An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- Wiley Open Access Collection


A guide to genome‐wide association analysis and post‐analytic interrogation
1 Department of Mathematics and Statistics, Mount Holyoke College, South Hadley, MA, U.S.A.
2 Department of Computer Science, University of Massachusetts, Amherst, MA, U.S.A.
3 Department of Biostatistics and Epidemiology, University of Massachusetts, Amherst, MA, U.S.A.
Muredach P. Reilly
4 Department of Medicine, University of Pennsylvania, Philadelphia, PA, U.S.A.
Andrea S. Foulkes
Associated data.
This tutorial is a learning resource that outlines the basic process and provides specific software tools for implementing a complete genome‐wide association analysis. Approaches to post‐analytic visualization and interrogation of potentially novel findings are also presented. Applications are illustrated using the free and open‐source R statistical computing and graphics software environment, Bioconductor software for bioinformatics and the UCSC Genome Browser. Complete genome‐wide association data on 1401 individuals across 861,473 typed single nucleotide polymorphisms from the PennCATH study of coronary artery disease are used for illustration. All data and code, as well as additional instructional resources, are publicly available through the Open Resources in Statistical Genomics project: http://www.stat-gen.org . © 2015 The Authors. Statistics in Medicine Published by John Wiley & Sons Ltd.
1. Introduction
This brief tutorial is intended as a learning and teaching tool, offering the fundamental computational skills for entry into the field of applied statistical genetics and bioinformatics. Unique from existing resources, this tutorial offers the framework as well as complete and extensible R scripts to perform a comprehensive genome‐wide association (GWA) analysis and post‐analytic interrogation. Familiarity at an elementary textbook level 1 , 2 , 3 with basic statistical and genetics concepts for GWA studies is assumed. The content of this tutorial builds and expands on several existing and highly recommended resources on genetic and statistical concepts, including 4 , 5 . We begin the analysis after genotyping calls are made and quality control and assurance measures are taken, as described, for example, in 6 , 7 . An alternative freely available software platform is plink , another toolset used for whole genome association analysis. Additional post‐analytic interrogation is also presented in this manuscript using the UCSC Genome Browser. A companion website is available for this tutorial through the Open Resources in Statistical Genomics (ORSG) project ( http://www.stat-gen.org ) with all R coding examples fully embedded in.Rmd files to be edited and weaved as dynamic and reproducible reports. A complete list of external resources is provided in Supplementary Information A.
The focus of this tutorial is on GWA analysis of common variants that involves testing association of each single nucleotide polymorphism (SNP) independently and subsequently characterizing findings through a variety of visual and analytic tools. In the rare variant setting, in which interest resides in investigating variations that are present in less than 1 % of the population, alternative techniques are needed that account for regional associations. The reader is referred to a rich literature that addresses rare variant analysis, including 8 , 9 , 10 . In the present manuscript, we focus on the analysis of data arising from population‐based GWA studies of unrelated individuals where primary interest resides in identifying associations between SNPs and a single binary, for example, case or control status or quantitative phenotype. Extensive methods and tools specific to family‐based investigations that account for within‐family correlation structures are also available (e.g., 11 , 12 ). Further extensions of the tools presented to censored survival or longitudinal outcomes can be achieved through application of an alternative modeling framework in the association analysis of step 7. The data used for illustration here are limited to the 22 autosomal chromosomes, and both typed and 1000 Genomes 13 imputed SNPs are considered as potential predictor variables. Post‐analytic interrogation of SNP‐level findings is an essential part of GWA analysis, and first steps, including mapping positive SNP findings to gene regions, are described herein. We note that there exists a large literature on alternative analytical paradigms for simultaneous analysis of multiple SNPs, including methods for gene‐based (e.g., 14 , 15 , 16 ) and pathway‐based (e.g., 17 , 18 ) analysis, as well as growing literature on gene–environment interaction analysis in the context of GWA studies 19 .
The PennCATH cohort data, arising from a GWA study of coronary artery disease (CAD) and cardiovascular risk factors based at University of Pennsylvania Medical Center 20 , are used throughout this tutorial as an illustrative example and have been made publicly available for training use to accompany the tutorial. In this study, a total of n = 3850 individuals were recruited between July 1998 and March 2003. A nested case‐control study of European ancestry severe angiographic CAD cases and angiographic normal controls were selected for genome‐wide genotyping. De‐identified data used in this tutorial are composed of n = 1401 individuals with genotype information across 861,473 SNPs. Corresponding clinical data, including age, sex, high‐density lipoprotein (HDL)‐cholesterol, low‐density lipoprotein cholesterol, triglycerides, and CAD status are available as well. HDL‐cholesterol, low‐density lipoprotein cholesterol and triglycerides are all quantitative traits that are well‐described cardiovascular disease risk factors. Notably, PennCATH is one of the core GWA studies nested within the Coronary ARtery DIsease Genome‐wide Replication And Meta‐analysis (CARDIoGRAM) consortium meta‐data and serves as a representative regional population with no admixture 20 , 21 .
Genome‐wide association analysis strategies typically include four broadly defined components: (i) data pre‐processing; (ii) new data generation; (iii) statistical analysis; and (iv) post‐analytic interrogation. A primary goal of these investigations is identifying and characterizing the association among SNPs and measures of disease progression or disease outcomes. In Sections 2 – 5 in the succeeding paragraphs, we present the key aspects of each of the core analytic components, including a description of attributes of the data, application of relevant software tools, and guidance on interpretation of findings. An overall summary of the analytic approach we follow is provided in Figure 1 . Notably, this figure highlights multiple stages within each of the four broadly defined components of analysis. The resultant ten steps are as follows: (1) reading data into R to create an R object; (2) SNP‐level filtering (part 1); (3) sample‐level filtering; (4) SNP‐level filtering (part 2); (5) principal component analysis (PCA); (6) imputation of non‐typed genotypes; (7) association analysis of typed SNPs; (8) association analysis of imputed data; (9) integration of imputed and typed SNP results; and (10) visualization and quality control of association findings. Further data interrogation using external resources is also discussed. In the following sections, we elaborate on each of these steps. Notably, this workflow is typical for analysis of a single GWA study and may be modified in the context of a large collaborative meta‐analysis involving the combination of multiple studies requiring harmonization. Additional detail on the analysis pipeline in this context is provided in Section 6 where we also present a broader contemporary context and additional available resources.

