Advertisement

A review on deep learning in medical image analysis
- Regular Paper
- Published: 04 September 2021
- Volume 11 , pages 19–38, ( 2022 )
Cite this article

- S. Suganyadevi ORCID: orcid.org/0000-0001-6096-7510 1 ,
- V. Seethalakshmi ORCID: orcid.org/0000-0001-6518-6350 1 &
- K. Balasamy ORCID: orcid.org/0000-0003-0973-5698 2
34k Accesses
170 Citations
Explore all metrics
Ongoing improvements in AI, particularly concerning deep learning techniques, are assisting to identify, classify, and quantify patterns in clinical images. Deep learning is the quickest developing field in artificial intelligence and is effectively utilized lately in numerous areas, including medication. A brief outline is given on studies carried out on the region of application: neuro, brain, retinal, pneumonic, computerized pathology, bosom, heart, breast, bone, stomach, and musculoskeletal. For information exploration, knowledge deployment, and knowledge-based prediction, deep learning networks can be successfully applied to big data. In the field of medical image processing methods and analysis, fundamental information and state-of-the-art approaches with deep learning are presented in this paper. The primary goals of this paper are to present research on medical image processing as well as to define and implement the key guidelines that are identified and addressed.
Similar content being viewed by others
A survey on Image Data Augmentation for Deep Learning

Artificial intelligence in disease diagnosis: a systematic literature review, synthesizing framework and future research agenda

Brain tumor detection and classification using machine learning: a comprehensive survey
Avoid common mistakes on your manuscript.
1 Introduction
All deep learning applications and related artificial intelligence (AI) models, clinical information, and picture investigation may have the most potential element for making a positive, enduring effect on human lives in a moderately short measure of time [ 1 ]. The computer processing and analysis of medical images involve image retrieval, image creation, image analysis, and image-based visualization [ 2 ]. Medical image processing had grown to include computer vision, pattern recognition, image mining, and also machine learning in several directions [ 3 ]. Deep learning is one methodology that is commonly used to provide the accuracy of the aft state. This opened new doors for medical image analysis [ 4 ]. Deep learning applications in healthcare addresses wide variety of issues, including cancer screening to infection monitoring to personalized advice for treatment [ 5 ].Today, a massive quantity of data is placed on different data sources such as radiological imaging, genomic sequences, as well as pathology imaging, at one's disposal of physicians [ 6 ]. We are all in a state of flux methods, however, to turn all this information into usable information, PET (positron emission tomography), X-ray, CT (computed tomography), fMRI (functional MRI), DTI (diffusion tensor imaging), and MRI (magnetic resonance imaging are the typical modalities used for medical imagery [ 7 , 8 ].
Deep learning involves learning patterns in data structures using neural networks of many convolutional nodes of artificial neurons. [ 9 , 10 ]. An artificial neuron is a type of cell that takes several inputs, basically works with a calculation, and returns the result similar to a biological neuron [ 11 , 12 , 13 , 14 , 15 ]. This simple calculation takes a linear source regular expression shape preceded by an activation function which is nonlinear [ 16 ]. The sigmoid conversion, ReLU (rectified linear unit) and their variants, and tanh (hyperbolic tangent) are examples of several commonly used nonlinear activation functions of a network [ 17 , 18 , 19 , 20 , 21 ]. The growth origins of deep learning can be traced back to the return to Warren McCulloch and Walter Pitts (1943). Besides the advancement of the ImageNet (2008), backpropagation model (1961), AlexNet (2010), (CNN) convolutional neural network model (1978), and (LSTM) long short-term memory (1996) [ 22 ]. GoogleNet is a search engine (winner of the ILSVRC 2013 issue) commenced by Google [ 23 ] in 2014, which introduced the notion of start-up modules that dramatically reduced the computing of CNN. In 2014, Google launched GoogleNet (winner of the challenge for ILSVRC 2014) [ 23 ] that incorporated the idea of start-up modules which significantly lowered CNN’s computational complexity. CNN architecture is composed of multiple layers that use a differentiable function to transform the input volume into an output volume (e.g., holding the class scores). Essentially, deep learning is a reincarnation of the artificial neural network, in which artificial neurons are stacked. Features of the network in CNN [ 22 , 23 ] are created by converting kernels into layers with outputs from previous layers. On the input images, the kernels in the first invisible layer carryout convolutions [ 24 , 25 ]. Although early hidden layers capture shapes, curves, and edges, as well as more abstract and complex features are captured by deeper hidden layers. Figure 1 shows the different types of learning processes available for the CNN networks [ 26 ].
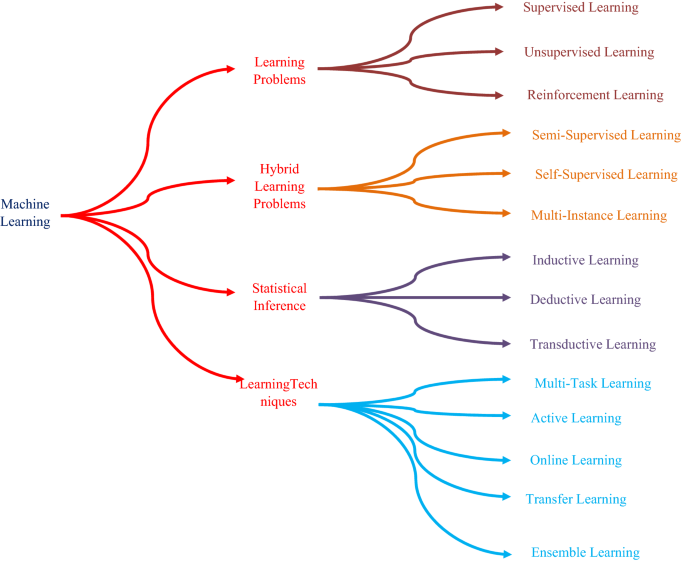
Types of learning
1.1 Types of learning
The followings are the 14 sorts of learning that we should be acquainted with as an AI specialist.
Learning problems
1. Supervised learning
2. Unsupervised learning
3. Reinforcement learning
Hybrid learning problems
4. Semi-supervised learning
5. Self-supervised learning
6. Multi-instance learning
Statistical inference
7. Inductive learning
8. Deductive inference
9. Transductive learning
Learning techniques
10. Multi-task learning
11. Active learning
12. Online learning
13. Transfer learning
14. Ensemble learning
An investigation has been done one by one in the accompanying segments.
1.1.1 Learning problems
1.1.1.1 supervised learning.
A problem in which a model is used to learn a representation between input examples and a target variable is represented by supervised learning [ 26 ]. Supervised learning problems are known as systems where the training data contains examples of input vectors and the target vectors that correspond to them. There are two major types of problems with supervised learning: classification involving detection of regression and a class mark involving detection of a significant value [ 27 ]. Classification is represented as a supervised problem of learning which requires the prediction of a class label. Regression is a problem of supervised learning involving predicting a numerical label [ 28 ].
There can be one or more input variables in classification and regression problems, and input process parameters can be in any data format, such as numerical and categorical data [ 28 ]. The MNIST is a handwriting digit dataset with handwritten digit images as inputs (pixel data) that will be an example of a classification problem [ 29 ]. Some machine learning algorithms are known as supervised machine learning algorithms because they are designed for supervised machine learning problems. Decision trees and support vector machines are two examples of it [ 29 , 30 ].
Algorithms are related to as supervised because, when an input data is given, they learn by making predictions, and those models are controlled and improved by an approach that can help determine the outcome [ 31 ]. Some methods can be perfectly suited for classification (e.g., logistic regression) or regression (e.g., linear regression), while some are employed for both types of problems with minor modifications (such as artificial neural networks) [ 32 , 34 ].
1.1.1.2 Unsupervised learning
Unsupervised learning identifies some difficulties involving in the use of data relationship model that describes or removes data relationships. Unsupervised learning works in comparison with supervised learning, only the input data is used, with no outputs or target variables [ 33 ]. As such, unsupervised learning close to supervised learning doesn’t have an instructor to correct the model. There are several ways of unsupervised learning, but they have two key issues which a practitioner frequently encounters: clustering involves grouping the data and estimating range, which entails a summary of data distribution. Clustering is represented as an unsupervised problem of learning which requires finding data for classes [ 33 , 34 , 35 , 36 , 37 , 38 , 39 ].
Estimation of density is termed as an unsupervised problem of learning that requires summarizing the data distribution. K-Means is a clustering technique in operation where k corresponds to the cluster centres to be found in the data [ 40 ]. A density neural network is Kernel Density Estimation, which uses small groups of closely related data samples to estimate the distribution of new points in the problem space [ 34 , 35 , 36 , 37 , 38 ]. To learn about the trends even in information, clustering as well as density estimation can be performed. Additional unsupervised approaches can also be used, such as visualization involving various forms of methods for graphing or plotting results, as well as projection methods involving decreasing those data’s dimensionality [ 41 ].
Visualization is an unsupervised problem of learning involving the development of data plots. Data visualization is a methodology for helping people recognizes vast quantities of data by using a variety of standardized and interactive visuals within a particular context [ 42 ]. The information is often viewed in a narrative style, which highlights patterns, trends, and associations that would otherwise go overlooked [ 43 ].
Projection is an unsupervised problem of learning that requires the development of lower-dimensional data representations [ 44 ]. Random projection is a more computationally effective dimensionality reduction approach than principal component analysis [ 45 ]. It is often used in datasets of too many dimensions for principal component analysis to be computed directly.
1.1.1.3 Reinforcement learning
Reinforcement learning is a set of challenges in which an individual must learn to use feedback to work in a given context [ 46 ]. It is identical to supervised learning, even though feedback may be delayed, and since the model is systematically noisy, it has some responses from which to learn, which finds it challenging for the entity and model to link causality [ 42 , 47 ]. Deep reinforcement learning, Q-learning, and temporal-difference learning are some common examples of reinforcement learning algorithms [ 48 ].
1.1.2 Hybrid learning problems
1.1.2.1 semi-supervised learning.
It is supervised learning where there are a few classified instances and a lot of unlabelled instances in that training data [ 48 ]. The purpose of a semi-supervised learning model is to use all the available data efficiently, not all labelled information like in the supervised learning technique [ 49 ]. It could include the use of unsupervised methods like clustering and density estimation, or it could be inspired by them to make effective use of unlabelled data [ 49 , 50 ]. Upon discovery of groups or patterns, supervised strategies or supervised learning ideas will be used for marking the unlabelled instances or add labels with unlabelled later on, these descriptions were used to make accurate predictions [ 51 ].
This category covers many concerns like audio data (automated speech recognition), text data (natural language processing), and image data, and these concerns will not be smoothly solved with traditional supervised learning techniques [ 34 , 51 ].
1.1.2.2 Self-supervised learning
The self-supervised learning system needs only unlabelled data to formulate a pretext learning assignment, such as predicting context or image rotation, for which a target objective can be calculated without supervision [ 52 ]. Self-supervised learning algorithms such as autoencoders are a good example. This is a type of neural network that is used to create a compact or compressed input sample representation [ 52 , 53 ]. They do this using a model that includes an encoder and a decoder component separated by a bottleneck that represents the input's internal compact representation [ 54 ]. These autoencoder models are educated by providing the input as both input and target output, forcing the model to reproduce the input by first encoding it to a compressed representation and then decoding it back to the original [ 53 ]. The decoder is removed after training, and the encoder is used to generate compact input representations as desired. Autoencoders have historically been used for the reduction of dimensionality or learning of features [ 54 ].
Self-supervised learning is often exemplified by generative adversarial networks, or GANs [ 54 , 55 ]. These are generative models that are most frequently used to generate synthetic photographs using only a collection of unlabelled examples from the target domain [ 55 ].
1.1.2.3 Multi-instance learning
In multi-instance learning, an entire set of examples is labelled as containing or not containing an example of a class, but individual members of the collection are not marked [ 56 ].
1.1.3 Statistical inference
The term inference refers to the process of arriving at a conclusion or making a decision. In machine learning, designing a model and making a prediction are both examples of inference [ 56 ]. There are several inference paradigms which can be used to explain how certain machine learning algorithms work or how to solve such learning problems. Approaches to learning include inductive, deductive, and transductive learning, as well as inference. Induction is the analysis of a general model based on specific instances [ 57 , 58 ]. The deduction method uses formula to make predictions. Making assumptions based on specific examples is known as transduction [ 58 ].
1.1.3.1 Inductive learning
To assess the result, inductive learning involves using proof. Inductive learning refers to using particular situations, e.g. specific to general, to decide general outcomes [ 59 ]. Many algorithms learn from particular past precedents through a process called inductive reasoning, in which general rules (the model) are taught (the data) [ 59 , 60 ]. It is an induction approach adapted to a machine learning model. The model is a generalization of the concrete examples in the training dataset. The training data is used to create a model or hypothesis about the problem, and the model is presumed to carry over fresh unknown data later [ 60 ].
1.1.3.2 Deductive inference
To evaluate concrete results, the use of general concepts is referred to as deduction or deductive inference. We can better understand induction by contrasting it to inference. A deduction is the polar opposite of induction [ 61 ]. In the same way that induction progresses from the person to the general, deduction progresses from the general to the specific [ 62 ]. Induction is a bottom-up form of reasoning that uses the evidence available as proof for an outcome, while deduction is a top-down method of reasoning that seeks to fulfil all premises before determining the result [ 63 ]. The algorithm can be used to make predictions before we use induction to suit a model on a training dataset, in the sense of machine learning [ 64 , 65 , 66 , 67 , 68 ]. The model is employed as a deductive method.
1.1.3.3 Transductive learning
Transduction or transductive learning is a term used in statistical learning theory to describe the process of predicting specific examples from domain [ 69 ]. It differs from induction, which involves learning universal rules, which is based on concrete examples [ 70 ]. In the model of estimating the value of a function at a given point of interest, a new definition of inference is defined [ 71 ]. Notice that when one would like to get the best outcome from a limited amount of knowledge, this principle of inference arises [ 72 ]. The k-nearest neighbours algorithm is a classic example, where the transductive algorithm uses it directly each time a prediction is required, instead of modelling the training data [ 3 , 47 , 72 ].
