prediction of S-palmitoylation sites in proteins 482. DSG2 expression is low in colon cancer and correlates with poor survival 483. Deep sparse transfer learning for remote smart tongue diagnosis [J] 484. A hybrid method for classification of physical action using discrete wavelet transform and artificial neural network 485. cncRNAdb: a manually curated resource of experimentally supported RNAs with both protein-coding and noncoding function 486. Design of an epitope-based peptide vaccine against the SARS-CoV-2: a vaccine-informatics approach 487. Upregulation of peroxisome proliferator-activated receptor-a and the lipid metabolism pathway promotes carcinogenesis of ampullary cancer 488. HISNAPI: a bioinformatic tool for dynamic hot spot analysis in nucleic acid–protein interface with a case study 489. Mesenchymal stromal cells provide hepatic support after extended hepatectomy by modulating thrombospondin-1/TGF-ß 490. PySmash: Python package and individual executable program for representative substructure generation and application 491. Exploring transcriptional switches from pairwise, temporal and population RNA-Seq data using deepTS 492. CAMAMED: a pipeline for composition-aware mapping-based analysis of metagenomic data 493. NCMCMDA: miRNA–disease association prediction through neighborhood constraint matrix completion 494. Allogeneic hematopoietic stem cell transplantation in leukocyte adhesion deficiency type I and III 495. Justification of the Choice of Signal Processing Method and Its Implementation in the Digital Part of the Receiver for Radar Stations 496. TRlnc: a comprehensive database for human transcriptional regulatory information of lncRNAs 497. Biomedical data and computational models for drug repositioning: a comprehensive review 498. ?????????????? 499. Comprehensive characterization of alternative splicing in renal cell carcinoma 500. Clinically relevant updates of the HbVar database of human hemoglobin variants and thalassemia mutations 501. Transcriptome analysis of cepharanthine against a SARS-CoV-2-related coronavirus 502. Toward a gold standard for benchmarking gene set enrichment analysis 503. Web-Based Base Editing Toolkits: BE-Designer and BE-Analyzer 504. Multigene editing: current approaches and beyond 505. Guía docente 200630-FBIO-Fundamentos de Bioinformática 506. Identification of Potential Gene and MicroRNA Biomarkers of Acute Kidney Injury 507. Further promotion of “the JSH plan for the future” conscious of new normal after/with COVID-19: message from the new president of the Japanese Society of … 508. Predicting drug-induced hepatotoxicity based on biological feature maps and diverse classification strategies 509. The international nucleotide sequence database collaboration 510. Integrating multi-network topology for gene function prediction using deep neural networks 511. PyConvU-Net: a lightweight and multiscale network for biomedical image segmentation 512. Recent advances in user-friendly computational tools to engineer protein function 513. Interpretable detection of novel human viruses from genome sequencing data 514. Comprehensive fundamental somatic variant calling and quality management strategies for human cancer genomes 515. DeepCPP: a deep neural network based on nucleotide bias information and minimum distribution similarity feature selection for RNA coding potential prediction 516. LncRNA LIFR-AS1 promotes proliferation and invasion of gastric cancer cell via miR-29a-3p/COL1A2 axis 517. Application of deep learning methods in biological networks 518. Molecular dynamics simulations for genetic interpretation in protein coding regions: where we are, where to go and when 519. Genome Resource: Ralstonia solanacearum Phylotype II Sequevar 1 (Race 3 Biovar 2) Strain UW848 From the 2020 US Geranium Introduction 520. Genome-wide expression profiling of long non-coding RNAs and competing endogenous RNA networks in alopecia areata [J] 521. Diagnostic and prognostic value of thymidylate synthase expression in breast cancer 522. The functional determinants in the organization of bacterial genomes 523. Inferring microenvironmental regulation of gene expression from single-cell RNA sequencing data using scMLnet with an application to COVID-19 524. From ArrayExpress to BioStudies 525. AntiCP 2.0: an updated model for predicting anticancer peptides 526. Exploration of natural compounds with anti-SARS-CoV-2 activity via inhibition of SARS-CoV-2 Mpro 527. The COVID-19 Pandemic Vulnerability Index (PVI) Dashboard: Monitoring county-level vulnerability using visualization, statistical modeling, and machine … 528. How do we share data in COVID-19 research? A systematic review of COVID-19 datasets in PubMed Central Articles 529. Systemic effects of missense mutations on SARS-CoV-2 spike glycoprotein stability and receptor-binding affinity 530. Progress and challenge for computational quantification of tissue immune cells 531. Closing the circle: current state and perspectives of circular RNA databases 532. The Compensation of Radiation-Induced Losses in the Fiber Optic Communication Line in Its Operation Mode 533. A comprehensive integrated drug similarity resource for in-silico drug repositioning and beyond 534. A Comprehensive Analysis Identified Hub Genes and Associated Drugs in Alzheimer’s Disease 535. GENCODE 2021 536. Network analyses in microbiome based on high-throughput multi-omics data 537. Different molecular enumeration influences in deep learning: an example using aqueous solubility 538. Integrated omics analysis reveals the alteration of gut microbe–metabolites in obese adults 539. Archaeal roots of intramembrane aspartyl protease siblings signal peptide peptidase and presenilin 540. RICORD: A Precedent for Open AI in COVID-19 Image Analytics 541. A new graph-based clustering method with application to single-cell RNA-seq data from human pancreatic islets 542. Discovery of G-quadruplex-forming sequences in SARS-CoV-2 543. Algorithm optimization for weighted gene co-expression network analysis: accelerating the calculation of Topology Overlap Matrices with OpenMP and SQLite 544. Exploring associations of non-coding RNAs in human diseases via three-matrix factorization with hypergraph-regular terms on center kernel alignment 545. QSAR-assisted-MMPA to expand chemical transformation space for lead optimization 546. Regulatory Assessment of Off-Target Changes and Spurious DNA Insertions in Gene-Edited Organisms for Agri-Food Use 547. DNSS2: improved ab initio protein secondary structure prediction using advanced deep learning architectures 548. DPCMNE: detecting protein complexes from protein-protein interaction networks via multi-level network embedding 549. PanACoTA: A modular tool for massive microbial comparative genomics 550. Peptides: Molecular and Biotechnological Aspects. Biomolecules 2021, 11, 52