Genome‐wide association (GWA) analysis workflow. GWA analysis is composed of 10 essential steps that fall into four broadly defined categories as illustrated in this figure. Additional detail on the structure of the data files, particularly the relationship of the.ped and.map files with the.bim,.bed, and.fam files, is provided in Figure 2 . This workflow is based on a single GWA analysis and may be modified in the context of a large collaborative meta‐analysis involving the combination of multiple GWA studies that require harmonization. Additional detail on typical modifications in this context is provided in Section 6 . *Substructure, also referred to as population admixture and population stratification, refers to the presence of genetic diversity (e.g., different allele frequencies) within an apparently homogenous population that is due to population genetic history (e.g., migration, selection, and/or ethnic integration).
2. Data pre‐processing
In the example we present, samples were genotyped using the Affymetrix 6.0 GeneChip and provided to us in.CEL format. The Birdseed calling algorithm, which is based on an expectation‐maximization type algorithm 20 , was applied to generate genotypes and confidence scores for each sample at every SNP. In turn, PERL and unix scripts were used to convert these to.ped and.map files. While R can read.ped and.map files, it is generally preferable to first convert these two files to.bim,.bed, and.fam files. This can be carried out using PLINK and is preferable as the conversion of the.ped file to a.bed file, a binary formatted file, results in a substantial reduction in file size. In the following texts, we describe the elements of each file type mentioned and their interrelatedness. A visual representation of the data files is provided in Figure 2 .
- .ped and.map files: The.ped file contains information on each study participant including family ID, participant ID, father ID, mother ID, sex, phenotype, and the full typed genotype. Here, each SNP is bi‐allelic (i.e., only two nucleotides are observed at any given SNP across study participants) and coded as a pair of nucleotides (A, C, T, or G). Notably, the ordering in the pair is non‐informative in the sense that the first alleles listed for each of the two SNPs are not necessarily on the same chromosome. The.map file contains a row for each SNP with rsNumber (SNP) and corresponding chromosome (chr) and coordinate (BPPos) based on the current genome build.
- .bim,.bed, and.fam files: The.bim file contains the same information as the.map file as well as the two observed alleles at each SNP (A1 and A2) from the.ped file. It contains a row for each SNP and six columns, containing information for the chromosome number, rsNumber, genetic distance, position identifier, allele 1, and allele 2. The.bed file contains a binary version of the genotype data. This is the largest of the three files because it contains every SNP in the study, as well as the genotype at this SNP for each individual. The.fam file contains the participant identification information, including a row for each individual and six columns, corresponding the same columns described for the.ped file with the exception of the genotype data. Note that not all of these columns contain unique information. That is, in a population‐based study of unrelated individuals, ‘family ID number’y and ‘individual ID number’ will be the same.
- Clinical data file: An additional ascii.txt or.csv file is typically available, which includes clinical data on each study subject. The rows of this file represent each subject, and the columns correspond to available covariates and phenotypes. There may be redundancies in this file and the data contained in the columns labeled ‘sex’ and ‘phenotype’ in the.fam file.