1.1.4 Learning techniques
1.1.4.1 multi-task learning.
Multi-task learning is a technique for improving generalization by combining details from a variety of tasks (which can be seen as soft constraints placed on the parameters) [ 73 ]. When there is an abundance of labelled input data for one task that can be shared with another task with much less labelled data, multi-task learning can be a useful approach to problem-solving [ 74 , 75 ].
A multi-task learning problem, for example, can include the same input patterns that can be used for many different outputs or supervised learning issues [ 76 ]. In this configuration, each output can be predicted by a different part of the model, allowing the core of a model to generalize the same inputs for each task [ 75 ].
1.1.4.2 Active learning
Active learning is often a methodology in which a model will ask a human user operator questions during the learning process to solve uncertainty [ 77 ]. Active learning is a form of supervised learning that aims to produce the same or better results than so-called passive supervised learning, even if the model’s data is more efficient [ 78 ]. The central principle behind active learning is that allowing a machine learning algorithm to choose the data from which it learns allows it to achieve greater precision with fewer training labels [ 79 , 80 ]. An active learner will raise questions, which will typically take the form of unlabelled instances of information that will be labelled by an oracle [e.g., annotator (human)] [ 81 ]. Active learning is a valuable tool when there is little data available and collecting or labelling new data is expensive [ 82 , 83 ]. The active learning process allows for domain sampling to be oriented in a way that decreases the number of samples while increasing the model’s effectiveness [ 84 ].
1.1.4.3 Online learning
Machine learning is typically carried out offline, meaning we have a batch of data and we refine an equation [ 85 ]. However, we need to conduct online learning if we have streaming data, so we can update our estimates when each new data point arrives rather than waiting until the end (which may never occur) [ 86 ]. Online learning is useful because, over time, the data can alter rapidly [ 87 , 88 , 89 ]. It is also useful for applications, involving a broad set of knowledge that, even if changes are incremental, is continuously increasing [ 90 ].In general, online learning aims to eliminate the inconsistency, which is how well the model performed relative to how well if all the knowledge available was available as a batch, it should have performed [ 91 ]. The so-called stochastic or online gradient descent used to suit an artificial neural network is one instance of online learning [ 92 ].
The fact that stochastic gradient descent minimizes generalization error is easiest to see in the online training situation, where examples or mini lots are taken from a data stream [ 93 ].
1.1.4.4 Transfer learning
It is a form of learning in which a model learns on one problem and then is used as the reference point for another activity [ 94 ]. This is a good solution for challenges where there was a process that is close to the main problem and the associated task necessitates a large amount of data [ 95 ]. Transfer learning varies from multi-task training, in that the tasks are learned sequentially, while multi-task training seeks desirable performance from a single model on all tasks at the same time in comparison. For example, the image classification process, in which a prediction model will be learned with a broad set of images, such as an artificial neural network, and while training on even a simpler, more specific dataset, like cats and dogs, model weights may be used as a preliminary step [ 94 ]. The characteristics that the model has already learned about the larger mission, like retrieving lines and also patterns, would help for another task.
1.1.4.5 Ensemble learning
Ensemble learning is a method in which at least two modes fit into similar information and coordinate the forecasts from each one [ 96 ]. In contrast to any individual model, the goal of ensemble learning is to accomplish better execution with the group of models [ 97 ]. This incorporates the view of how to build models utilized in the group and how likely fuse the individuals from the outfit's forecasts [ 98 , 99 , 100 , 101 ].
Ensemble learning is an important method for creating prescient abilities in a pain point and lessening the vulnerability of stochastic learning calculations, like artificial neural organizations. Bootstrap, weighted normal, and stacking (stacked speculation) are a few instances of regular group learning calculations (Bagging) [ 103 ].
2 Deep learning architectures
During the recent twenty years, we have been furnished with deep learning models that have dramatically increased the type and number of problems that could be solved by neural networks [ 101 ]. Deep learning is not a solitary method, yet rather a class of calculations and geographies that can be applied to a wide assortment of issues [ 102 , 103 ]. Connectionist structures have existed for over 70 years, but they have been brought to the frontline of artificial intelligence by modern architectures and GPUs (graphical processing units). Figure 2 shows the general architecture of neural networks [ 102 ].
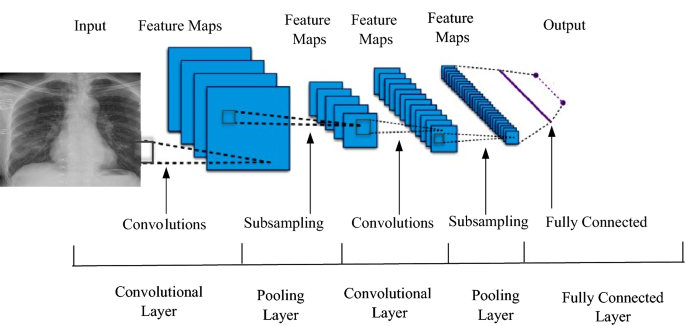
General architecture of neural network and deep learning
Although deep learning techniques are not new, because of the intersection of deeply layered neural networks and the use of GPUs to accelerate their execution, it is experiencing exponential development. In this article, comparison is made among different architectures of deep learning models [ 103 , 104 ]. General deep learning architecture contains the following layers: input layers, convolution and fully connected layers, sequence layers, activation layers, normalization, dropout, and cropping layers, pooling, and un pooling layers, combination layers, object detection layers, generative adversarial network layers, output layers [ 101 , 102 , 103 , 104 , 105 , 106 , 107 , 108 ].
Network’s secret sauce is the hidden layer(s). Because of their nodes/neurons, they may model complex data. Since the true values of their nodes are unknown in the training dataset, they are hidden. In fact, we only have access to the input and output [ 100 , 101 , 102 , 103 , 104 , 105 , 106 , 107 , 108 , 109 , 110 ]. At least one hidden layer exists in any neural network. No law says you must multiply the number of inputs by N. The ideal number of hidden units could easily be less than the number of inputs [ 111 ]. We can use several hidden units if you have a lot of training examples, but with little data, often only two hidden units will suffice.
2.1 Deep neural network (DNN)
In this architecture, at least two layers are there that allow nonlinear complexities. Classification and regression can be carried out here. The advantage of this model is generally used because of its great accuracy [ 104 ]. The drawback is that the method of training will not be easy since the error is transmitted back to the past layer and also becomes low. Also, the model's learning behaviour is too late [ 105 ].
2.2 Convolutional neural network (CNN)
This model could be best suited for 2D data. This network consists of a convolutional filter for transforming 2D to 3D which is quite strong in performance and is a rapid learning model. For classification process, it needs a lot of labelled data [ 54 , 69 , 106 ]. However, CNN faces issues, such as local minima, slow rate of convergence, and intense interference by humans. After AlexNet's great success in 2012, CNNs have been increasingly used to enhance the efficacy of human clinicians in medical image processing [ 107 ].
2.3 Recurrent neural network (RNN)
RNNs have the ability for recognizing the sequences. The weights of the neurons are spread through all measures. There are many variants such as LSTM, BLSTM, MDLSTM, and HLSTM [ 110 , 111 , 112 , 113 , 114 , 115 ]. This includes state-of-the-art accuracies in character recognition, speech recognition, and some other natural language processing-related problems. Learning sequential events can model time conditions [ 116 ]. The disadvantage is that this method has more issues because of gradient vanishing and this architecture is in need of big datasets [ 117 ].
2.4 Deep conventional-extreme learning machine (DC-ELM)
It blends CNN's strength and ELM's rapid preparation. To viably digest significant level attributes from input pictures, it utilizes various substitute convolution layers and pooling layers [ 118 ]. The preoccupied highlights are then taken care of to an ELM classifier, which prompts improved after effects of speculation with quicker learning speed [ 3 ]. In the last hidden layer, deep conventional-extreme learning machine was used to implement stochastic pooling to significantly reduce the dimensionality of functions, saving a lot of training time and computational resources [ 117 ].
2.5 Deep Boltzmann machine (DBM)
A DBM (deep Boltzmann machine) is a three-layer generative model. It is similar to a deep belief network but instead allows bidirectional connections in the bottom layers. Its energy function is as an extension of the energy function of the RBM is shown in Eq. ( 1 ).
DBM with N hidden layers; Unidirectional connections are made among all hidden layers. Top-down feedback for more accurate inference integrates ambiguous results [ 119 , 120 ]. Optimization of a parameter is difficult for a big dataset.
2.6 Deep belief network (DBN)
Deep Belief Networks are a graphical portrayal that is fundamentally generative; all the potential qualities that can be produced for the current situation are created. It is a combination of likelihood and measurements with neural organizations and AI [ 110 ]. Deep belief networks comprise a few layers with values, where the layers have a relationship, however not the qualities. The essential target is to assist the machine with characterizing the information into different classifications. The drawback of this architecture is, initialization process makes the training expensive [ 110 , 112 ].
2.7 Deep autoencoder (DAN)
Applicable in the unsupervised learning process, this could be helpful for dimensionality reduction and feature extraction. Here the number of inputs is equal to the number of outputs [ 2 ]. The advantage of the model is it does not need labelled data. Various kinds of autoencoders such as denoising autoencoder, sparse autoencoder, Conventional autoencoder is needed for robustness. Here it needs to give pre-training step, but training could be vanished [ 3 , 4 , 5 ].
Generally, autoencoder [ 6 ] consists of both encoder and decoder, which can be defined as \(\Phi \) and \(\Psi \) shown in Eq. ( 2 ).

2.8 Deep stacking networks (DSN)
A deep stacking network, also termed as deep convex network, is the final architecture [ 7 ]. A deep stacking network is separate from conventional deep learning systems in which it is essentially a deep collection of individual networks, each with its hidden layers, even though it consists of a deep network. This architecture model is a response to one of the deep learning issues: the trouble of preparing [ 8 ]. The intricacy of preparing is expanded dramatically by each layer in a deep learning design, so the DSN sees preparing not as a solitary issue but rather as a progression of individual preparing issues [ 9 ].
2.9 Long short-term memory/gated recurrent unit networks (LSTM/GRU)
The gated recurrent unit network was invented in 1997 by Hoch Reiter and Schimdhuber; however, it was filled in ubiquity lately as an RNN engineering for different applications [ 4 , 5 ]. The LSTM pulled out from ordinary neuron-based neural association models and rather introduced the possibility of a memory cell [ 5 ]. The memory cell can hold its motivation for a short or long time as a segment of its data sources, which allows the phone to review what is huge and not just its last enlisted worth [ 7 ]. In 2014, an improvement of the LSTM was introduced with the gated recurrent unit. This model has two entryways, discarding the yield entrance present in the LSTM model [ 8 ]. For certain applications, the GRU has execution like the LSTM, yet being simpler techniques fewer loads and speedier execution [ 9 ].
The GRU joins two entryways: an update doorway and a reset entryway. The updated entryway exhibits the measure of the past cell substance to keep up. The reset entryway describes how to meld the new commitment with the past cell substance [ 10 ]. A GRU can show a standard RNN just by setting the reset entryway to 1 and the update doorway to 0. Different kinds of architectures are used for wide variety of applications that is mentioned in Table 1 [ 11 ].
3 Development frameworks
It is possible to implement these deep learning architectures, but it can take time to start from scratch, and they will require time to refine and mature [ 12 ]. Fortunately, many open-source platforms can be used to more quickly implement deep learning algorithms. These frameworks support Java, R programming language, Python and, C/C++ [ 13 ].
Tensor flow It began as an internal Google project called Google Brain in 2011. As an open-source deep learning neural network that can run over several CPUs and GPUs, it was publicly released in 2017 [ 2 , 3 , 4 ]. It is used for training neural networks, similar to human learning and reasoning, to identify and decode patterns and connections. It gives C++ interface and Python [ 2 , 3 , 4 , 5 ].
Caffe In 2014, Berkeley Artificial Intelligence Research (BAIR) was developed. This provides Python and C++ interface, and academic research became popular. Using convolution nets, it is a deep learning system [ 4 ].
Caffe 2 As a commercial successor to Caffe, Facebook launched it in 2017. It was designed to resolve the scalability of Caffe's problems and to make it lighter [ 4 , 5 ]. It enables distributed computing, implementation, and quantized computations to be carried out. It provides Python and C++ interface [ 6 ].
ONNX Facebook and Microsoft recently announced the Open Neural Network exchange in September 2017 [ 2 , 3 , 4 ]. ONNX is an interchange format intended to make it easy to pass deep learning models between the frameworks used to construct them [ 5 ]. This initiative could make different frameworks easier for developers to use [ 4 , 5 ].
Torch Torch is an open-source machine learning library, a scientific computing platform, and a scripting language based on the Lua programming language [ 6 ]. Torch is used by IBM, Yandex, Idiap Research Institute and Facebook AI Research Group. As open-source software, Facebook has published a collection of extension modules (PyTorch) [ 4 , 5 ].
Keras It is high-level programming that can run on top of Theano and Tensor flow [ 4 , 5 ], and it seems as an interface. While not as weak as other structures, Keras is especially famous for its rapid growth. Using popular networks and evaluating networks algorithms and layers, it has been described as an entry point for new users’ deep learning.
MatConvNet It is commonly used Mat lab’s Deep Learning Library [ 6 ].
Theano It is a Python library for deep implementation [ 5 ]. Developing and promoting learning strategies by MILA, Montreal University.
Deeplearning4j Deeplearning4j is a common deep learning platform that focuses on Java technology but also includes application programming interfaces for other languages including Clojure, Scala, and Python [ 6 ]. Delivered under the Apache permit, the stage offers help for RNN, RBMs, CNN, and DBNs. Deeplearning4j additionally gives dispersed equal variants (enormous information preparing structures) that work with Apache Hadoop and Spark [ 15 , 16 , 17 , 18 , 19 , 20 , 21 , 22 , 23 , 24 , 25 , 26 , 27 , 28 , 29 , 30 , 31 , 32 , 33 , 34 , 35 , 36 , 37 , 38 , 39 , 40 , 41 , 42 , 43 ]. It has been applied to several issues, including financial sector fraud detection, recommendation systems, image recognition, and cyber security (detection of network intrusion). For GPU optimization, the system integrates with CUDA and could be distributed by using OpenMP and Hadoop [ 20 ].