Research Topics Computer Science
Bioinformatics
Related Posts:
- Computational Geometry Research Topics ideas
- Dynamic Networks Research Topics ideas
- Research Topics Ideas of Information Privacy
- Research Topics Ideas of Operating Systems
- Probabilistic algorithms Research Topics Ideas
- Research Topics ideas and Areas of communication protocols
Translational Informatics
Welcome to the future
ISBDS: Project ideas and templates
Independent Study in Biomedical Informatics (ISBDS)
This document provides ideas for research projects, and links to research plan templates, which are partially completed plans. Template files are available via the ISBDS course GitHub repository . For ISBDS, a research plan template can vary within biomedical science topics, but definitely includes a specific data source, overall problem statement, and methodological approach. Students will be required to complete the template to comprise a preliminary research plan for approval prior to registration. Advisors are invited to contribute research plan templates in their areas of interest and expertise, which may be based on the generic ISBDS Research Plan Template .
General Suggestions
- Review and analysis of an important public dataset.
- Review and analysis of an important public informatics tool.
- Reproducing and extending a published analysis.
- Building a database from public sources for a biomedical topic of interest.
- Adapt approaches, projects, and learning objectives from an existing, MOOC or other online course (e.g. Coursera , edX , Johns Hopkins , Indiana , Stanford , Hasso Plattner ), with or without completing the course.
- Respond to an online data science challenge (e.g. Kaggle ).
- Building an online app for researchers, clinicians, or patients.
- Create or improve an open source software package.
Bioinformatics
- Network Analysis in Systems Biology (coursera.org)
- Target Illumination GWAS Analytics (TIGA); see paper and repository .
- Knowledge Graph Analytics Platform (KGAP); see paper and repository .
- STRING: functional protein association networks
- Systems Biology; Metabolic engineering for synthetic biology.
- Structure to function.
- GTEx Portal
- Sequence alignment.
Cheminformatics
- PubChem analysis, descriptive or predictive
- ChEMBL analysis, descriptive or predictive
- DrugCentral analysis, descriptive or predictive
- Badapple analysis, descriptive or predictive
Drug Discovery
- Bioactivity prediction by machine learning (see https://predictor.ncats.io/ , https://atomscience.org/ , https://drugcentral.org/Redial , https://deepchem.io/ ).
- TEMPLATE: Homology Modeling (adapted from Intro to Biocomputing Unit 2 Assignment 1 and Assignment 2 )
- TEMPLATE: Virtual Screening (adapted from Intro to Biocomputing Unit 3 Assignment 1 and Assignment 2 )
- Chemical Predictive Modeling (Abhik Seal).
- Knime for Cheminformatics (Abhik Seal).
Medical Informatics
- OHDSI (Observational Health Data Sciences and Informatics): replicate, vary or extend published studies .
- Open Medical Record System (OpenMRS) The global OpenMRS community works together to build the world’s leading open source enterprise electronic medical record system platform. https://wiki.openmrs.org/
- Clinical Data Analysis in R (Abhik Seal)
Computational modeling
- Tumor modeling
- Bacterial infection modeling
- Blood Sugar Regulation
- Viral transmission modeling
Public Health & Epidemiology
- Public Health: Big Cities Health Coalition (BCHC) and Big Cities Health Inventory (BCHI)
- Healthcare Cost and Utilization: HCUP-US Databases
- HealthData.gov
- SEER-Medicare Health Outcomes Survey (SEER-MHOS) Linked Data Resource Surveillance, Epidemiology & End Results.
- Medicare Provider Utilization and Payment Data https://www.cms.gov/Research-Statistics-Data-and-Systems/Statistics-Trends-and-Reports/Medicare-Provider-Charge-Data/Physician-and-Other-Supplier.html
- CORD-19 , COVID-19 Open Research Dataset (CORD).
- WHO Coronavirus (COVID-19) Dashboard | WHO Coronavirus (COVID-19) Dashboard With Vaccination Data .
- Johns Hopkins Coronavirus Resource Center .
- EMBL-EBI COVID-19 Data Portal
Fitness, Wellness, & Health
- The Open Artificial Pancreas System project OpenAPS.org is an open and transparent effort to make safe and effective basic Artificial Pancreas System (APS) technology widely available to more quickly improve and save as many lives as possible and reduce the burden of Type 1 diabetes. OpenAPS means basic overnight closed loop APS technology is more widely available to anyone with compatible medical devices who is willing to build their own system .
- ResearchKit & CareKit from Apple. CareKit allows developers to build apps that leverage a variety of customizable modules. CareKit apps will let users regularly track care plans, monitor their progress, and share their insights with care teams. CareKit is open source, developers can build upon existing modules and contribute new code to help users world wide create a bigger—and better—picture of their health.
Natural language processing (NLP) and text mining
- PubMed named entity recognition (NER); see JensenLab Tools including Tagger .
- Twitter sentiment analysis
- Clustering by topic modeling
- See code and projects from Jason Timm ,
Databases and datasets
- MHEALTH Dataset Data Set body motion and vital signs.
- Kaggle (over 50,000 public datasets and 400,000 public notebooks),.
- Aggregate Analysis of ClincalTrials.gov (AACT) Database | Clinical Trials Transformation Initiative ,
- Hetionet – An integrative network of biomedical knowledge assembled from 29 different databases of genes, compounds, diseases, and more. The network combines over 50 years of biomedical information into a single resource, consisting of 47,031 nodes (11 types) and 2,250,197 relationships (24 types).
- ROBOKOP (Reasoning Over Biomedical Objects linked in Knowledge Oriented Pathways): Robokop is a biomedical reasoning system that interacts with many biomedical knowledge sources to answer questions. Robokop is one of several prototype systems under active development with NIH NCATS.
- Drug Central , online drug compendium.
- Illuminating the Druggable Genome (IDG) : Pharos and Target Central Resource Database (TCRD) .
- The openFDA FDA Adverse Event Reporting System (FAERS) is a database that contains information on adverse event and medication error reports submitted to FDA.
- New Mexico Decedent Image Database
- Embase , a highly versatile, multipurpose and up-to-date biomedical research and literature database.