Genome‐wide association data files. GWA data files are typically organized into either.ped and.map files or.bim,.bed, and.fam files. Plink converts.ped and.map files into.bim,.bed, and.fam files. The later set is substantially smaller because the.bed file contains a binary version of the genotype data. R can read in either set of files although the later is preferable.
We begin by installing packages and setting up global parameters in R. This tutorial utilizes several packages available from Bioconductor, an open‐source bioinformatic software repository. Of these, we make the most use of snpStats , which includes functions to read in various formats of genotype data and carry out quality control, imputation, and association analysis. SNPRelate is also well utilized and includes functions for sample‐level quality control and computationally efficient principal component (PC) calculation. Other packages include functionalities for data visualization ( ggplot2 , LDheatmap , postgwas ), data manipulation ( plyr ), and parallel processing ( doParallel ), as well as their dependencies.

We additionally specify the parameters used in the data processing and analysis. Of particular note, we set the location of the GWA data set (available at https://www.mtholyoke.edu/courses/afoulkes/Data/GWAStutorial/ ) and specify input and output files.

2.1. Reading and formatting data in R (step 1)
In order to read the.fam,.bim, and.bed files in R, we use the read.plink() function in the Bioconductor snpStats package. The genotype slot of the resulting list contains an SnpMatrix object, which is a matrix of genotype data with a column for each SNP and a row for each study participant.

The clinical data ( GWAStutorial_clinical.csv ) can be read in the familiar way as a comma delimited text file.

Finally, we subset the data at this stage to include only individuals who have data available in both the genotype and phenotype files.

In the data example provided, genotype information is available for 861,473 typed SNPs across n = 1401 individuals with available phenotype data.
As illustrated in Figure 1 , once we have read in the the genotype and clinical information, we are ready to proceed with the next steps of the GWA data pre‐processing. This involves two stages of filtering the data, at SNP and sample levels, respectively. Each of these is described in more detail in the succeeding texts, accompanied by the appropriate R code for implementation. We note again that the order of analysis may vary depending on whether a single GWA analysis is being performed (as described herein) or the analyst is preparing results to be incorporated into a larger meta‐analysis that requires data harmonization across multiple studies. In the latter case, the following filtering steps (steps 2, 3, and 4) may be excluded or performed centrally after analysis (steps 7 and 8) as summary level data are combined across studies.
2.2. Single nucleotide polymorphism‐level filtering – part 1 (step 2)
The second data pre‐processing step involve removing (also referred to as ‘filtering’) SNPs that will not be included analysis. Well‐described reasons for this exclusion include large amounts of missing data, low variability, and genotyping errors (e.g., 22 ). Typically, SNP‐level filtering based on a large amount of missing data and lower variability is performed first. This is followed by sample‐level filtering (see step 3 in the succeeding texts), and finally, SNP‐level filtering based on possible genotyping errors (see step 4 in the succeeding texts) is performed. The rationale for this is that both sample‐level relatedness and substructure (for which we filter in step 3) can influence the Hardy–Weinberg equilibrium (HWE) criterion (step 4) used for filtering SNPs based on genotyping errors. An iterative procedure that repeats the SNP and sample‐level filtering until no additional samples are removed is also common. In our setting, however, no samples are filtered, deeming this loop unnecessary.
- SNP‐level filtering: call rate. The call rate for a given SNP is defined as the proportion of individuals in the study for which the corresponding SNP information is not missing. In the following example, we filter using a call rate of 95 % , meaning we retain SNPs for which there is less than 5 % missing data. More stringent cut points (e.g., less than 5 % ) may be employed in smaller sample settings.
- SNP‐level filtering: minor allele frequency (MAF). A large degree of homogeneity at a given SNP across study participants generally results in inadequate power to infer a statistically significant relationship between the SNP and the trait under study. This can occur when we have a very small MAF so that the large majority of individuals have two copies of the major allele. Here, we remove SNPs for which the MAF is less than 1 % . In some instances, particularly small sample settings, a cut point of 5 % is applied.
We filter simultaneously on call rate and MAF using the following script.