Distributed deep learning IBM distributed deep learning (DDL) is a library that interfaces with driving structures like Tensor Flow and Caffe, nicknamed the jet engine of deep learning DDL can be utilized over bunches of workers and many GPUs to accelerate deep learning calculations [ 35 ]. By indicating ideal ways that the subsequent information should take between GPUs, DDL streamlines the correspondence of neuron computations [ 37 ]. Deep learning libraries provided by Microsoft including MXnet, Microsoft Cognitive Toolkit, Paddle Paddle, SciKit-Learn, Matlab, Pandas, Numpy, cuDNN, NVIDIA TensorRT, NVIDIA DIGITS, Jupyter Notebook, etc., are the other popular libraries, frameworks, and tools that are popular among developers [ 38 ].
4 Process involved in medical image analysis
Medical image computation is highly associated with the field of medical imaging, but it depends on the computational analysis of images, not their acquisition [ 35 , 36 , 37 , 38 , 39 , 40 , 41 , 42 , 43 , 44 , 45 , 46 , 47 , 48 , 49 , 50 , 51 , 52 , 53 , 54 , 55 , 56 , 57 , 58 , 59 , 60 , 61 , 62 , 63 , 64 , 65 , 66 , 67 , 68 , 69 , 70 , 71 , 72 , 73 , 74 , 75 , 76 ]. The techniques can be grouped into many broad categories: segmentation of images, image registration, physiological modelling based on images, and others. These techniques can be discussed in the following section. Figure 3 shows the processes that are involved in medical image analysis [ 96 ].
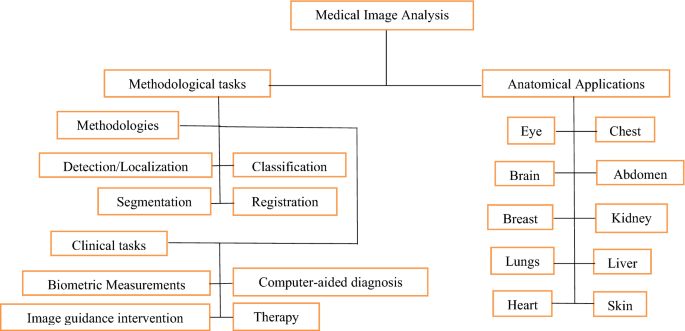
- Medical image analysis
4.1 Deep learning networks based on goals and architecture type
Both processing steps that are used for quantitative measurements, as well as abstract representations of medical images, are used in image analysis [ 96 , 97 ]. These measures necessitate prior knowledge of the images' meaning and content, which must be incorporated into the algorithms at a high degree of abstraction. Figure 4 shows the taxonomy of literature review [ 96 ].
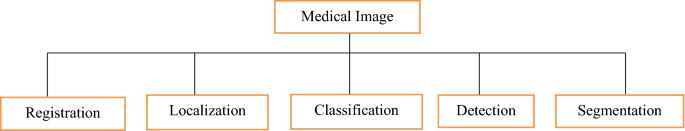
Taxonomy of literature review
4.1.1 Image registration
Goal Find [ 3 ] coordinates for transformation to align different images of the same thing, such as an MRI and CT scan, or two scans taken at various locations punctuates in time and general deep learning method used here is deep regression networks and deep learning networks (DNN) [ 6 ]. Generally, medical image analysis methods can be grouped into many categories which are shown in Fig. 4 ; this image registration is also one of the methods for the analysis purpose [ 9 ]. It is feasible to depict image registration, otherwise called image mapping, fusion, or warping, as the way toward adjusting at least two images. The motivation behind an image registration framework is to track down the ideal change in the image data [ 12 ]. Image registration is the method of converting various image datasets into one matched coordinate system with matched imaging content which has significant implications in the medical field [ 10 , 11 ]. For image processing, image registration is a critical stage in which useful information is transmitted in more than one photograph. This means images obtained at various times, from different points of view, or by the various sensors will be harmonizing. The exact fusion of useful data from at least two images is therefore very critical [ 10 ]. Deep FLASH is a new network presented here with effective medical learning preparation and inference for learning-based registration of images. Unlike established approaches, that from training data in the high elevation learns spatial transformations. From training data in the high elevation, it learns spatial conversions [ 12 ]. Developed a new image registration method using dimensional imaging, fully in a low dimensional band limited space, the network [ 13 ]. This significantly decreases the computational expense and memory footprint of costly inference and training to achieve this, and complex-valued operations and representations of neural architectures were introduced, which provide key components for learning-based registration models and create an explicit loss function of transformation fields fully characterized in a band-restricted space with much fewer parameterizations [ 14 ].
In various clinical applications medical image registration is used (Hiba A. Mohammed), image fusion (Fatma El-Zahraa Ahmed El-Gamal, Mohammed Elmogy, Ahmed Atwan), learning-based image registration (Jian Wang, Miaomiao Zhang), (Grant Haskins1, Uwe Kruger1, Pingkun Yan1) and image reconstruction [ 11 ]. The registration of medical images is an enormous theme that can be ordered from different perspectives. Registration approaches might be separated from the info picture point of view into interpatient, intrapatient (for e.g. same-day or else different), multimodal, unimodal, and enlistment [ 12 ]. Registration from the perspective of the twisting model, it is feasible to characterize strategies as unbending, relative, and deformable methods [ 13 ]. Registration techniques can be assembled from the point of view of the district of interest (ROI) as per anatomical destinations like cerebrum, liver, lung, and so forth. From the viewpoint of the picture pair measurement, enrolment strategies can be part into 3-dimensional to 3-dimensional, 3-dimensional to 2-dimensional and 2-dimensional to 2-dimensional/3-dimensional [ 14 ]. Deep learning-based methodologies of image registration as per its techniques, highlights, and ubiquity in seven gatherings, including (1) reinforcement learning-based strategies, (2) deep strategies dependent on similitudes, (3) predicting managed change, (4) unmonitored change prediction, (5) generative adversarial network in the enrolment of clinical pictures, (6) deep learning utilized to check enrolments and (7) also another strategy focused on learning [ 15 , 16 , 17 ].
There are many freely available tools [ 19 ] and toolkits for medical image registration, such as ANTs [ 24 ] and Simple ITK [ 25 ]. These methods and techniques are typically recorded by iteratively upgrading the transformational variables until a pre-defined metric of consistency is achieved [ 27 ]. Those techniques suggest ascendancy efficiency. Nevertheless, the standard was restricted by its poor procedure for registration [ 29 ].
4.1.2 Object localization
Goal: Recognize where organs or other organs are located in space (2 and 3D) or in time, landmarks or objects (video/4D) and general deep learning method used here is to identify the intersection of interest in using separate CNNs with each 2D plane running a 3D image [ 18 ]. Localization [ 19 ] for biological architectures would be a fundamental prerequisite for different medical image investigation initiatives. Generally, medical image analysis methods can be grouped into many categories which are shown in Fig. 4 , and this image localization is also one of the methods for analysis purpose [ 20 ]. Localization for the radiologist can be a hassle-free operation, or it is typically a challenging job for neural networks which are susceptible to variations in the medical data images caused by discrepancies with the process of obtainment of images, differences in structure, and pathology between patients. In medical image processing [ 13 ], the localization of anatomic structures is necessary for several activities. By defining their existence with 2-dimensional image data slices in ConvNet, here a technique is proposed for one or more anatomical systems in a 3-dimensional medical image of data localization automatically [ 14 , 15 ]. A ConvNet is equipped to look for sagittal slices, axial and coronal retrieved from a 3-dimensional image of the anatomical structure of interest. To allow the ConvNet to examine slices of different sizes, spatial pyramid pooling has been used [ 24 ]. After combining the recognition, three-dimensional convolution layers are generated by combining the output of the ConvNet in all slices. In the experiment, 100 CT scan images of the abdomen, 200 chest CT image scans, and 100 heart CTA (angiography) were used [ 25 ]. In chest CT scan image, aortic arch, the localized ascending aorta, descending aorta became visible, as were the left cardiac ventricle, CT scans of the lungs, and cardiac CTA scan image, the left cardiac ventricle, CT scans of the lungs and the liver [ 10 , 11 , 12 , 13 ]. Localization has been analysed by measuring the distances between manually and automatically specified distances bounding box centroids and walls for reference. The best outcomes have been obtained with the localization and detection of a system from established limits; for example, aortic arch is well defined and it’s too worst while the border of the system is not clearly shown for example liver [ 14 ]. Here they proposed a novel strategy confinement, for the most part, appropriate to clinical pictures in which the articles can be recognized from the foundation fundamentally dependent on element contrasts then planned another on the global and underlying levels, CRF system to additional difference and value possibilities, which represent the greater logical data between areas [ 15 ]. A scanty coding-based arrangement solution for premium district recognition with discriminative word references was also suggested as a second assessment for more precise location labelling [ 16 ]. Here the article limitation technique is assessed with 2 clinical imaging applications: sore disparity on thoracic PET-CT pictures, and cell division with minuscule pictures than those assessments show better when contrasting with as of late revealed approaches [ 17 ].
4.1.3 Classification and detection
4.1.3.1 exam classification.
Goal: Categorize a picture of a diagnostic exam as absent/present or normal/Abnormal illness and general deep learning technique here is CNN (Convolutional Neural Networks), in particular, CNNs are pre-trained on natural images [ 95 ]. Generally, medical image analysis methods can be grouped into many categories as shown in Fig. 4 , this image classification and detection are the methods for the purpose of analysis [ 96 ].
4.1.3.2 Object classification
Goal: Classify an entity that has been pre-identified (such as a Chest CT nodule) to one of two or more classes and general Deep Learning Method here is Multi-Stream CNN and additional Methods are SAE (Sparse Auto-Encoders), RBM (Restricted Boltzmann Machines), CSA (Convolutional Sparse Auto-Encoders) [ 15 , 16 , 17 ].
4.1.3.3 Classification algorithms
Classification Algorithms have a simple capability. We anticipate the objective data class by dissecting the preparation of datasets [ 16 , 17 , 18 , 19 , 20 ]. We utilize the preparation dataset to improve limit conditions which could be used to decide each target class. When those limit circumstances were resolved, the following assignment is used to foresee the objective data class [ 21 , 22 , 23 , 24 , 25 ]. Then the entire cycle is referred to as the classification technique.
4.1.3.4 Algorithm types related to classification
Figure 5 shows the various kinds of algorithms that are used in classification process [ 38 ]. The most important distinction between regression and classification is that while regression predicts continuous quantities, classification predicts discrete class labels. The two types of machine learning algorithms also have certain similarities and distinctions [ 39 , 40 ].
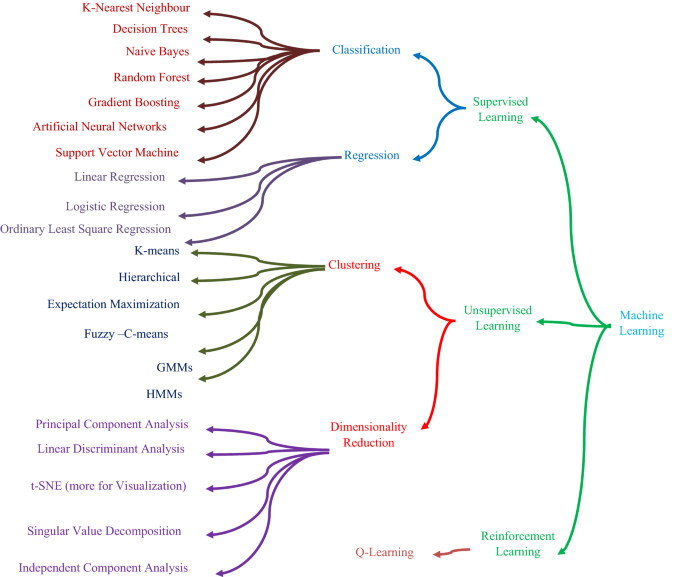
Classification algorithms
Deep learning could recognize patterns in visual inputs and determine class labels in an image [ 30 , 31 , 32 , 33 ]. A convolutional neural network (CNN) or unique CNN frameworks like AlexNet, VGG, inception, and ResNet are the most popular deep learning architectures used for image processing. Figure 6 shows the evolution in deep learning techniques [ 35 , 36 , 37 , 38 , 39 , 40 , 41 , 42 , 43 , 44 , 45 , 46 , 47 , 48 , 49 , 50 , 51 , 52 , 53 , 54 ].

Evolution in deep learning techniques
4.1.3.5 Essential terminologies in classification algorithms
Classifier A classifier is an algorithm that assigns a particular category to the data it receives [ 10 ].
Classification model It will foresee the new data's class labels and divisions [ 10 , 11 ].
Feature Feature is represented as an observable attribute of a mechanism that is being observed [ 10 , 11 , 12 , 13 , 14 , 15 ].
Binary classification Binary classification is a classification task that has two results. Gender classification either female or male is an example [ 10 , 11 , 12 , 13 , 14 , 15 ].
Multi-class classification Classification of more than 2 classifications is referred to as multi-class classification. Each data is allocated to only one targeted classification label in multi-class classification. For example, an animal maybe a dog or cat, and not both together [ 17 ].
Multi-label classification A classification task in which each item is assigned to a collection of target labels is known as multi-label classification (more than 1 data class). For example, a news article may be about games, an individual, and a place all at once [ 13 , 14 , 15 , 16 , 17 , 18 , 19 , 20 ].
Arrangement of infections [ 19 ] utilizing deep learning advancements on clinical pictures has picked up a ton of foothold over the most recent couple of years. For neuroimaging, the significant focal point of 3Dimensional deep learning has been on distinguishing infections with some anatomical pictures [ 21 ]. A few investigations had not identify dementia and also shows some variations from various imaging models such as practical Magnetic Resonance Imaging, DTI [ 22 ]. AD (Alzheimer's disease) will be the most widely recognized type in dementia, typically connected to the neurotic amyloid affidavits, primary decay, and some metabolic varieties with the science in the mind [ 23 ]. The ideal conclusion of Alzheimer’s disease assumes a significant job to block the movement of the infection.