- Previous Article
- Next Article
Cover Image

Introduction to bioinformatics
Skills required for bioinformatics, further reading, author information, a beginner’s guide to bioinformatics.
- Split-Screen
- Article contents
- Figures & tables
- Supplementary Data
- Peer Review
- Open the PDF for in another window
- Cite Icon Cite
- Get Permissions
Krutik Patel; A beginner’s guide to bioinformatics. Biochem (Lond) 28 April 2023; 45 (2): 11–15. doi: https://doi.org/10.1042/bio_2022_136
Download citation file:
- Ris (Zotero)
- Reference Manager
Bioinformatics has revolutionized the modern life sciences and has become a component of many undergraduate training courses and post-graduate research projects. As such, we are seeing more bioinformatics and programming aspects within undergraduate training and so it is important to understand what bioinformatics is, why it is a necessity in modern research and how young academics can begin their journey as bioinformaticians. This article outlines the broad spectrum of what bioinformatics is used for within research labs and provides several resources for beginners to learn how to code and perform bioinformatic tasks.
When imaging a bioinformatician, you may think of a dishevelled individual sitting in a dark room, frantically writing code and speaking in a complex and unintelligible jargon. While this caricature may often be portrayed, bioinformatics has become an integral part of research and is commonly found as a component of undergraduate and post-graduate programs. Bioinformatics or computational biology (which are the same thing) has quickly jumped from a specialist term describing an elite group of biologists that swapped their pipettes for Python and Pearl to a more generic description which encapsulates any form of computational analysis on biological data. These skills were utilized by higher education institutes during the pandemic as undergraduate and post-graduate students were often asked to undertake alternative capstone projects completely based online. Bioinformatic skills are also heavily required within the research environment as the volume of data being generated by labs is increasing, so the demand is growing for those with the skills to process and analyse large quantities of data.
The demand of bioinformaticians can be reflected by the increasing number of institutions in the UK which enrol prospective students into biology courses with bioinformatic modules or entirely computational post-graduate training courses. Requirements for bioinformatics masters’ courses often require a 2:1 in a relevant bachelor’s degree. Several drivers have influenced this increase in bioinformatics-related courses such as a greater appreciation of how skills in programming can hasten research outcomes, a data-boom in biology which requires specialist analysis, an ever-increasing demand to increase automation and efficiency in research and industry and the potential benefit machine learning approaches have to offer research. These and other factors are influencing the landscape of modern research and are clearly explained by the references below, which are recommended as additional reading to those interested in the subject.
How does bioinformatics work?
During an undergraduate capstone project, academic staff may instruct their students to perform bioinformatics – which is becoming an increasingly vague term. So, first, I believe it is important to understand what bioinformatics is. At its heart, bioinformatics is the use of computation to understand more about biology and should not be thought of as a replacement to traditional laboratory research – rather a synergistic partner. Another way to phrase it would be to use data science approaches to ask questions in the life sciences.
To help visualize the question ‘what is bioinformatics’, we can try to frame how bioinformatics works in research. Many questions from life science come from the natural world/ in vivo – this can include any range of topics such as investigating the molecular causes of diseases, contrasting virulence of different pathogens or researching the origins of human evolution. Researchers could then replicate and test what was seen in the natural world in the laboratory using in vitro techniques such as a western blot to measure the effect a gene mutation has on the protein expression level of a known oncogenic gene, or infecting cells with a pathogen and contrasting the viability between infected cells and non-infected cells. A bioinformatician would then use what has been measured in vitro to inform computational analysis ( in silico ). The analysis itself can range from measuring samples in parallel to reduce manual effort, running specialist software for quality control or simply producing graphs in a popular coding language like Python or R . The goal of the analysis could be to generate testable hypothesis which can be validated back in the laboratory, and this in turn would lead to learning something new about the natural world ( Figure 1 ).