In the data example provided, we filter 203,287 SNPs based on call rate <0.95 and/or MAF <0.01.
2.3. Sample‐level filtering (step 3)
The third stage of data pre‐processing involves filtering samples, that is, removing individuals who we select to be excluded from analysis. Criteria for sample‐level filtering are generally based on missing data, sample contamination, correlation (for population‐based investigations), and racial, ethnic, or gender ambiguity or discordance. Each of these is described later. Additional detail on sample‐level filtering is available in, for example, 22 .
- Sample‐level filtering: call rate. Similar to SNP‐level filtering based on call rate, we exclude individuals who are missing genotype data across more than a pre‐defined percentage of the typed SNPs. This proportion of missingness across SNPs is referred to as the sample call rate, and we apply a threshold of 95 % . That is, individuals who are missing genotype data for more than 5 % of the typed SNPs are removed. A new reduced dimension SnpMatrix genotype object is created, which incorporates this filter.
- Sample‐level filtering: heterozygosity. Heterozygosity refers to the presence of each of the two alleles at a given SNP within an individual. This is expected under HWE to occur with probability 2∗ p ∗(1 − p ), where p is the dominant allele frequency at that SNP (assuming a bi‐allelic SNP). Excess heterozygosity across typed SNPs within an individual may be an indication of poor sample quality, while deficient heterozygosity can indicate inbreeding or other substructure in that person 23 . Thus, samples with an inbreeding coefficient | F |=(1 − O / E ) > 0.10 are removed, where O and E are respectively the observed and expected counts of heterozygous SNPs within an individual. Note that we calculate the expected counts for each individual based on the observed SNPs for that individual.
We filter on sample call rate and heterozygosity simultaneously in the following script:

Because the PennCATH data provided are pre‐filtered, no additional samples are filtered based on an inbreeding coefficient | F |>0.10.
We begin by applying linkage disequilibrium (LD) pruning using a threshold value of 0.2, which eliminates a large degree of redundancy in the data and reduces the influence of chromosomal artifacts 6 . This dimension reduction step is commonly applied prior to both IBD analysis and PCA, applied in the succeeding texts for ancestry filtering, and results in large computational savings.

This reduces the number of SNPs from 658,186 at the end of step 2 to 72,812. Next, we calculate pairwise IBD distances to search for sample relatedness. A strategy is employed that iteratively removes subjects with the highest number of pairwise kinship coefficients >0.1.

In our example, none of the samples are filtered based on the IBD kinship coefficient >0.10.
No additional samples are filtered based on visual inspection of PCA plots. Again, we expect this as the PennCATH data provided are pre‐filtered.

2.4. Single nucleotide polymorphism‐level filtering – part 2 (step 4)

We filter out an additional 1,296 SNPs based on HWE p < 1×10 −6 in CAD controls. This results in 656,890 typed SNPs to be considered in the association analysis.
3. New data generation
After completion of SNP and sample‐level filtering, we generate two new types of data prior to performing our statistical analysis. The first are PCs that are intended to capture information of latent population substructure that is typically not available in self‐reported race and ethnicity variables. The second are genotypes of untyped SNPs that may have a functional relationship to the outcome and therefore provide additional power for identifying association. Each of these is described in more detail in the succeeding texts.
3.1. Creating principal components for capturing population‐substructure (step 5)
Substructure, also referred to as population admixture and population stratification, refers to the presence of genetic diversity (e.g., different allele frequencies) within an apparently homogenous population that is due to population genetic history (e.g., migration, selection, and/or ethnic integration). PCs based on observed genotype data capture information on substructure and are straightforward to generate using the snpgdsPCA() function in the SNPRelate package based on the full genotype data. Notably, we again apply LD pruning prior to the PCA. Typically, the first 10 PCs are considered as possible confounders. This number is routine, but arbitrary, and one alternative is to select the number of PCs based on a pre‐defined proportion of variability that they explain. The λ ‐statistic is typically used to evaluate whether inclusion of PCs is necessary. This is described further in step 10 (quantile–quantile (Q–Q) plots and the λ ‐statistic).