For analysis and disease diagnosis [ 16 ], radiologists may need to consult medical archives for similar clinical cases. It is difficult to retrieve the relevant variety of diseases and imaging modalities, clinical cases are automatically, reliably, and correctly taken from the substantial medical image collection [ 17 ]. A powerful and reliable method for medical image classification, modality classification was used in this study, which can be used to extract clinical data from vast medical repositories. The method was created by combining the transfer learning principle with a pre-trained ResNet50 model for optimized feature retrieval and also classification with TLRN-LDA (Linear Discriminant Analysis) [ 18 ]. In Image CLEF benchmark (31-class image dataset), the evolved technique gives 88 percent of average accuracy in classification that is up to 11 percent higher related to the current state-of-the-art methods with the same image datasets [ 19 ]. Furthermore, hand-crafted features were extracted for comparison in this study [ 19 , 20 ].
Transfer learning [ 17 ] is the viable component that can give a promising arrangement transferring information from nonexclusive article acknowledgment errands to area explicit undertakings. Here utilized a deep convolutional neural network, called DeTraC (Decompose, Transfer, Compose) with the characterization of Covid-19 +ve cases of chest X-ray data [ 18 ]. DeTraC can able to manage any kind of abnormalities with the picture dataset by examining its image class limits utilizing image class decay system [ 19 ]. Then the exploratory outcomes indicated that the ability of DeTraC with the discovery of Covid 19 +ve cases from complete picture datasets taken from few clinics in all over the globe. More exactness of 94 percent (True positive rate of 100percent) has been accomplished with DeTraC the identification of +ve Covid-19 X-ray data from ordinary cases, and also extreme intense respiratory condition scenarios [ 20 ].
Here [ 18 ] played out various levelled arrangements utilizing this HMIC (Hierarchical Medical Image characterization) method. This utilizes heaps of deep learning technique models for giving specific perception at every degree of medical image orders [ 19 , 20 ]. For verification and testing purpose, medical image is categorized into 3 classifications at the parent level (histologically typical controls, Celiac disease and environmental enteropathy) [ 21 ]. Then for the kid level, 4 classes (I, IIIa, IIIb, and IIIC) of celiac disease severity are arranged [ 22 ].
Table 2 describes the summary of previous research results associated with image classification [ 50 , 51 , 52 , 53 , 54 , 55 , 56 , 57 , 58 , 59 , 60 , 61 , 62 , 63 , 64 , 65 , 66 , 67 , 68 , 69 , 70 , 71 , 72 , 73 , 74 , 75 , 76 , 77 , 78 , 79 , 80 ]. Several deep learning architectures produced different accuracy levels that are described in Fig. 7 [ 55 , 56 , 57 , 58 , 59 , 60 , 61 , 62 , 63 , 64 , 65 , 66 , 67 , 68 , 69 , 70 , 71 , 72 , 73 , 74 , 75 , 76 , 77 , 78 , 79 , 80 , 81 , 82 , 83 , 84 , 85 ].
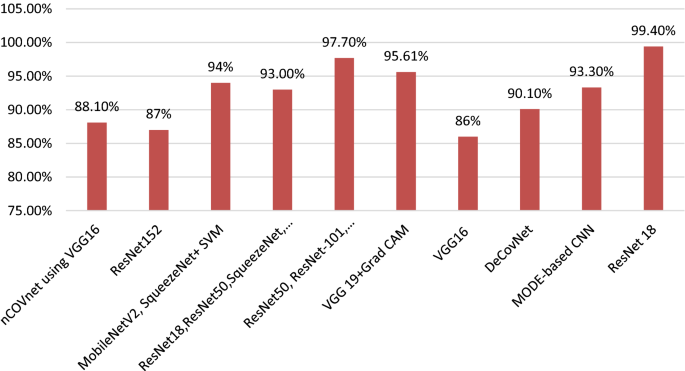
Deep learning algorithms with accuracy
4.1.4 Object Detection
Goal: Identify a lesion or other interesting entity an image inside an image, and general deep learning method here is CNN and CNN with multi-stream [ 85 ].
4.1.5 Segmentation
4.1.5.1 substructure/organ segmentation.
Goal: Identify the curvature of the organ or its interior interest, enabling volume to be quantitatively analysed shape and form, such as for the heart or brain, and general deep learning methods are (recurrent neural network) RNN, CNNs, fCNNs (neural networks that are fully convolutionary) network [ 90 , 91 ]. Generally, medical image analysis methods can be grouped into many categories which are shown in Fig. 4 , and this image segmentation is also one of the methods for analysis purpose [ 92 ].
4.1.5.2 Lesion segmentation
Goal: Integration of object detection and organ/organ recognition segmentation of substructures and general deep learning method is Multi-Stream CNN [ 5 , 6 , 7 ].
[ 7 ] Medical image segmentation plays an essential role in numerous applications of computer-aided diagnostic systems. New medical image processing algorithms are being applied through the enormous investment, and advancement of microscopy, ultrasound, computed tomography (CT), dermoscopy, magnetic resonance imaging (MRI), and positron emission tomography and X-ray is examples of medical imaging modalities [ 8 ]. A 3D (three-dimensional) and or 2D (two-dimensional) image data is automatically or semi-automatically detected by medical image segmentation. Picture segmentation is the process by which a digital image is separated into multiple pixels. The prior objective of segmentation is to make it clearer and turn medical image representation into a meaningful subject. Because of the high variability in the photographs, segmentation is a hard job [ 19 ]. In recent days, AI and machine learning calculations have been encouraging radiologists in the division of clinical pictures, for example, bosom disease mammograms, cerebrum tumours, mind sores, skull stripping, and so forth division not as zeroing in on explicit locales in the clinical picture, additionally helps master radiologists in quantitative evaluation, and arranging further treatment [ 20 ]. A few analysts have added with the utilization of 3D convolutional neural networks in clinical picture division [ 21 ].
5 Trends and challenges
With the various CNN-based deep neural networks created, a significant result was achieved on ImageNet Challenger, the most significant image classification and segmentation challenge in the image analysing area. The key benefit of CNN over its predecessors is that it identifies essential features without the need for human intervention [ 27 ]. Various classification models are discussed in Fig. 6 , produced different performance metrics such as precision, sensitivity/recall, specificity, accuracy, f-measure, and receiver operating characteristic (ROC) curves. Based on these performance metrics, we can evaluate the best CNN model [ 28 ].
We have come across the issues of imbalanced data, lack of confidence interval, and lack of properly annotated data so much in the recent deep learning related Medical Imaging literature that it’s easy to label it the fundamental challenge that the medical imaging field is currently experiencing in completely exploring deep learning advances [ 29 ].The number of samples and patients in the public databases currently available for medical imaging tasks are limited, except few datasets. Medical imaging datasets are too limited when compared to datasets for general computer vision issues, which usually range from a few hundred thousand to millions of annotated photos [ 30 ]. But on the other hand, we can see a growing trend in the medical imaging community to follow the practices of the wider pattern recognition community, to learn deep models end-to-end. The wider community, on the other hand, has typically embraced such activities based on the availability of large-scale annotated datasets, which is a crucial prerequisite for inducing accurate deep models [ 31 ].As a result, it's still unclear how well end-to-end qualified models can perform medical image analysis tasks without over- fitting to the training datasets. We have developed some elementary data augmentation techniques such as image flipping, padding, principal component analysis (PCA), image cropping, and adversarial training. However, these algorithms are not as advanced as the GAN for enhancing datasets [ 32 , 33 , 34 , 35 , 36 ].
Another major obstacle could be the use of black boxes; the legal ramifications of black-box functionality could be a deterrent since healthcare professionals would not rely on it. Who might be held liable if the outcome was unfavourable? Due to the sensitivity of this region, the hospital might be hesitant to use a black-box system, which would allow the hospital to track that a specific result came from an optometrist [ 35 ]. Trying to unlock the black box is a major research subject, and deep learning scientists are working to solve it [ 36 ].
Furthermore, due to complex data structures, teaching deep learning models is an extremely expensive endeavour. They sometimes necessitate high-end GPUs and hundreds of computers, which drives up the price for consumers [ 43 ].
Since the increased complexity of several layers necessitates a high computational load, there by the training performance suffers. Enhanced activation functions, cost function architecture, and drop-out approaches have all been used to combat vanishing gradient and over-fitting issues [ 47 ]. Using highly parallel hardware, such as GPUs and batch normalization, the issue of the high computational load has been addressed. We have also mentioned deep learning architectures in Table 1 related to corresponding applications from earlier days to till now [ 48 ]. The development of an interdisciplinary data pool is made possible by the availability of a vast volume of electronic medical record data. Machine learning extracts information from large amounts of data and generates output that can be used for individual outcome prediction and clinical decision-making [ 16 ]. This could pave the way for personalized medicine (also known as precision medicine), in which each person’s genetic, environmental, and lifestyle factors are considered for disease prevention, treatment, and prognosis [ 17 ].
6 Conclusion
Medical imaging is a key technology that bridges scientific and societal needs and can provide an important synergy that may contribute to advances in each of the areas. Our survey has illuminated the current state of the art based on the recent scientific literature from 120 medical imaging research papers which may be beneficial to radiologists worldwide. In addition to finding that the ResNet architecture typically has the highest performance, we also covered the current challenges, major issues, and future directions.
LeCun Y, Bengio Y, Hinton G (2015) Deep learning. Nature 521(7553):436–444. https://doi.org/10.1038/nature14539
Article Google Scholar
Razzak MI, Naz ZS, Zaib A (2018) Deep learning for medical image processing: overview, challenges and the future. Classification in BioApps: automation of decision making. Springer, Cham, Switzerland, pp 323–350. https://doi.org/10.1007/978-3-319-65981-7_12
Pang S, Yang X (2016) Deep Convolutional Extreme learning Machine and its application in Handwritten Digit Classification. Hindawi Publ Corp Comput Intell Neurosci 2016:3049632. https://doi.org/10.1155/2016/3049632
Chollet F et al (2015) Keras. https://github.com/fchollet
Zhang Y, Zhang S et al (2016) Theano: A Python framework for fast computation of mathematical expressions, arXiv e-prints, abs/1605.02688. http://arxiv.org/abs/1605.02688
Vedaldi A, Lenc K (2015) Matconvnet: convolutional neural networks for matlab. In: Proceedings of the 23rd ACM international conference on Multimedia. ACM, pp 689–692. https://doi.org/10.1145/2733373.2807412
Guo Y, Ashour A (2019) Neutrosophic sets in dermoscopic medical image segmentation. Neutroscophic Set Med Image Anal 11(4):229–243. https://doi.org/10.1016/B978-0-12-818148-5.00011-4
Merjulah R, Chandra J (2019) Classification of myocardial ischemia in delayed contrast enhancement using machine learning. Intell Data Anal Biomed Appl, pp 209–235. https://doi.org/10.1016/B978-0-12-815553-0.00011-2
Oliveira FPM, Tavares JMRS (2014) Medical Image Registration: a review. Comput Methods Biomech Biomed Eng pp 73–93. https://doi.org/10.1080/10255842.2012.670855
Wang J, Zhang M (2020) Deep FLASH: an efficient network for learning-based Medical Image Registration. In: Proceedings of 2020 IEEE/CVF conference on computer vision and pattern recognition (CVPR), pp 4443–4451. https://doi.org/10.1109/cvpr42600.2020.00450
Fu Y, Lei Y, Wang T, Curran WJ, Liu T, Yang X (2020) Deep learning in medical image registration: a review. Phys Med Biol 65(20). https://doi.org/10.1088/1361-6560/ab843e
Haskins G, Kruger U, Yan P (2020) Deep learning in medical image registration: a survey. Mach Vis Appl 31:1–18. https://doi.org/10.1007/s00138-020-01060-x
De Vos BD, Wolterink JM, Jong PA, Leiner T, Viergever MA, Isgum I (2017) ConvNet-based localization of anatomical structures in 3D medical images. IEEE Trans Med Imaging 36(7):1470–1481. https://doi.org/10.1109/TMI.2017.2673121
Song Y, Cai W, Huang H et al (2013) Region-based progressive localization of cell nuclei in microscopic images with data adaptive modelling. BMC Bioinformatics 14:173. https://doi.org/10.1186/1471-2105-14-173
Sharma H, Jain JS, Gupta S, Bansal P (2020) Feature extraction and classification of chest X-ray images using CNN to detect pneumonia. 2020 In: Proceedings of the 10th international conference on cloud computing, data science & engineering (confluence), pp 227–231. https://doi.org/10.1109/Confluence47617.2020.9057809
Hassan M, Ali S, Alquhayz H, Safdar K (2020) Developing intelligent medical image modality classification system using deep transfer learning and LDA. Sci Rep 10(1):12868. https://doi.org/10.1038/s41598-020-69813-2
Abbas A, Abdelsamea MM, Gaber MM (2021) Classification of COVID-19 in chest X-ray images using DeTraC deep convolutional neural network. Appl Intell 51:854–864. https://doi.org/10.1007/s10489-020-01829-7
Kowsari K, Sali R, Ehsan L, Adorno W et al (2020) Hierarchical medical image classification, a deep learning approach. Information 11(6):318. https://doi.org/10.3390/info11060318