How bioinformatics contributes to biology. Figure made with BioRender. Summary of how bioinformatics works within research. Questions from the natural world can be replicated within a lab and then analysed using computational techniques. Bioinformatic analysis can direct the validatory tests which would need to be conducted within a laboratory setting assess the processed and learn new information.
Why is bioinformatics necessary in biological sciences?
Bioinformatics supports every aspect in modern biological research. In some ways bioinformatics is not a useful term for the vast array of different tasks that can be performed for uses in biology ( Figure 2 ). Illustrated in Figure 2 is a portfolio which shows the range of tasks from distinct disciplines within biological research that require an expert in bioinformatics. Building machine learning models to make predictions or classifications based on biological data is a novel method of using large complex datasets to find patterns which cannot be detected without the use of powerful algorithms. Such machine learning tasks are becoming a popular avenue with the increasing availability of big data in biology. Big data refers to the exponentially expanding amount of biological data (e.g., sequencing data, gene expression data, population level data), which bioinformaticians must work with. Running a DNA sequencing workflow to detect genetic variations has become the optimal method of diagnosing patients with rare diseases, and this is shown by the success of the 100,000 Genomes project in the UK, which has identified many novel rare diagnoses. Building evolutionary trees based on ancient DNA samples is a necessary step to accurately understand species evolution and this can be powered by intelligent algorithms. Contrasting disease and healthy control samples to identify biological markers is a heavily researched area for complex conditions such as cancers and neurodegenerative disorders. Finally, graphical representation of large data such as clustering graphs (e.g., heatmaps) to identify similarities between biological samples is useful and can lead to efficient methods of identifying patterns. In each of these examples the specific context is less important than the overall theme of using data science approaches to unearth novel insights from biological information.

Examples of tasks which can be classed as bioinformatics. Figure made with BioRender. Several bioinformatic techniques include building machine learning models, sequencing DNA samples, building evolutionary trees, contrasting expression data between disease and control groups and visualizing data.
Now that we have a better grasp of what bioinformatics is, we go through the skills required to become a bioinformatician – either for a project or for career development.
Bioinformatics is centred around programming
Coding is at the core of bioinformatics, as bioinformaticians are expected to produce specialist scripts – lines of code to perform functions for specific tasks. The two most well-used programming languages in modern biological research labs are R and Python , and both have their merits.
R has a bigger biological community and much of this is driven by the Bioconductor project which contains thousands of biology-based tools written in R . With many ready-to-use applications, the burden of labour falls less towards script development, making R very useful for shorter projects such as capstone or master research projects. Another advantage of R is its popular integrated development environment (IDE) Rstudio , which provides a very accessible layout for scripting. The major disadvantages of relying on R is that many of the error messages are difficult to decipher and that tools can become out-of-date quite quickly (<2 years).
On the other hand, Python also has tools for biological use and, in contrast to R -based tools, can often continue functioning for decades; the error messages in Python are generally more easily understandable; and Python is faster than R making it more useful for larger datasets. Another advantage of Python is that it has many complete end-to-end and user-friendly machine learning-based tools that contain many algorithms, machine learning processes and structures available for use, such as scikit-learn and keras , which are becoming more popular for use in biology. However, working in Python means that researchers must write more extensive scripts compared to R . Additionally, Python has many more options for IDEs than R , such as Spyder , Jupyter Notebook and Pycharm , and it may take time to find an ideal one. Pycharm is great for experienced coders, and Jupyter is great for learning – making it an excellent choice for your first python project. My personal preference is Spyder , as in my opinion, its design is well suited for analysing complex biological datasets.
Overall, both R and Python have their advantages and most tasks like those shown in Figure 2 can be performed with either. There are distinct advantages when using R such as the many ready-to-use tools which can make scripting tasks simpler. But, in biological research right now, Python is currently far more useful for machine learning-based tasks.
Resources to begin coding
There are several ways to increase knowledge about coding. Courses are a great way to begin, such as those promoted by Datacamp . Even watching free tutorials on YouTube can be an effective learning strategy to learn programming and more about bioinformatic concepts or tools. Some brilliant channels include StatQuest which goes over statistical and bioinformatic application in short videos and the Bioinformatics-along learning videos which are a collection of hour-long videos going over bioinformatic tool use. I have provided a short playlist of helpful videos for beginners. Another good resource are the people around you – do not be shy about asking for help or saying you do not know how to perform certain tasks. However, by far the best method is to get stuck in and learn by doing. To get started, create a small project which consists of uncomplicated tasks which can be performed in excel, e.g., create a bar graph from some data or calculate the mean of each row in a table and try them out in R , Python or any other language you are interested in learning. Many interesting datasets to begin coding can be freely downloaded from kaggle . For more guided project ideas, there are several resources online, such as blogs by Datacamp ( R ) or Upgrad ( Python ). If you get stuck, look online. Most coding-related questions have probably been asked and are probably on a forum on the web. Good forums for bioinformatics-related issues include SEQanswers and Biostars , and most coding issues may have answers written in Stack Overflow .
Resources to begin a machine learning project
Like the points made above, there are good courses to begin learning how to begin a machine learning project on sites such as Datacamp . Again, a brilliant resource is kaggle , which contains downloadable datasets, readymade projects aimed at beginners, and has regular challenges for beginners and more advanced programmers. The biochemical society also runs online courses in R and Python . Simple machine learning projects could include creating a model to differentiate between cats and dogs or predicting the preferred coffee choices from staff at a school. The skills learnt through such training can be transferred to creating a model to differentiate cancer and non-cancerous tissue images or creating a model to predict genes important in neurodegenerative diseases.
Another part of machine learning is the theory of why certain practices are more efficient than others and a good resource to better understand this is towards data science, which houses blogs from talented data scientists, the blogs containing the code and rationale behind the code and are all clearly written for both experts and non-experts. For those that prefer videos, the Andrew Ng’s Stanford lecture course is available on YouTube and is great for those who want to grasp theory and begin developing applicable ideas.
Further skills important in bioinformatics
There are several further qualities which can be very helpful in bioinformatics. First, as expanded on in this article, hands-on experience in coding is very useful as it shows aptitude for creating scripts and performing programming tasks. Creating a portfolio of scripts you have created is a good way to show evidence of your skills. Such scripts could be stored as a public repository in Github , which would also be an impressive resource to have in your CV. Also, there is a big advantage in understanding the fundamentals of the biological questions you are interested in answering using bioinformatics. Being a bioinformatician does not mean one should ignore or be abstinent to the biological questions being researched. This is quite a misconception and, in all honesty, not understanding the data makes the computational tasks more difficult because there is little or no use in performing analysis or creating machine learning models which mis-interprets the research question. Another skill is to be flexible and understanding that bioinformatics has become an umbrella term which covers a wide range of interdisciplinary elements including programming, software development, maths, statistics and much more ( Figure 3 ) and so to work in the field one must develop the skills to be flexible. Furthermore, it is also important to know about current trends in the field. Machine learning/AI, big data techniques and multi-omics (combined DNA, RNA and/or protein) analysis are currently the major themes in the frontline of modern bioinformatics and there is a need for skilled individuals to research and work in these areas.

Showing that bioinformatics is an interdisciplinary field. Figure made with BioRender. Bioinformatics is an umbrella term for multiple disciplines within biology.
Further reading on the current demand on bioinformatics in life science
Attwood, Teresa K., et al. "A global perspective on evolving bioinformatics and data science training needs." Briefings in Bioinformatics20.2 (2019): 398-404. https://doi.org/10.1093/bib/bbx100
Gauthier, Jeff, et al. "A brief history of bioinformatics." Briefings in bioinformatics20.6 (2019): 1981-1996. https://doi.org/10.1093/bib/bby063
Kanehisa, Minoru, and Peer Bork. "Bioinformatics in the post-sequence era." Nature genetics33.3 (2003): 305-310. https://doi.org/10.1038/ng1109
Bioinformatics/coding resources
Bioconductor - Home. Resource with over 2000 working bioinformatics tools made to work in R .
Learn R, Python & Data Science Online | DataCamp. Full of programming and machine learning courses for a range of skill levels.
StatQuest with Josh Starmer - YouTube. YouTube channel for bioinformatic and statistical concepts.
Simon Cockell - YouTube. YouTube channel for longer bioinformatic tutorials.
Author curated playlist of several YouTube videos which could be very helpful to new bioinformaticians.
Kaggle: Your Machine Learning and Data Science Community. Resource to begin machine learning practice and datasets to use can be found here. Find Open Datasets and Machine Learning Projects | Kaggle. – Personal preference for free datasets.
10 interesting R project ideas and links to data sources by DataCamp.
42 interesting Python project ideas and links to data sources by UpGrad.
Towards Data Science. Machine learning blogs to gather theory and coding examples.
Bioinformatics Answers (biostars.org). Forum for bioinformatics queries.
Forums - SEQanswers. Forum for specifically sequencing related queries
Stack Overflow - Where Developers Learn, Share, & Build Careers. Forum for coding queries
Stanford CS229: Machine Learning Course, Lecture 1 - Andrew Ng (Autumn 2018) - YouTube. Lecture course on machine learning and practical advice.
Github is a free and useful method of keeping track of code and projects. Very attractive on a CV.