Singh SP, Wang L, Gupta S, Goli H, Padmanabhan P, Gulyas B (2020) 3D deep learning on medical images: a review. Sensors 20(18):5097. https://doi.org/10.3390/s20185097
Shen D, Wu G, Suk H (2017) Deep learning in medical image analysis. Annu Rev Biomed Eng 19:221–248. https://doi.org/10.1146/annurev-bioeng-071516-044442
Wang SH, Phillips P, Sui Y, Bin L, Yang M, Cheng H (2018) Classification of Alzheimer’s disease based on eight-layer convolutional neural network with leaky rectified linear unit and max pooling. J Med Syst 42(5):85. https://doi.org/10.1007/s10916-018-0932-7
Krizhevsky A, Sulskever I, Hinton GE (2012) ImageNet classification with deep convolutional neural networks. Adv Neural Inf Process Syst 60(6):84–90. https://doi.org/10.1145/3065386
Szegedy C, Liu W, Jia Y, Sermanet P, Reed S et al (2015) Going deeper with convolutions. In: Proceedings of the IEEE Computer Society conference on computer vision and pattern recognition; IEEE Computer Society, 7(12), pp 1–9
Lowekamp BC, Chen DT, Ibanez L, Blezek D (2013) The design of SimpleITK. Front Neuroinform 45(7). https://doi.org/10.3389/fninf.2013.00045
Avants B, Tustison N, Song G (2009) Advanced Normalization Tools (ANTS). The Insight Journal. http://hdl.handle.net/10380/3113
Rahmat T, Ismail A, Sharifah A (2019) Chest X-ray image classification using faster R-CNN. Malays J Comput 4(1):225–236. https://doi.org/10.24191/mjoc.v4i1.6095
Jain G, Mittal D, Takur D, Mittal MK (2020) A deep learning approach to detect Covid-19 coronavirus with X-ray images. Elsevier-Biocybern Biomed Eng 40(4):1391–1405. https://doi.org/10.1016/j.bbe.2020.08.008
Togacar M, Ergen B, Comert Z (2020) COVID-19 detection using deep learning models to exploit social mimic optimization and structured chest X-ray images using fuzzy color and stacking approaches. Elsevier-Comput Biol Med 121:103805. https://doi.org/10.1016/j.compbiomed.2020.103805
Minaee S, Kafieh R, Sonka M, Yazdani S, Soufi GJ (2020) Deep-COVID: predicting COVID-19 from chest X-ray images using deep transfer learning. Elsevier-Med Image Anal 65:101794. https://doi.org/10.1016/j.media.2020.101794
Apostolopoulos ID, Mpesiana TA (2020) Covid-19: automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Springer-Phys Eng Sci Med 43(2):635–640. https://doi.org/10.1007/s13246-020-0865-4
Panwar H, Gupta PK, Siddiqui MK, Morales-Menendez R, Singh V (2020) Application of deep learning for fast detection of COVID-19 in X-rays using nCOVnet. Chaos Solitons Fractals 138(3):109944. https://doi.org/10.1016/j.chaos.2020.109944
Ouchicha C, Ammor O, Meknassi M (2020) CVDNet: a novel deep learning architecture for detection of coronavirus (Covid-19) from chest X-ray images. Elsevier-Chaos Solitons Fractals 140(5):110245. https://doi.org/10.1016/j.chaos.2020.110245
Sethy PK, Behera SK, Ratha PK (2020) Detection of coronavirus disease (COVID-19) based on deep features and support vector machine. Int J Math Eng Manag Sci 5(4):643–651. https://doi.org/10.33889/IJMEMS.2020.5.4.052
Jaiswal AK, Tiwari P, Kumar S, Gupta D, Khanna A, Rodrigues JJ (2019) Identifying pneumonia in chest X-rays: a deep learning approach. Measurement 145(2):511–518. https://doi.org/10.1016/j.measurement.2019.05.076
Civit-Masot J, Luna-Perejon F, Dominguez Morales M, Civit A (2020) Deep learning system for COVID-19 diagnosis aid using X-ray pulmonary images. Appl Sci 10(13):4640. https://doi.org/10.3390/app10134640
Punn NS, Sonbhadra SK, Agarwal S (2020) COVID-19 epidemic analysis using machine learning and deep learning algorithms. https://doi.org/10.1101/2020.04.08.20057679
Dahl GE, Yu D, Deng L, Acero A (2012) Context-dependent pre-trained deep neural networks for large-vocabulary speech recognition. IEEE Trans Audio Speech Lang Process 20(1):30–42. https://doi.org/10.1109/TASL.2011.2134090
Krizhevsky A, Sutskever I, Hinton GE (2012) ImageNet classification with deep convolutional neural networks. Adv Neural Inf Process Syst 25(2):1097–1105. https://doi.org/10.1145/3065386
Silver D, Huang A, Maddison C et al (2016) Mastering the game of Go with deep neural networks and tree search. Nature 529:484–489. https://doi.org/10.1038/nature16961
Mnih V et al (2015) Human-level control through deep reinforcement learning. Nature 518:529–533. https://doi.org/10.1038/nature14236
Abràmoff MD, Lou Y, Erginay A, Clarida W, Amelon R, Folk JC, Niemeijer M (2016) Improved automated detection of diabetic retinopathy on a publicly available dataset through integration of deep learning. Invest Ophthalmol Vis Sci 57(13):5200–5206. https://doi.org/10.1167/iovs.16-19964
Akram SU, Kannala J, Eklund L, Heikkila J (2016) Cell segmentation proposal network for microscopy image analysis. In: Proceedings of the Deep Learning in Medical Image Analysis (DLMIA). Lecture Notes in Computer Science, 10 0 08, pp 21–29. https://doi.org/10.1007/978-3-319-46976-8_3
Ballin A, Karlinsky L, Alpert S, Hasoul S, Ari R, Barkan E (2016) A region based convolutional network for tumor detection and classification in breast mammography. In: Proceedings of the deep learning in medical image analysis (DLMIA). Lecture Notes in Computer Science, 10 0 08, pp 197–205. https://doi.org/10.1007/978-3-319-46976-8_21
Alansary A et al (2016) Fast fully automatic segmentation of the human placenta from motion corrupted MRI. In: Proceedings of the medical image computing and computer-assisted intervention. Lecture Notes in Computer Science, 9901, pp 589–597. https://doi.org/10.1007/978-3-319-46723-8_68
Albarqouni S, Baur C, Achilles F, Belagiannis V, Demirci S, Navab N (2016) Aggnet: deep learning from crowds for mitosis detection in breast cancer histology images. IEEE Trans Med Imaging 35:1313–1321. https://doi.org/10.1109/TMI.2016.2528120
Anavi Y, Kogan I, Gelbart E, Geva O, Greenspan H (2015) A comparative study for chest radiograph image retrieval using binary texture and deep learning classification. In: Proceedings of the IEEE engineering in Medicine and Biology Society, pp 2940–2943. https://doi.org/10.1109/EMBC.2015.7319008
Anavi Y, Kogan I, Gelbart E, Geva O, Greenspan H (2016) Visualizing and enhancing a deep learning framework using patients age and gender for chest X-ray IMAGE Retrieval. In: Proceedings of the SPIE on medical imaging, 9785, 978510
Andermatt S, Pezold S, Cattin P (2016) Multi-dimensional gated recurrent units for the segmentation of biomedical 3D-data. In: Proceedings of the deep learning in medical image analysis (DLMIA). Lecture Notes in Computer Science, 10 0 08, pp 142–151
Anthimopoulos M, Christodoulidis S, Ebner L, Christe A, Mougiakakou S (2016) Lung pattern classification for interstitial lung diseases using a deep convolutional neural network. IEEE Trans Med Imaging 35(5):1207–1216. https://doi.org/10.1109/TMI.2016.2535865
Antony J, McGuinness K, Connor NEO, Moran K (2016) Quantifying radio graphic knee osteoarthritis severity using deep convolutional neural networks. arxiv: 1609.02469
Apou G, Schaadt NS, Naegel B, Forestier G, Schönmeyer R, Feuerhake F, Wemmert C, Grote A (2016) Detection of lobular structures in normal breast tissue. Comput Biol Med 74:91–102. https://doi.org/10.1016/j.compbiomed.2016.05.004
Arevalo J, Gonzalez FA, Pollan R, Oliveira JL, Lopez MAG (2016) Representation learning for mammography mass lesion classification with convolutional neural networks. Comput Methods Programs Biomed 127:248–257. https://doi.org/10.1016/j.cmpb.2015.12.014
Baumgartner CF, Kamnitsas K, Matthew J, Smith S, Kainz B, Rueckert D (2016) Real-time standard scan plane detection and localisation in fetal ultrasound using fully convolutional neural networks. In: Proceedings of the medical image computing and computer-assisted intervention. Lecture Notes in Computer Science, 9901, pp 203–211. https://doi.org/10.1007/978-3-319-46723-8_24
Balasamy K, Ramakrishnan S (2019) An intelligent reversible watermarking system for authenticating medical images using wavelet and PSO. Clust Comput 22(2):4431–4442. https://doi.org/10.1007/s10586-018-1991-8
Bengio Y (2012) Practical recommendations for gradient-based training of deep architectures. Neural networks: tricks of the trade. Lecture Notes in Computer Science, 7700, Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-35289-8_26
Bengio Y, Courville A, Vincent P (2013) Representation learning: a review and new perspectives. IEEE Trans Pattern Anal Match Intell 35(8):1798–1828. https://doi.org/10.1109/TPAMI.2013.50
Bengio Y, Lamblin P, Popovici D, Larochelle H (2007) Greedy layer-wise training of deep networks. In: Proceedings of the advances in neural information processing systems, pp 153–160
Bengio Y, Simard P, Frasconi P (1994) Learning long-term dependencies with gradient descent is difficult. IEEE Trans Neural Networks 5(2):157–166. https://doi.org/10.1109/72.279181
Benou A, Veksler R, Friedman A, Raviv TR (2016) Denoising of contrast enhanced MRI sequences by an ensemble of expert deep neural networks. In: Proceedings of the deep learning in medical image analysis (DLMIA). Lecture Notes in Computer Science, 10 0 08, pp 95–110. https://doi.org/10.1146/annurev-bioeng-071516-044442
BenTaieb A, Hamarneh G (2016) Topology aware fully convolutional networks for histology gland segmentation. In: Proceedings of the medical image computing and computer-assisted intervention. Lecture Notes in Computer Science, 9901, pp 460–468. https://doi.org/10.1007/978-3-319-46723-8_53
BenTaieb A, Kawahara J, Hamarneh G (2016) Multi-loss convolutional networks for gland analysis in microscopy. In: Proceedings of the IEEE international symposium on biomedical imaging, pp 642–645. https://doi.org/10.1109/ISBI.2016.7493349
Bergstra J, Bengio Y (2012) Random search for hyper-parameter optimization. J Mach Learn Res 13(10):281–305
MathSciNet MATH Google Scholar
Birenbaum A, Greenspan H (2016) Longitudinal multiple sclerosis lesion segmentation using multi-view convolutional neural networks. In: Proceedings of the deep learning in medical image analysis (DLMIA). Lecture Notes in Computer Science, 10 0 08, pp 58–67. https://doi.org/10.1007/978-3-319-46976-8_7
Cheng X, Zhang L, Zheng Y (2015) Deep similarity learning for multimodal medical images. Comput Methods Biomech Biomed Eng pp 248–252. https://doi.org/10.1080/21681163.2015.1135299
Cicero M, Bilbily A, Colak E, Dowdell T, Gray B, Perampaladas K, Barfett J (2017) Training and validating a deep convolutional neural network for computer-aided detection and classification of abnormalities on frontal chest radiographs. Invest Radiol 52(5):281–287. https://doi.org/10.1097/RLI.0000000000000341
Ertosun MG, Rubin DL Automated grading of Gliomas using deep learning in digital pathology images: a modular approach with ensemble of convolutional neural networks. In: AMIA annual symposium proceedings, pp 1899–908
Guo Y, Gao Y, Shen D (2016) Deformable MR prostate segmentation via deep feature learning and sparse patch matching. IEEE Trans Med Imaging 35(4):1077–1089. https://doi.org/10.1109/TMI.2015.2508280
Guo Y, Wu G, Commander LA, Szary S, Jewells V, Lin W, Shen D (2014) Segmenting Hippocampus from infant brains by sparse patch matching with deep-learned features. In: Proceedings of the medical image computing and computer-assisted intervention. Lecture Notes in Computer Science, 8674, pp 308–315. https://doi.org/10.1007/978-3-319-10470-6_39
Han XH, Lei J, Chen YW (2016) HEp-2 cell classification using K-support spatial pooling in deep CNNs. In: Proceedings of the deep learning in medical image analysis (DLMIA). Lecture Notes in Computer Science, 10 0 08, pp 3–11. https://doi.org/10.1007/978-3-319-46976-8_1
Haugeland J (1985) Artificial intelligence: the very idea. The MIT Press, Cambridge. ISBN: 0262081539
Google Scholar
Havaei M, Davy A, Warde Farley D, Biard A, Courville A, Bengio Y, Pal C, Jodoin PM, Larochelle H (2016) Brain tumor segmentation with deep neural networks. Med Image Anal 35:18–31. https://doi.org/10.1016/j.media.2016.05.004
Havaei M, Guizard N, Chapados N, Bengio Y (2016) HeMIS: hetero-modal image segmentation. In: Proceedings of the medical image computing and computer-assisted intervention. Lecture Notes in Computer Science, 9901, pp 469–477. https://doi.org/10.1007/978-3-319-46723-8_54
He K, Zhang X, Ren S, Sun J (2015) Deep residual learning for image recognition. arxiv: 1512.03385
Janowczyk A, Madabhushi A (2016) Deep learning for digital pathology image analysis: a comprehensive tutorial with selected use cases. J Pathol Inform 7:29. https://doi.org/10.4103/2153-3539.186902
Jia Y, Shelhamer E, Donahue J, Karayev S, Long J, Girshick R, Guadarrama S, Darrell T (2014) Caffe: convolutional architecture for fast feature embedding. In: Proceedings of the twenty-second ACM international conference on multi-media, pp 675–678. https://doi.org/10.1145/2647868.2654.889
Kainz P, Pfeiffer M, Urschler M (2015) Semantic segmentation of colon glands with deep convolutional neural networks and total variation segmentation. arxiv: 1511.06919
Kallen H, Molin J, Heyden A, Lundstr C, Astrom K (2016) Towards grading gleason score using generically trained deep convolutional neural networks. In: Proceedings of the IEEE international symposium on biomedical imaging, pp 1163–1167. https://doi.org/10.1109/ISBI.2016.7493473
Balasamy K, Suganyadevi S (2021) A fuzzy based ROI selection for encryption and watermarking in medical image using DWT and SVD. Multimedia Tools Appl 80:7167–7186. https://doi.org/10.1007/s11042-020-09981-5
Lekadir K, Galimzianova A, Betriu A, Vila MDM, Igual L, Rubin, DL, Fernandez E, Radeva P, Napel S (2017) A convolutional neural network for automatic characterization of plaque composition in carotid ultrasound. IEEE J Biomed Health Inform 21(1):48–55. https://doi.org/10.1109/JBHI.2016.2631401
Li R, Zhang W, Suk HI, Wang L, Li J, Shen D, Ji S (2014) Deep learning based imaging data completion for improved brain disease diagnosis. Med Image Comput Comput-Assist Interv 17(Pt 3):305–312. https://doi.org/10.1007/978-3-319-0443-0_39
Li W, Manivannan S, Akbar S, Zhang J, Trucco E, McKenna SJ (2016) Gland segmentation in colon histology images using hand-crafted features and convolutional neural networks. In: Proceedings of the IEEE international symposium on biomedical imaging, pp 1405–1408. https://doi.org/10.1109/ISBI.2016.7493530
Miao S, Wang ZJ, Liao R (2016) A CNN regression approach for real-time 2D/3D registration. IEEE Trans Med Imaging 35(5):1352–1363. https://doi.org/10.1109/TMI.2016.2521800
Moeskops P, Viergever MA, Mendrik AM, Vries LSD, Benders MJNL, Isgum I (2016) Automatic segmentation of MR brain images with a convolutional neural network. IEEE Trans Med Imaging 35(5):1252–1262. https://doi.org/10.1109/TMI.2016.2548501
Pinaya WHL, Gadelha A, Doyle OM, Noto C, Zugman A, Cordeiro Q, Jackowski AP, Bressan RA, Sato JR (2016) Using deep belief network modelling to characterize differences in brain morphometry in schizophrenia. Nat Sci Rep 6:38897. https://doi.org/10.1038/srep38897
Plis SM, Hjelm DR, Salakhutdinov R, Allen EA, Bockholt HJ, Long JD, Johnson HJ, Paulsen JS, Turner JA, Calhoun VD (2014) Deep learning for neuroimaging: a validation study. Front Neurosci 8:229. https://doi.org/10.3389/fnins.2014.00229
Poudel RPK, Lamata P, Montana G (2016) Recurrent fully convolutional neural networks for multi-slice MRI cardiac segmentation. arxiv: 1608.03974
Prasoon A, Petersen K, Igel C, Lauze F, Dam E, Nielsen M (2013) Deep feature learning for knee cartilage segmentation using a triplanar convolutional neural network. In: Proceedings of the medical image computing and computer-assisted intervention. Lecture Notes in Computer Science, 8150, pp 246–253. https://doi.org/10.1007/978-3-642-40763-5_31
Rajkomar A, Lingam S, Taylor AG, Blum M, Mongan J (2017) High-throughput classification of radiographs using deep convolutional neural networks. J Digit Imaging 30:95–101. https://doi.org/10.1007/s10278-016-9914-9
Ravi D, Wong C, Deligianni F, Berthelot M, Andreu Perez J, Lo B, Yang GZ (2017) Deep learning for health informatics. IEEE J Biomed Health Inform 21(1):4–21. https://doi.org/10.1109/JBHI.2016.2636665
Ravishankar H, Prabhu SM, Vaidya V, Singhal N (2016) Hybrid approach for automatic segmentation of fetal abdomen from ultrasound images using deep learning. In: Proceedings of the IEEE international symposium on biomedical imaging, pp 779–782. https://doi.org/10.1109/ISBI.2016.7493382
Sahiner B, Chan HP, Petrick N, Wei D, Helvie MA, Adler DD, Goodsitt MM (1996) Classification of mass and normal breast tissue: a convolution neural network classifier with spatial domain and texture images. IEEE Trans Med Imaging 15(5):598–610. https://doi.org/10.1109/42.538937
Samala RK, Chan HP, Hadjiiski LM, Cha K, Helvie MA (2016) Deep-learning convolution neural network for computer-aided detection of microcalcifications in digital breast tomosynthesis. In: Proceedings 9785, medical imaging, computer-aided diagnosis, 97850Y. https://doi.org/10.1117/12.2217092
Samala RK, Chan HP, Hadjiiski L, Helvie MA, Wei J, Cha K (2016) Mass detection in digital breast tomosynthesis: deep convolutional neural network with transfer learning from mammography. Med Phys 43(12):6654. https://doi.org/10.1118/1.4967345 .
Sarraf S, Tofighi G (2016) Classification of Alzheimer’s disease using fMRI data and deep learning convolutional neural networks. arxiv: 1603.08631
Schaumberg AJ, Rubin MA, Fuchs TJ (2016) H&E-stained whole slide deep learning predicts SPOP mutation state in prostate cancer. arxiv: 064279. https://doi.org/10.1101/064279
Suganyadevi S, Shamia D, Balasamy K (2021) An IoT-based diet monitoring healthcare system for women. Smart Healthc Syst Des Secur Priv Asp. https://doi.org/10.1002/9781119792253.ch8
Spampinato C, Palazzo S, Giordano D, Aldinucci M, Leonardi R (2016) Deep learning for automated skeletal bone age assessment in X-ray images. Med Image Anal 36:41–51. https://doi.org/10.1016/j.media
Balasamy K, Shamia D (2021) Feature extraction-based medical image watermarking using fuzzy-based median filter. IETE J Res 1–9. https://doi.org/10.1080/03772063.2021.1893231
Stern D, Payer C, Lepetit V, Urschler M (2016) Automated age estimation from hand MRI volumes using deep learning. In: Proceedings of the medical image computing and computer-assisted intervention. Lecture Notes in Computer Science, 9901, pp 194–202. https://doi.org/10.1007/978-3-319-46723-8_23
Suk HI, Shen D (2013) Deep learning based feature representation for AD/MCI classification. In: Proceedings of the medical image computing and computer-assisted intervention. Lecture Notes in Computer Science, 8150, pp 583–590. https://doi.org/10.1007/978-3-642-40763-5_72
Sun W, Seng B, Zhang J, Qian W (2016) Enhancing deep convolutional neural network scheme for breast cancer diagnosis with unlabeled data. Comput Med Imaging Gr 57:4–9. https://doi.org/10.1016/j.compmedimag
Sun W, Zheng B, Qian W (2016) Computer aided lung cancer diagnosis with deep learning algorithms. In: Proceedings of the SPIE medical imaging, 9785, 97850Z. https://doi.org/10.1117/12.2216307
Teikari P, Santos M, Poon C, Hynynen K (2016) Deep learning convolutional networks for multiphoton microscopy vasculature segmentation. arxiv: 1606.02382
Tran PV (2016) A fully convolutional neural network for cardiac segmentation in short axis MRI. arxiv: 1604.00494.abs/1604.00494
Xie Y, Xing F, Kong X, Su H, Yang L (2015) Beyond classification: structured regression for robust cell detection using convolutional neural network. In: Proceedings of the medical image computing and computer-assisted intervention. Lecture Notes in Computer Science, 9351, pp 358–365. https://doi.org/10.1007/978-3-319-24574-4_43
Xie Y, Zhang Z, Sapkota M, Yang L (2016) Spatial clockwork recurrent neural network for muscle perimysium segmentation. In: Proceedings of the international conference on medical image computing and computer-assisted intervention. Lecture Notes in Computer Science, 9901. Springer, pp 185–193. https://doi.org/10.1007/978-3-319-46723-8_22
Xu T, Zhang H, Huang X, Zhang S, Metaxas DN (2016) Multimodal deep learning for cervical dysplasia diagnosis. In: Proceedings of the medical image computing and computer-assisted intervention. Lecture Notes in Computer Science, 9901, pp 115–123. https://doi.org/10.1007/978-3-319-46723-8_14
Krishnasamy B, Balakrishnan M, Christopher A (2021) A genetic algorithm based medical image watermarking for improving robustness and fidelity in wavelet domain. Intelligent data engineering and analytics. Advances in intelligent systems and computing, 1177. Springer, Singapore. https://doi.org/10.1007/978-981-15-679-1_27
Xu Z, Huang J (2016) Detecting Cells in one second. In: Proceedings of the medical image computing and computer-assisted intervention. Lecture Notes in Computer Science, 9901, pp 676–684. https://doi.org/10.1007/978-3-319-46723-8_78
Yang D, Zhang S, Yan Z, Tan C, Li K, Metaxas D (2015) Automated anatomical landmark detection on distal femur surface using convolutional neural network. In: Proceedings of the IEEE international symposium on biomedical imaging, pp 17–21. https://doi.org/10.1109/isbi.2015.7163806
Yang H, Sun J, Li, H, Wang L, Xu Z (2016) Deep fusion net for multi-atlas segmentation: Application to cardiac MR images. In: Proceedings of the medical image computing and computer assisted intervention. Lecture Notes in Computer Science, 9901, pp 521–528. https://doi.org/10.1007/978-3-319-46723-8_60
Wang S, Yao J, Xu Z, Huang J (2016) Subtype cell detection with an accelerated deep convolution neural network. In: Proceedings of the medical image computing and computer-assisted intervention. In: Lecture Notes in Computer Science, 9901, pp 640–648. https://doi.org/10.1007/978-3-319-46723-8_74
Xu Y, Mo T, Feng Q, Zhong P, Lai M, Chang E (2014) Deep learning of feature representation with multiple instance learning for medical image analysis. In: Proceedings of the IEEE international conference on acoustics, speech and signal processing (ICASSP), pp 1626–1630. https://doi.org/10.1109/ICASSP.2014.6853873
Yang X, Kwitt R, Niethammer M (2016) Fast predictive image registration. In: Proceedings of the deep learning in medical image analysis (DLMIA). Lecture Notes in Computer Science, 10 0 08, pp 48–57. https://doi.org/10.1007/978-3-319-46976-8_6
Yao J, Wang S, Zhu, X, Huang J (2016) Imaging biomarker discovery for lung cancer survival prediction. In: Proceedings of the medical image computing and computer-assisted intervention. Lecture Notes in Computer Science, 9901, pp 649–657. https://doi.org/10.1007/978-3-319-46723-8_75
Zhao J, Zhang M, Zhou Z, Chu J, Cao F (2017) Automatic detection and classification of leukocytes using convolutional neural networks. Med Biol Eng Comput 55(8):1287–1301. https://doi.org/10.1007/s11517-016-1590-x .
Zhang Q, Xiao Y, Dai W, Suo J, Wang C, Shi J, Zheng H (2020) Deep learning based classification of breast tumors with shear-wave elastography. Ultrasonics 72:150–157. https://doi.org/10.1016/j.ultras.2016.08.004
Zhang H, Li L, Qiao K, Wang L, Yan B, Li L, Hu G (2016) Image prediction for limited-angle tomography via deep learning with convolutional neural network. arxiv: 1607.08707
Zeiler MD, Fergus R (2014) Visualizing and understanding convolutional networks. In: Proceedings of the European conference on computer vision, pp 818–833
Yu L, Yang X, Chen H, Qin J, Heng P (2017) Volumetric convnets with mixed residual connections for automated prostate segmentation from 3D MR images. In: Proceedings of the thirty-first AAAI conference on artificial intelligence, pp 66–72
Download references
Author information
Authors and affiliations.
Department of ECE, KPR Institute of Engineering and Technology, Coimbatore, India
S. Suganyadevi & V. Seethalakshmi
Department of IT, Dr. Mahalingam College of Engineering and Technology, Coimbatore, India
K. Balasamy
You can also search for this author in PubMed Google Scholar
Corresponding author
Correspondence to S. Suganyadevi .
Additional information
Publisher's note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Reprints and permissions
About this article
Suganyadevi, S., Seethalakshmi, V. & Balasamy, K. A review on deep learning in medical image analysis. Int J Multimed Info Retr 11 , 19–38 (2022). https://doi.org/10.1007/s13735-021-00218-1
Download citation
Received : 02 April 2021
Revised : 06 August 2021
Accepted : 09 August 2021
Published : 04 September 2021
Issue Date : March 2022
DOI : https://doi.org/10.1007/s13735-021-00218-1
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Deep learning
- Image classes
- Find a journal
- Publish with us
- Track your research
Reviewing Machine Learning and Image Processing Based Decision-Making Systems for Breast Cancer Imaging
Affiliations.
- 1 Modeling, Simulation and Data Analysis, Mohammed VI Polytechnic University, Benguerir, Morocco.
- 2 Modeling, Simulation and Data Analysis, Mohammed VI Polytechnic University, Benguerir, Morocco. [email protected].
- 3 Software Project Management Research Team, ENSIAS, Mohammed V University in Rabat, Rabat, Morocco. [email protected].
- PMID: 33404910
- DOI: 10.1007/s10916-020-01689-1
Breast cancer (BC) is the leading cause of death among women worldwide. It affects in general women older than 40 years old. Medical images analysis is one of the most promising research areas since it provides facilities for diagnosis and decision-making of several diseases such as BC. This paper conducts a Structured Literature Review (SLR) of the use of Machine Learning (ML) and Image Processing (IP) techniques to deal with BC imaging. A set of 530 papers published between 2000 and August 2019 were selected and analyzed according to ten criteria: year and publication channel, empirical type, research type, medical task, machine learning techniques, datasets used, validation methods, performance measures and image processing techniques which include image pre-processing, segmentation, feature extraction and feature selection. Results showed that diagnosis was the most used medical task and that Deep Learning techniques (DL) were largely used to perform classification. Furthermore, we found out that classification was the most ML objective investigated followed by prediction and clustering. Most of the selected studies used Mammograms as imaging modalities rather than Ultrasound or Magnetic Resonance Imaging with the use of public or private datasets with MIAS as the most frequently investigated public dataset. As for image processing techniques, the majority of the selected studies pre-process their input images by reducing the noise and normalizing the colors, and some of them use segmentation to extract the region of interest with the thresholding method. For feature extraction, we note that researchers extracted the relevant features using classical feature extraction techniques (e.g. Texture features, Shape features, etc.) or DL techniques (e. g. VGG16, VGG19, ResNet, etc.), and finally few papers used feature selection techniques in particular the filter methods.
Keywords: Breast Cancer. Machine learning. Image processing. Structured literature review. Deep learning.