Krutik Patel recently completed his PhD in bioinformatics at Newcastle University and has since been employed as a research associate/bioinformatician at Newcastle University. His interests are in applying data science techniques for interesting questions in biology and developing software for researchers. Email: [email protected] .
Get Email Alerts
- Online ISSN 1740-1194
- Print ISSN 0954-982X
- Submit Your Work
- Language-editing services
- Recommend to Your Librarian
- Accessibility
- Sign up for alerts
- Sign up to our mailing list
- Biochemical Society Membership
- About Biochemical Society
- Publishing Life Cycle
- Biochemical Society Events
- Sponsored award winners
- About Portland Press
- Portland Press Tel
- +44 (0)20 3880 2795
- Portland Press Company no. 02453983
- Biochemical Society Tel
- +44 (0)20 3880 2793
- Email: [email protected]
- Biochemical Society Company no. 00892796
- Registered Charity no. 253894
- VAT no. GB 523 2392 69
- Privacy and cookies
- © Copyright 2024 Portland Press
This Feature Is Available To Subscribers Only
Sign In or Create an Account
Numbers, Facts and Trends Shaping Your World
Read our research on:
Full Topic List
Regions & Countries
Publications
- Our Methods
- Short Reads
- Tools & Resources
Read Our Research On:
Internet & Technology
6 facts about americans and tiktok.
62% of U.S. adults under 30 say they use TikTok, compared with 39% of those ages 30 to 49, 24% of those 50 to 64, and 10% of those 65 and older.
Many Americans think generative AI programs should credit the sources they rely on
Americans’ use of chatgpt is ticking up, but few trust its election information, whatsapp and facebook dominate the social media landscape in middle-income nations, sign up for our internet, science, and tech newsletter.
New findings, delivered monthly
Electric Vehicle Charging Infrastructure in the U.S.
64% of Americans live within 2 miles of a public electric vehicle charging station, and those who live closest to chargers view EVs more positively.
When Online Content Disappears
A quarter of all webpages that existed at one point between 2013 and 2023 are no longer accessible.
A quarter of U.S. teachers say AI tools do more harm than good in K-12 education
High school teachers are more likely than elementary and middle school teachers to hold negative views about AI tools in education.
Teens and Video Games Today
85% of U.S. teens say they play video games. They see both positive and negative sides, from making friends to harassment and sleep loss.
Americans’ Views of Technology Companies
Most Americans are wary of social media’s role in politics and its overall impact on the country, and these concerns are ticking up among Democrats. Still, Republicans stand out on several measures, with a majority believing major technology companies are biased toward liberals.
22% of Americans say they interact with artificial intelligence almost constantly or several times a day. 27% say they do this about once a day or several times a week.
About one-in-five U.S. adults have used ChatGPT to learn something new (17%) or for entertainment (17%).
Across eight countries surveyed in Latin America, Africa and South Asia, a median of 73% of adults say they use WhatsApp and 62% say they use Facebook.
5 facts about Americans and sports
About half of Americans (48%) say they took part in organized, competitive sports in high school or college.
REFINE YOUR SELECTION
Research teams, signature reports.
The State of Online Harassment
Roughly four-in-ten Americans have experienced online harassment, with half of this group citing politics as the reason they think they were targeted. Growing shares face more severe online abuse such as sexual harassment or stalking
Parenting Children in the Age of Screens
Two-thirds of parents in the U.S. say parenting is harder today than it was 20 years ago, with many citing technologies – like social media or smartphones – as a reason.
Dating and Relationships in the Digital Age
From distractions to jealousy, how Americans navigate cellphones and social media in their romantic relationships.
Americans and Privacy: Concerned, Confused and Feeling Lack of Control Over Their Personal Information
Majorities of U.S. adults believe their personal data is less secure now, that data collection poses more risks than benefits, and that it is not possible to go through daily life without being tracked.
Americans and ‘Cancel Culture’: Where Some See Calls for Accountability, Others See Censorship, Punishment
Social media fact sheet, digital knowledge quiz, video: how do americans define online harassment.
1615 L St. NW, Suite 800 Washington, DC 20036 USA (+1) 202-419-4300 | Main (+1) 202-857-8562 | Fax (+1) 202-419-4372 | Media Inquiries
Research Topics
- Email Newsletters
ABOUT PEW RESEARCH CENTER Pew Research Center is a nonpartisan fact tank that informs the public about the issues, attitudes and trends shaping the world. It conducts public opinion polling, demographic research, media content analysis and other empirical social science research. Pew Research Center does not take policy positions. It is a subsidiary of The Pew Charitable Trusts .
© 2024 Pew Research Center
Summer construction projects underway
Installations, repairs, renovations, and refreshes to go full speed ahead to welcome new and returning students in august.

Carlos Ortiz/RIT
Construction projects continue on the RIT campus this summer, including major work on residence halls. The summer weather and less populated campus mean more work to repair, replace, or refresh is done in a few short weeks to welcome new and returning students back to campus in August.
Summer construction projects are full speed ahead at RIT, including numerous improvements to greet students in August, such as new roofing, air conditioning, and refreshes in residence halls.

Construction projects continue on the RIT campus this summer, including major work on residence halls.
In its third, and most ambitious year of a five-year plan, this summer’s residence hall renovations include masonry repairs on Kate Gleason Hall and Residence Halls A , B, and C ; Eugene Colby Hall , Helen Fish Hall and Fredricka Douglass Sprague Perry Hall will be getting new roofs; refreshes and fire alarm work will be done in Colby Hall and Gleason Hall; and Sol Heumann Hall will be getting new air conditioning.
In addition, Frances Baker Hall and Residence Halls A, B and C will be getting new room doors and card access.
“A lot of work is going to happen in a very short amount of time,” said John Moore , associate vice president for Facilities Management Services at RIT. “We will try to be completed by Aug. 9 so the move-in is flawless. That’s the highest priority we have, so move-in can be awesome.”
Although most students won’t be on campus over the summer, the residence halls will be occupied periodically by visitors, including participants in the Genius Olympiad.
“We still have our annual cleaning of all the rooms and need to make way for these summer guests,” Moore said. “There’s so much activity over there that the timing of being ready is super critical. FMS has a lot of people working every day to make sure all of this can get done, and get done in time. We’ve partnered with Residence Life and RIT Housing to make sure we haven’t missed anything. We’ve been planning this for months.”

Construction projects continue on the RIT campus this summer, including new roofs on some apartments in University Commons.
Additional work is being done in other housing areas, including roof replacements this summer and over the next three summers in University Commons ; some units will receive kitchen and bathroom renovations; air conditioning is being installed over two years in Perkins Green apartments; and some foundation repairs, kitchen replacements and shower renovations are being done on a few Riverknoll apartments.
Other summer projects include:
Frank Ritter Ice Arena , which was used as a temporary storage and study area in recent years, will now be converted to its new use: a state-of-the-art indoor training facility available for all 25 intercollegiate athletic teams. Artificial turf will be installed inside the former ice arena this summer. The project is expected to be completed this fall.
George H. Clark Gymnasium : The original floor, which is 50 years old, is being replaced and is expected to be completed by the first week of August.
Gannett Hall , Gosnell Hall , and Booth Hall : Projects are underway to add air conditioning to the remaining areas not cooled. These projects will be completed over the next two summers. This work includes replacement of air handling equipment, replacement of duct work above ceilings, and other work that cannot be competed when classes are in session.
Student Innovation Hall : Renovations will be made to the first floor to create space for the RIT Service Center , Information and Technology Services , and Institutional Research to move there to make space for a home for the School of Performing Arts on the first floor of George Eastman Hall , possibly in 2026.
And work continues on three large projects on campus:
Music performance theater : The theater, which broke ground in September, will feature a 750-seat theater primarily to be used for musical theater productions. It will have two balconies and a historic pipe organ as its centerpiece. The estimated completion is December 2025, and like other venues on campus, it will be available for public use.