- Breast* / diagnostic imaging
- Image Processing, Computer-Assisted*
- Machine Learning
- Magnetic Resonance Imaging
- Mammography

We apologize for the inconvenience...
To ensure we keep this website safe, please can you confirm you are a human by ticking the box below.
If you are unable to complete the above request please contact us using the below link, providing a screenshot of your experience.
https://ioppublishing.org/contacts/
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- Biomed Res Int

Review of Breast Cancer Pathologigcal Image Processing
Ya-nan zhang.
1 School of Computer Science and Technology, Harbin University of Science and Technology, Harbin 150080, China
2 HRG International Institute (Hefei) of Research and Innovation, Hefei 230000, China
Chang-yi LI
Breast cancer is one of the most common malignancies. Pathological image processing of breast has become an important means for early diagnosis of breast cancer. Using medical image processing to assist doctors to detect potential breast cancer as early as possible has always been a hot topic in the field of medical image diagnosis. In this paper, a breast cancer recognition method based on image processing is systematically expounded from four aspects: breast cancer detection, image segmentation, image registration, and image fusion. The achievements and application scope of supervised learning, unsupervised learning, deep learning, CNN, and so on in breast cancer examination are expounded. The prospect of unsupervised learning and transfer learning for breast cancer diagnosis is prospected. Finally, the privacy protection of breast cancer patients is put forward.
1. Introduction
Breast cancer is one of the most common malignant tumors. According to Chinese Women's survey, breast cancer is the most common malignant tumor in Chinese women, and the incidence rate is increasing year by year. The key to reduce the mortality of breast cancer is early diagnosis and treatment. At present, mammography is the most commonly used method to detect breast cancer. However, because of the huge amount of data and the poor imaging features of early breast cancer, early diagnosis is very difficult. With the development of image processing technology and early diagnosis technology, image processing of breast pathology has become an important way of early diagnosis of breast cancer, which mainly includes the study of masses, calcifications, and breast density. One of the main manifestations of breast cancer in breast mammography is mass. The basic steps of pathological image processing are as follows: first is image preprocessing, which includes removing background, marker, pectoral muscle and noise, and breast segmentation and image enhancement. Secondly, the region of interest is found by a basic image processing method. Then, features that can represent the quality, such as texture features and morphological features, are extracted. Finally, the tumor and normal tissue were separated according to the extracted features. Another manifestation of breast cancer on X-ray images is a large breast density [ 1 ].
The object of breast pathology image processing is a variety of medical images with different imaging mechanisms. The types of medical imaging widely used in clinic include X-ray imaging (X-CT), magnetic resonance imaging (MRI), nuclear medicine imaging (NMI), and ultrasonic imaging (UI). X-ray imaging (X-CT), mainly through X-ray tomography, such as head tomography, is used to diagnose cerebral vascular diseases and intracranial hemorrhage. X-ray tomography has a good effect in the diagnosis of traumatic skull and facial fractures. Magnetic resonance imaging (MRI) is a noninvasive imaging technique, which can produce three-dimensional anatomical images [ 2 ]. Nuclear medicine imaging (NMI) is based on the difference of radioactivity concentration inside and outside organs or between different parts of organs [ 3 , 4 ]. UI was used to observe the shape, location, size, number and scope of the mass, and the activity of abdominal organs. The edge echo, capsule, smoothness, wall thickness, and halo were observed. According to the clinical data, the symptoms of breast cancer can be adjusted by means of X-ray examination, which is identified by X-ray mammography. MRI results of tumor detection are higher than the actual value, while CT results of tumor detection are lower than the actual value. Dynamic enhanced CT examination of breast lesions can better overcome the situation of missed detection.
Medical image acquisition and interpretation are the basis of breast cancer diagnosis based on medical imaging. In recent years, the speed and resolution of image acquisition have been greatly improved. However, image diagnosis is limited by the doctor's experience, ability, and other subjective factors, and its ability of replication and promotion is limited. In order to minimize the dependence on doctors, image processing technology is applied to medical imaging processing. Medical image processing includes lesion detection, image segmentation, image registration, and image fusion. In addition to clinical diagnosis, medical image processing also plays an important auxiliary role in medical teaching, operation planning, operation simulation, and various medical researches [ 5 , 6 ].
2. Detection of Breast Cancer
Detection of breast cancer is mainly based on detection methods, image processing of lesion detection, lesion location matching, and extraction of lesion feature values. Breast cancer detection can detect candidate lesion location by supervised learning or classical image processing. The combination of image processing and Convolutional Neural Networks (CNN) is one of the successful examples of deep learning in recent years. The CNN is applied in breast cancer image analysis, mapping the input layer, pool layer, modified linear unit, all link layer, and output layer, respectively, and predicting the information represented by medical images. For example, Setio et al. extracted the features of pulmonary nodules in nine different directions of 3D chest CT scanning, selected the appropriate candidate as the center, and classified the candidates through CNN [ 3 , 7 – 11 ]. Ross et al. decomposed the 3D image into 2D patches and then rotated the 2D patches randomly to get the “2.5D” view. The CNN was used to detect the early features of cancer from the 2.5D view. The combination of deep learning and image processing greatly improves the accuracy of lesion detection [ 12 ], while it is difficult to achieve high accuracy by using nondeep learning classifiers such as support vector machine. The accuracy of CNN algorithm depends on the training of initial markers by experts and needs a wide range of case coverage. Therefore, the promotion of CNN in the field of medical image processing is constrained by resources “transfer learning” that can reduce the dependence of CNN on initial marker training to a certain extent [ 13 , 14 ], but the application of transfer learning itself is limited, so it is difficult to find the application conditions of transfer learning between medical images of human organs.
Early diagnosis of breast cancer can be detected by tumor markers. Tumor markers are substances produced and secreted by tumor cells during growth and reproduction. When these substances reach a certain amount, they can be extracted from breast images. The early feature values of breast cancer can be identified by using SIFT (scale invariant feature transform) or HOG (Histogram of Oriented Gradient) and so on to provide support for the early diagnosis of breast cancer. The combination of image processing technology and reinforcement learning technology can reduce the dependence on a human doctor's experience to the greatest extent. Image processing technology is used to process two-dimensional slices, and then reinforcement learning is combined to set the enhancement target. Through the judgment of each discrete two-dimensional slice, the optimal decision strategy is found, so as to maximize the benefit of judging the pathological correctness of the whole group of two-dimensional slices. Through the analysis and processing of two-dimensional slice image, the segmentation, extraction, three-dimensional reconstruction, and three-dimensional display of human breast, surrounding soft tissue and lesion are realized. After the calibration of features, the reinforcement learning is used to quantitatively analyze the lesion and the region around the breast. Combined with the revenue target, the learning is carried out through continuous attempts. The goal is to obtain the maximum revenue value. Based on the reinforcement learning method, it is found that breast cancer does not need to know how to produce correct breast cancer recognition action. Reinforcement learning relies on its own learning, constantly trying and making mistakes and constantly recording the maximum value of income in the process of trial and error until the method of finding the maximum value of income is found.
3. Breast Cancer Image Segmentation
Based on the given feature factors, the medical image segmentation compares the similarity of feature factors between images and divides the image into several regions. The objects of medical image segmentation mainly include cells, tissues, and organs. The region-based segmentation method is mainly based on the spatial local features of the image, such as gray, texture, and other pixel statistical characteristics. The boundary-based segmentation method mainly uses gradient information to determine the boundary of the target. For example, the fast marching algorithm and the medical image segmentation method of watered transform can segment the image quickly and accurately [ 15 ].
In recent years, with the development of other emerging disciplines, image segmentation technology develops rapidly, and new methods generated by interdisciplinary emerge in endlessly. Some new image segmentation techniques have been developed for breast cancer detection, such as the method based on statistics, the method based on fuzzy theory, the method based on neural network, the method based on wavelet analysis, the model-based snake model (dynamic contour model), and the combination optimization model. Although new segmentation methods have been proposed, the results are not ideal. At present, the research focus is a knowledge-based segmentation method; that is, some prior knowledge is introduced into the segmentation process by some means, so as to constrain the computer segmentation process, so that the segmentation results can be controlled within the range we can understand without going too far [ 16 ]. For example, when the gray value of the tumor in the liver is very different from that of the normal liver, the tumor and the normal liver will not be regarded as two independent tissues. All of the above methods have their own limitations. The results are good in specific scenarios, but not ideal beyond specific scenarios. Because the boundaries of internal organs, muscles, blood vessels, and other organs are usually very complex, the diseased areas of organs are mixed with normal tissues, and the differences between the gray levels and boundaries of diseased areas and normal tissues are relatively small, it is very difficult to identify these organs and diseased areas in medical images, and the existing image segmentation algorithms can not complete the task of image segmentation independently. Human intervention is also needed [ 17 ]. Medical image segmentation is quite different from image segmentation in other fields. The effect of existing classical algorithms in medical image segmentation is not good. It is still necessary to continue to study in improving the accuracy, speed, adaptability, and robustness of image segmentation [ 18 ]. The image segmentation method based on prior knowledge can well control the segmentation boundary of the image. For example, in the segmentation of intrahepatic mass, the image segmentation method based on prior knowledge can recognize intrahepatic mass and normal liver by gray value. However, image segmentation based on prior knowledge requires a large number of prior data. The more prior data, the more accurate the results. For example, Ghesu et al., based on 2891 times of cardiac ultrasound data, used deep learning and edge space learning to detect and segment cardiac ultrasound images [ 17 , 18 ]. Parameter space exploration and data sparsity are important factors to improve the efficiency of medical image segmentation. Brosch et al. proposed a 3D deep convolution coder network through convolution and deconvolution to segment multiple sclerosis brain lesions and normal brain regions [ 19 ]. Data normalization and data enhancement techniques are applied to image enhancement and core regions of suspected tumors in brain tumor segmentation research and achieved good results [ 20 ]. The research of medical image segmentation methods has the following remarkable characteristics: it is difficult for any single existing image segmentation algorithm to achieve satisfactory results for general images, so more attention should be paid to the effective combination of multiple segmentation algorithms. Due to the complexity of human anatomical structure and the systematicness of function, although there have been studies on the methods of automatic segmentation of medical images to distinguish the required organs and tissues or find the lesion area, the existing software packages generally can not complete the automatic segmentation, and the manual intervention of anatomy is still needed [ 21 ]. At present, it is impossible for computer to complete the task of image segmentation, so the human-computer interactive segmentation method has gradually become the focus of research. The research of new segmentation methods mainly focuses on automatic, accurate, fast, adaptive, and robust features. The comprehensive utilization of classical segmentation technology and modern segmentation technology is the development direction of medical image segmentation technology in the future [ 22 , 23 ].
4. Breast Cancer Image Registration
Image registration is the first mock exam of image fusion. In breast cancer clinical diagnosis, multiple modes or modes of image registration and fusion are needed. More information can help doctors give more accurate diagnosis [ 24 ]. In the clinical diagnosis process of breast cancer, medical image registration mainly locates reference points in two or more images; through spatial location transformation, such as rotation, the reference point is located in a coordinate system. Registration requires that the mapping of points between images is one-to-one correspondence; that is, each point in an image space has corresponding points in another image space, or in the sense of medical diagnosis, the points in the image can be accurately or approximately accurately corresponded [ 25 – 27 ]. Registration can be divided into two types: based on external features and based on internal features. Registration based on image internal features is noninvasive and traceable, which is the focus of research on registration algorithm [ 28 ].
There are two major categories of medical registration research for breast cancer: (1) a deep learning network is used to estimate the similarity of two images and drive iterative optimization, and (2) the depth regression network is directly used to predict the conversion parameters. The former only uses deep learning for similarity measurement and still needs the traditional registration method for iterative optimization. It does not give full play to the advantages of deep learning, takes a long time, and is difficult to achieve real-time registration. Therefore, only the latter is studied and discussed, and the conclusion is limited to this kind of nonrigid registration method. Based on supervised learning, there are two ways to obtain tags: (1) the traditional classical registration method is used for registration, and the deformation field is used as tags. (2) The original image is simulated as a fixed image, the deformed image as a moving image, and the simulated deformation field as a label. Based on unsupervised learning, the registration pair is input into the network to obtain the deformation field, and the moving image is interpolated to obtain the registration image. The 3D image is similar to it. The 3D image is input into the network to obtain the deformation field (dx, dy, dz), and then, the registration image is obtained by interpolation. However, the medical image registration of breast cancer is still an unsolved classic problem. There are no universally recognized gold standard and no corresponding large database in this field. Deep learning methods have some successful cases in breast cancer image registration. There are usually several reasons: (1) the expert knowledge of the field is well utilized, (2) the data are properly preprocessed and processed by data enhancement, (3) a special network structure is designed for a single task, and (4) the appropriate super parameter optimization method is used: such as parameter adjustment based on intuition or the Bayesian method. There are still some difficulties and challenges in the field of breast cancer medical image registration: (1) there is a lack of large databases with precise annotation. (2) Specific tasks need guidance from experts in the field. (3) It is difficult to agree with the opinions of different experts in some ambiguous images. (4) The two classification models are too simple to be competent for more complex cases. (5) the difficulties of breast cancer medical image analysis still exist in images. Besides the analysis, we need to make full use of the information about other dimensions of the patient, like age, medical history, etc. (6) The slice-based neural network is having difficulty in using the location information of the corresponding anatomical structure in the original image, but the method of transferring the whole image into the neural network has corresponding disadvantages. Although Esteva et al. made amazing progress in dermatology in 2017 and Gulshan et al. in ophthalmology in 2016, they have achieved image classification models with higher accuracy than human experts in both fields. However, the essential reason for its success is that the above two problems are simple enough, and the data volume of ImageNet dataset is very large, so the existing model is applied to the above two problems and has achieved very good results. Usually, there is no such simple structure, and there is no effective network structure that can be used for 3D gray or multichannel image preprocessing. The advantage of unsupervised learning over supervised learning is that unsupervised learning does not need a large number of precisely labeled data. In addition, unsupervised learning imitates the way of human learning and can automatically perform the required tasks without special labeling or can deal with a large number of classification problems with little supervision. At present, the main methods of unsupervised learning are self-coding VAE and counter neural network Gan. Compared with accurately labeled breast cancer medical images, unlabeled breast cancer medical image data are easier to obtain. Unsupervised learning can directly use standard breast cancer medical images without any supervision, so it has more advantages. Finally, because deep learning is similar to black box model, the interpretation of breast cancer medical image domain requires a higher model, and further work is needed. At present, some work includes introducing Bayesian statistics into deep learning, which will help to measure the uncertainty of prediction.