Major construction projects, such as the music performance theater, continue this summer on the RIT campus, as well as additional projects throughout campus.
Tiger Stadium : The official groundbreaking of the new Tiger Stadium was held in April. The $30 million, 38,828-square-foot facility will provide a state-of-the-art modern home for the men’s and women’s lacrosse and men’s and women’s soccer programs. The facility will seat 1,180, with additional capacity in the hospitality room, along with standing room. Amenities for the new stadium include team locker rooms; a training room with two large hot and cold tubs, taping tables, and exam tables; media suite; concession area; hospitality room with glass viewing wall; and an outdoor concourse.
It is expected to be completed in the fall of 2025.
Research building : Work continues on a new research building on the west side of campus. The building will be the host to several new research spaces for science, engineering, and technology. The building is expected to be open in late fall of 2024, with research beginning in the spring of 2025.
“These are big projects, and very important for the university,” Moore said. “The theater is a strategic project and will be the crown jewel for our Performing Arts program. The stadium will be a strategic initiative for recruiting athletes and certainly will add impressiveness to the campus. And we’re already recruiting high-level researchers to come to RIT to be part of our new research building.”
No major roadwork is expected on campus this summer, although some minor repairs will be made to parking lots and roadways, Moore said.
Recommended News
June 3, 2024

Engineering faculty and cardiologist collaborate to design heart pump assessment prototype
Researchers at RIT are developing technology that will be able to determine the lifespan of a heart valve with more precision.

Meet Mia White, the Littleton native about to make history with Deaf Women’s National Team
The Denver Post highlights the athletic career of Mia White '20 (management), defender on the U.S. Women's Deaf Soccer Team.

U.S. Deaf Women's National Team using Colorado showcase as platform to reach kids
KMGH-TV talks to Mia White '20 (management) and her teammates about how they intend to inspire a new generation of deaf children in athletics.

RIT to offer new pathway for earning MS in sustainable systems and MBA degrees
The Rochester Business Journal talks to Amit Batabyal, the Arthur J. Gosnell Professor of Economics and interim head of the Department of Sustainability, and Bill Dresnack, associate professor in the Department of Finance and Accounting, about the collaborative degree.
Pregnancy and Early Childhood

- Drug use during pregnancy can affect the health of a pregnant person and their child. For example, a pregnant person’s use or misuse of opioids can cause a newborn infant to experience withdrawal symptoms, a condition known as neonatal opioid withdrawal syndrome. Overdose deaths are also rising among women during and after pregnancy.
- Treatment for a substance use disorder during pregnancy such as behavioral interventions and medication for opioid use disorder reduces health risks, including preterm delivery and low birth weight. Treatment also helps people with substance use disorders stay employed, take care of their children, and engage with their families and communities. However, pregnant people with substance use disorders often face challenges when seeking treatment, including fear, stigma and access to care.
- NIDA plays a leading role in the HEALthy Brain and Child Development (HBCD) Study , which seeks to better understand how drug use during pregnancy interacts with genetics and other biological influences to affect a child’s mental and physical health over time.
The HEALthy Brain and Child Development Study
The study explores how parental use of opioids and other environmental factors affect a child’s brain and development
Latest from NIDA

More than 321,000 U.S. children lost a parent to drug overdose from 2011 to 2021

Overdose deaths increased in pregnant and postpartum women from early 2018 to late 2021

Innovative projects answer NIDA’s challenge to implement substance use prevention in primary care
Find more information about pregnancy, early childhood and substance use.
- Learn more about medications during pregnancy at the Centers for Disease Control and Prevention website.
- Read about alcohol use during pregnancy from the National Institute on Alcohol Abuse and Alcoholism.
- Read about the NIH’s Helping to End Addiction Long-term (HEAL) Initiative .
- For information on exposure to drugs and chemicals while breastfeeding, see the National Library of Medicine’s Drugs and Lactation Database (LactMed) .