Multimodality medical image registration is a new direction of breast cancer registration, such as nonrigid multimodal medical image registration based on structure representation of PCANet [ 29 ]. PCANet can automatically learn intrinsic features from a large number of medical images through multilevel linear and nonlinear transformation, which has better information entropy than the artificial feature extraction method. Multilevel image features extracted from each layer of PCANet can effectively represent multimodal images. The fusion of medical image registration technology and informatics theory opens a new idea for medical image registration of breast cancer. For example, the principle of maximum information entropy is applied to image registration, which can maximize the diversity of information and retain the main information without neglecting the secondary information [ 30 – 35 ]. Three-dimensional multimode image registration is a new direction of medical image registration. It has more information than two-dimensional image and can support a doctor's diagnosis more effectively. In addition, some new image registration algorithms, such as image recognition of breast cancer based on topology, feature points are extracted from existing breast cancer images. They are combined into a matching area with a certain topological structure as the matching template. In the breast images to be matched, regions with similar topological structures are found; these regions may be breast cancer. The main steps of image recognition of breast cancer based on topology are as follows: (1) the first one is extracting feature points or feature regions of a specific scale and combining them into topological templates. (2) The topology of the image to be matched is extracted. (3) By comparing the topology in the image to be matched with the topology template, regions with similar topology are found. (4) The similarity between similar topology and feature points in topology template is compared, and the product of topology similarity and feature point similarity is regarded as the final similarity. The schematic diagram of image recognition based on topology structure is shown in Figure 1 . In Figure 1 , the left dotted line is the topology template, the middle dotted line is the topology extracted from the image to be matched, and the right dotted line is the region with similar topology [ 36 ].

Image recognition sketch based on topological structure.
Other methods such as algorithms based on wavelet transform, statistical parametric mapping algorithm, and genetic algorithm are also continuously integrated into breast cancer image registration. The combination of multiobjective optimization, reinforcement learning, and other methods with medical image registration is the future development direction of medical image registration.
5. Breast Cancer Image Fusion
Breast cancer image fusion extracts useful information from multiple images, filters redundant information, and improves the medical value of images. In general, image fusion from low to high is signal level fusion, data level fusion, feature level fusion, and decision level fusion.
- Signal level: at the lowest level, the unprocessed sensor output is mixed in the signal domain to produce a fused signal. The fused signal has the same form as the source signal, but its quality is better. The signal from the sensor can be modeled as random variables mixed with different correlated noises. In this case, fusion can be considered an estimation process, and signal level image fusion is the optimal concentration or distribution detection problem of signal to a large extent, which requires the highest registration in time and space.
- Pixel level: pixel level image fusion is the most basic fusion of the three levels. After pixel level image fusion, the obtained image has more detailed information, such as edge and texture extraction, which is conducive to the further analysis, processing, and understanding of the image. It can also expose the potential target, which is conducive to the operation of judging and identifying the potential target pixels. This method can save as much information as possible in the source image and increase the content and details of the fused image [ 37 ]. This advantage is unique and only exists in pixel level fusion. However, the limitations of pixel level image fusion can not be ignored, because it is to operate on pixels, so the computer has to process a large number of data, and the processing time will be relatively long, so the fused image can not be displayed in time and real-time processing can not be realized. In addition, in data communication, the amount of information is large, and it is easy to be affected by noise. In addition, if you directly participate in image fusion without strict image registration, the fused image will be blurred, and the target and details are not clear and accurate.
- Feature level: feature level image fusion is to extract the feature information from the source image. The feature information is the information of the target or the region of interest in the source image, such as edge, person, building, or vehicle. Then, the feature information is analyzed, processed, and integrated to get the fused image features. The accuracy of target recognition based on fused features is obviously higher than that of the original image. The image information is compressed by feature level fusion and then analyzed and processed by computer. Compared with pixel level, the memory and time consumed will be reduced, and the real-time performance of the required image will be improved. Feature level image fusion requires less accuracy of image matching than the first layer, and its computing speed is faster than the first layer. However, it extracts image features as fusion information, so it will lose a lot of detail features.
- Decision level: decision level image fusion is a cognitive-based method, which is not only the highest level of image fusion method but also the highest level of abstraction. Decision level image fusion is targeted [ 38 ]. According to the specific requirements of the problem, the feature information obtained from the feature level image is used, and then, the optimal decision is made directly according to certain criteria and the credibility of each decision, that is, the probability of the existence of the target. Among the three fusion levels, the calculation of decision level image fusion is the smallest, but this method has a strong dependence on the previous level, and the image is not very clear compared with the former two fusion methods. It is difficult to realize the decision level image fusion, but the noise has the least influence on the image transmission.
To sum up, data level fusion is the process of directly processing the collected data to obtain the fused image, which is the basis of high-level image fusion. Feature level fusion preserves the information contained in different images. Decision level fusion is the highest level of image fusion based on subjective needs. In breast medical image fusion, data level fusion is the main method. For example, multimodality medical image fusion is a technology that integrates multiple dimensions information. It can provide more comprehensive and accurate information for clinical detection of breast cancer [ 39 ]. The steps of image fusion are mainly divided into image data fusion and fusion image display. At present, the data fusion of breast image is mainly based on pixels, which process the image point by point and sum the gray values of the corresponding pixels of the two images. However, the image will be blurred to a certain extent after using this method. The fusion method based on breast image features needs to extract image features and do target segmentation and other processing on the image. The display of fusion image includes pseudocolor display, tomographic display, and three-dimensional display. Pseudocolor display takes an image as a benchmark and superimposes the gray and contrast features of the image to be fused with the benchmark image. The tomographic display method can display the fused three-dimensional data synchronously in cross-sectional, coronal, and sagittal images, which are convenient for the observer to diagnose. The three-dimensional display method, namely, three-dimensional reconstruction, is to display the fused breast data in the form of three-dimensional images, which can more intuitively observe the spatial anatomical position of the lesions. The earliest method of 3D reconstruction is back projection. At present, there are two common reconstruction methods: filtered back projection and convolution back projection. The information content of three-dimensional image is large, and the future three-dimensional image fusion technology will be a focus of image fusion research. With the development of interdisciplinary research, new image fusion methods are emerging. The application of wavelet transform, nonlinear registration based on finite element analysis, and artificial intelligence technology in breast image fusion will be the focus of image fusion research.
6. Forecast and Challenge
- The application of unsupervised and supervised learning in the field of breast cancer image processing: breast cancer image classification based on CNN is the mainstream classification method nowadays. Fine tuning of CNN parameters directly affects the final image processing results. Unsupervised learning will have more promising results in breast cancer image processing
- Transfer learning and fine tuning in the application of breast cancer image processing: transfer learning can partially alleviate the plight of not enough annotation data. In the process of transfer learning, a better plan is to use pretrained CNN as initialization of network and then carry out further supervision training. The annotated dataset is still a challenge in the field of breast cancer image processing
- Patients pay more and more attention to privacy protection. The privacy of breast cancer image data has attracted much attention. In the process of breast cancer image processing, it is necessary to improve the accuracy of recognition, help doctors to give diagnosis, and pay attention to the protection of patients' privacy, protect the original information of the image from unauthorized access, and diagnose the whole process of image information authorized access and make the image access trace checkable
7. Conclusion
Deep learning and reinforcement learning are the relatively close combination of machine learning algorithm and breast cancer image processing and have made considerable progress. There are obvious differences between breast cancer image and other fields such as noise reduction, grayscale transformation, target segmentation, and feature extraction. The traditional image processing method is not directly applied to breast cancer image processing. Accumulation of deep learning in image processing can not be directly transferred to breast cancer image processing. Reinforcement learning belongs to unsupervised learning, which is different from deep learning. It uses an incentive mechanism that does not need a large number of sample space. Compared with deep learning, reinforcement learning has a wider applicability and lower promotion cost. Moreover, reinforcement learning has achieved very good results in chess, man-machine game, and other fields, which is suitable for complex logic processing. The combination of reinforcement learning and medical image processing will play a greater role in the clinical detection and prediction of breast cancer.
Conflicts of Interest
The authors declare that they have no conflicts of interest.

IMAGES
VIDEO
COMMENTS
Abstract. Image Processing includes changing the nature of an image in order to improve its pictorial information for human interpretation, for autonomous machine perception. Digital image ...
Within the domain of image processing, a wide array of methodologies is dedicated to tasks including denoising, enhancement, segmentation, feature extraction, and classification. These techniques collectively address the challenges and opportunities posed by different aspects of image analysis and manipulation, enabling applications across various fields. Each of these methodologies ...
Significant evolution in deep learning took place in 2010, when software developers started using graphical processing units for general-purpose applications. From that date, the deep neural network (DNN) started progressive steps across different applications ranging from natural language processing to hyperspectral image processing. The convolutional neural network (CNN) mostly triggers the ...
With the recent advances in digital technology, there is an eminent integration of ML and image processing to help resolve complex problems. In this special issue, we received six interesting papers covering the following topics: image prediction, image segmentation, clustering, compressed sensing, variational learning, and dynamic light coding.
The literature review is conducted to synthesize and summarize existing research on image restoration. An overview of algorithms and methods is given. ... Image processing techniques use algorithms to process digital images. Machine learning and deep learning techniques consists of a learner that learns from data and then applies this knowledge ...
Machine learning, particularly deep learning (DL), has become a central and state-of-the-art method for several computer vision applications and remote sensing (RS) image processing. Researchers are continually trying to improve the performance of the DL methods by developing new architectural designs of the networks and/or developing new techniques, such as attention mechanisms. Since the ...
Systematic reviews start by defining a review protocol that considers the research objective being addressed and the methods to perform the study [34].In this research, we conducted a SLR to investigate the published studies in the context of COVID-19 and image processing, as a potential approach to disease diagnosis.
The main goal of this study is to present a literature review of the deployed methods of medical image processing in the context of the COVID-19 crisis. With this in mind, the studies available in reliable databases were retrieved, studied, evaluated, and synthesized. Based on the in-depth review of literature, this study structured a ...
Our systematic literature review collected many studies related to our search topic over the last five years, between 2017 and 2021, from the following databases: Elsevier, IEEE Xplorer, Springer, and Google Scholar. ... A Review of Image Processing Techniques Common in Human and Plant Disease Diagnosis. Symmetry. 2018; 10:270. doi: 10.3390 ...
In this article, we have summarized a brief review of recent research related to the underwater image processing and analysis. Previously representative methods are divided into seven distinct categories: Underwater image, enhancement, noise reduction, defogging, segmentation, saliency detection, color constancy and recovery/restoration.
This literature review suggests that designing a suitable image‐processing procedure is a prerequisite for a successful classification of remotely sensed data into a thematic map. Effective use of multiple features of remotely sensed data and the selection of a suitable classification method are especially significant for improving ...
All deep learning applications and related artificial intelligence (AI) models, clinical information, and picture investigation may have the most potential element for making a positive, enduring effect on human lives in a moderately short measure of time [].The computer processing and analysis of medical images involve image retrieval, image creation, image analysis, and image-based ...
Medical image processing and analysis techniques play a significant role in diagnosing diseases. Thus, during the last decade, several noteworthy improvements in medical diagnostics have been made based on medical image processing techniques. In this article, we reviewed articles published in the most important journals and conferences that used or proposed medical image analysis techniques to ...
Research protocol. Systematic reviews start by defining a review protocol that considers the research objective being addressed and the methods to perform the study [].In this research, we conducted a SLR to investigate the published studies in the context of COVID-19 and image processing, as a potential approach to disease diagnosis.
Medical image processing had grown to include computer vision, pattern recognition, image mining, and also machine learning in several directions [ 3 ]. Deep learning is one methodology that is commonly used to provide the accuracy of the aft state. This opened new doors for medical image analysis [ 4 ].
Reviewing Machine Learning and Image Processing Based Decision-Making Systems for Breast Cancer Imaging J Med Syst. 2021 Jan 4;45(1):8. doi: 10.1007/s10916-020-01689-1. ... This paper conducts a Structured Literature Review (SLR) of the use of Machine Learning (ML) and Image Processing (IP) techniques to deal with BC imaging. ...
Abstract. The term glaucoma refers to a group of heterogeneous diseases that cause the degeneration of retinal ganglion cells (RGCs). The degeneration of RGCs leads to two main issues: (i) structural changes to the optic nerve head as well as the nerve fiber layer, and (ii) simultaneous functional failure of the visual field.
4.2. Deepfake Image Detection. Unlike the detection of video deepfakes, which are sequences of images, deepfake image detection focuses on identifying a single image as a deepfake, as shown in Figure 9. For example, the study described in [ 91] proposed a system to detect deepfake human faces.
The objective is to study different methodologies of object counting and identify future research directions and to review the literature based on object counting problem using image processing. Object counting is a challenging problem in image processing. It is routinely carried out in different areas of industries, research institutes, laboratories, agriculture industries among others ...
Deep learning is used to address a wide range of challenging issues including large data analysis, image processing, object detection, and autonomous control. In the same way, deep learning techniques are also used to develop software and techniques that pose a danger to privacy, democracy, and national security. Fake content in the form of images and videos using digital manipulation with ...
The proposed image processing model first takes a fresh RGB image as an input which is collected from clinical laboratory. Thereafter, we convert the image from RGB to Gray and keep pixel values greater than 20 (threshold value) in order to binarized the gray scale image. From the literature review, we observed that the process of binarization ...
Pathological image processing of breast has become an important means for early diagnosis of breast cancer. Using medical image processing to assist doctors to detect potential breast cancer as early as possible has always been a hot topic in the field of medical image diagnosis. In this paper, a breast cancer recognition method based on image ...