IMAGES
VIDEO
COMMENTS
A recent study has found that the interest of researchers in these topics plateaued over after the early 2000s [1]. Besides the above mentioned hot topics, the following topics are considered demanding in bioinformatics. Cloud computing, big data, Hadoop. Machine learning. Artificial intelligence.
Best Project Ideas for Bioinformatics ... Analyzing genetic data to identify links between specific genes and diseases is a significant area of research. Projects in this category can have a real ...
Computational Methods for Analysis of DNA Methylation Data, Volume II. Pietro Di Lena. Christine Nardini. Matteo Pellegrini. 3,688 views. 6 articles. An innovative journal that provides a forum for new discoveries in bioinformatics. It focuses on how new tools and applications can bring insights to specific biological problems.
Bioinformatics is a field of study that uses computation to extract knowledge from biological data. It includes the collection, storage, retrieval, manipulation and modelling of data for analysis ...
Bioinformatics researchers integrate and manage the vast amounts of biological data now being generated, including genomic data. ... project is a collaborative effort to address the need for consistent descriptions of gene products across databases. ... Bioinformatics; Research Highlight June 12, 2020
In this video, we go over some of the main topics that will help you develop a research project using bioinformatics. For a detailed overview, please visit: ...
This is an editorial report of the supplements to BMC Bioinformatics that includes 6 papers selected from the BIOCOMP'19—The 2019 International Conference on Bioinformatics and Computational Biology. These articles reflect current trend and development in bioinformatics research.
Atom. RSS Feed. Computational biology and bioinformatics is an interdisciplinary field that develops and applies computational methods to analyse large collections of biological data, such as ...
Bioinformatics. Bioinformatics is an interdisciplinary field that intersects with biology, computer science, mathematics and statistics. It concerns itself with the development and use of methods and software tools for collecting and analyzing biological data.
Undergraduate and Masters Research. General Information. There are plenty of opportunities for Bioinformatics research projects at UCLA. This program is designed to help interested students find research projects related to Bioinformatics across campus. Typically, these projects are for credit; in exceptional circumstances they may offer funding.
Sample Grouping or individual as per experimental design, Group-wise OTU Clustering and abundance Report, OTU identification and taxonomic annotation Report (Sample Wise - Genius Level) and OTU Fasta file will be provided, Pie chart representation TOP 10 taxonomic classification; phylum to species-level. 5. SmallRNA Sequencing.
Here are five exciting machine learning projects for bioinformatics to help you understand the application of machine learning in healthcare, mainly bioinformatics. 1. Anti-Cancer Drug Efficacy Prediction. Predicting which patients are likely to benefit or not from a specific therapy is a significant concern in cancer treatment because ...
BMI/CS 776: Advanced Bioinformatics Project ideas Prof. Daifeng Wang 1/2 Overview In contrast to the homework, you are encouraged to use existing software and packages for the ... The goals of the class project are to gain experience working on a (small) bioinformatics research problem using real biological data. In order to encourage ...
Introduction. Because of the technological boom, life scientists are increasingly turning to high-throughput sequencing in their research programs and generating enormous volumes of data [].These projects are characterized by the use of specialized computational and tools to analyze the generated data, highlighting the need for interdisciplinary services and/or deep collaborations between ...
The interdisciplinary nature of bioinformatics makes it an ideal framework to develop activities enabling enquiry-based learning. We describe here the development and implementation of a pilot project to use bioinformatics-based research activities in high schools, called "Bioinformatics@school." It includes web-based research projects that students can pursue alone or under teacher ...
List of Bioinformatics Research Topics Ideas for. 1. Data access control in the cloud computing environment for bioinformatics 2. ... Cataloguing experimentally confirmed 80.7 kb-long ACKR1 haplotypes from the 1000 Genomes Project database 291. BoardION: real-time monitoring of Oxford Nanopore sequencing instruments 292. Augur: a bioinformatics ...
Independent Study in Biomedical Informatics (ISBDS) This document provides ideas for research projects, and links to research plan templates, which are partially completed plans. Template files are available via the ISBDS course GitHub repository . For ISBDS, a research plan template can vary within biomedical science topics, but definitely ...
It's one of your best bets if you have actual data to analyze (or download available datasets off the internet, e.g. NCBI). It can be difficult to pick up on at first; you'll have to do your own investigating, troubleshooting, etc., but it's extremely rewarding. It requires no prior knowledge in programming/coding. 5.
It involves a lot of data curation which is very important in microbial bioinformatics. Pick a bacterial species (maybe one that is present across multiple species) and collect as many assembled genomes from NCBI. Also look at SRA database for the un assembled genomes from published studies. Curate the data (genome completeness, quality, check ...
A beginner's guide to bioinformatics. Biochem (Lond) (2023) 45 (2): 11-15. Bioinformatics has revolutionized the modern life sciences and has become a component of many undergraduate training courses and post-graduate research projects. As such, we are seeing more bioinformatics and programming aspects within undergraduate training and so ...
In this video, we go over some of the main topics that will help you develop a research project using bioinformatics. For a detailed overview, please visit: ...
Hey looking for bioinformatics mini project ideas technical question I'm a newbie to bioinfo from biotechnology bg with zero knowledge of coding, and this is my final year project and I'm not at sure what to dive into, all I've learnt till in the past week are pymol, Pyrx, emboss and autodock. ... VFX & Research papers/books
Collins, in his work as head of the Human Genome Project, was one of the scientists who discovered the gene for cystic fibrosis. That discovery led to a breakthrough treatment for a disease that ...
Americans' Views of Technology Companies. Most Americans are wary of social media's role in politics and its overall impact on the country, and these concerns are ticking up among Democrats. Still, Republicans stand out on several measures, with a majority believing major technology companies are biased toward liberals. short readsApr 3, 2024.
The School of Communications has published the spring 2024 issue of the Elon Journal of Undergraduate Research in Communications, featuring student research on far-ranging topics such as the use of social media propaganda distributed during the Russo-Ukrainian War to a content analysis of Duolingo's brand communications success on TikTok.. This is the cover of the spring 2024 issue of the ...
Try flexing your python muscles by rewriting a part of biopython. You could pick a simple alignment algorithm or a simple toolkit for manipulating DNA/RNA sequences. You could also try and write a variant filtering tool, parsing and selecting variants from vcf/bcf files. A nice project idea that you can scale down might be a writing a pipeline ...
Major construction projects, such as the music performance theater, continue this summer on the RIT campus, as well as additional projects throughout campus. Tiger Stadium: The official groundbreaking of the new Tiger Stadium was held in April. The $30 million, 38,828-square-foot facility will provide a state-of-the-art modern home for the men ...
Highlights. Drug use during pregnancy can affect the health of a pregnant person and their child. For example, a pregnant person's use or misuse of opioids can cause a newborn infant to experience withdrawal symptoms, a condition known as neonatal opioid withdrawal syndrome. Overdose deaths are also rising among women during and after pregnancy.