3.2. Imputing non‐typed single nucleotide polymorphisms using 1000 Genomes data (step 6)
Typed SNPs measured using chip array technology typically capture approximately one‐million polymorphisms, which vary in at least 1 % of the general population. More generally, interest lies in analyzing association of genotypes of non‐typed SNPs with disease outcomes because functional (causal) variants may not be measured. Using the extensive externally derived resources on reference haplotypes and their LD structure, such as HapMap and 1000 Genomes data, we can impute the unmeasured genotype data. Three well‐described and recommended stand‐alone packages for SNP‐level imputation are IMPUTE2, MACH, and BEAGLE. Imputed genotypes can be reported as the ‘best guess’ genotype or as the posterior probability of each genotype at a given location on the genome. Importantly, the uncertainty in this estimation process needs to be accounted for in the association analysis, and thus, we distinguish between genotyped and imputed data henceforth. Methods that specifically account for the uncertainty in the imputed SNP data are described in step 8 later.
After imputation, a quality control step is performed to filter imputed data with high degrees of uncertainty. Common measures of uncertainty are the information content and R 2 25 . We apply an R 2 threshold of 0.7 for inclusion in association analysis where in this case, R 2 is the value association with the linear model regressing each imputed SNP on regional typed SNPs. This is described further in the snpStats package documentation. Additionally, we exclude SNPs at this stage with a MAF, after assignment of the highest posterior probability genotype, of less than 0.01. For the purpose of illustration, we use the snp.imputation() and impute.snps() functions in the R package snpStats to impute a limited set of 1000 Genome SNPs on the same chromosome (chromosome 16) as the genotyped SNPs identified as genome‐wide significant in the GWA association analysis (step 7). In practice, imputation is often performed across all chromosomes, resulting in up to 12.5 million typed and imputed SNPs on which association analysis can be performed 13 .

We then remove failed imputations, imputed SNPs with high degrees of associated uncertainty, and imputed SNPs with low estimated MAF.

This analysis results in 162,565 1000 Genomes imputed SNPs on chromosome 16 that are carried forward in step 8 for association analysis. We again emphasize that the uncertainty in imputation needs to be considered in the context of association analysis, and thus, these SNPs are considered separately from the typed SNPs analyzed in step 7.
4. Genome‐wide association analysis
4.1. association analysis of typed single nucleotide polymorphisms (step 7).
Association analysis typically involves regressing each SNP separately on a given trait, adjusted for patient‐level clinical, demographic, and environmental factors. The assumed underlying genetic model of association for each SNP (e.g., dominant, recessive, or additive) will impact the resulting findings; however, because of the large number of SNPs and the generally uncharacterized relationships to the outcome, a single additive model is typically selected. In this case and as illustrated in the code provided, each SNP is represented as the corresponding number of minor alleles (0, 1, or 2). Notably, coding SNP variables based on alternative models (e.g., dominant or recessive) is straightforward, and the association analysis described proceeds identically 26 , 27 . In practice, a Bonferonni‐corrected genome‐wide significance threshold of 5 × 10 −8 is used for control of the family‐wise error rate. This cutoff is based on research, suggesting approximately one‐million independent SNPs across the genome (e.g., 28 ), so tends be applied regardless of the actual number of typed or imputed SNPs under investigation.
In our data example, we use inverse normally transformed HDL‐cholesterol as the response, adjusting for age, sex, and the first 10 PCs. HDL‐cholesterol is a complex trait associated with cardiovascular disease, for which age and sex are established risk factors. These two covariates and the arbitrary choice of 10 PCs are routine for cardiovascular disease trait association studies (e.g., 20 , 26 , 27 ). Importantly, as in any model fitting procedure, it is essential to evaluate the appropriateness of model assumption and specifically the normality of the trait under study. Visual inspection of a histogram of HDL‐cholesterol (code provided but plot not shown) reveals some extreme values, and therefore, an inverse normal transformation is selected. Alternative transformations, such as the log‐transformation, may also be reasonable and have the advantage of maintaining the relative distance between observations. We do not emphasize this in the present tutorial as standard statistical modeling practice can be applied. The following code prepares the phenotype data for analysis.

Running a GWA analysis in parallel, that is, simultaneously across several cores, is recommended because of the large number of models that require fitting. Each core runs the GWA analysis for a subset of the SNPs, and when computation is completed across all cores, the output is returned to its original order. In the succeeding texts, we describe a cross‐platform approach to running the analysis in parallel on MAC, Unix, and Windows operating systems. The detectCores() function can be used to determine the available number of workers. To run the analysis in parallel, we use the dopar() function in the doParallel package, indicating the number of workers. The output of doPar() is an ascii text file. These are contained in the GWAA() function that we developed, and is available in Supplementary Information B of this manuscript.

In our setting, two genotyped SNPs in the cholesteryl ester transfer protein (CETP) gene region, rs1532625 and rs247617, are suggestive of association ( p < 5×10 −6 ) with respective p ‐values of 8.92 × 10 −8 and 1.25 × 10 −7 . CETP is a well‐characterized gene that has been associated previously with HDL‐C (e.g., 26 ). More information on these SNPs and the process of post‐analytic interrogation is provided in steps 9 and 10 later.
4.2. Association analysis of imputed data (step 8)
Several stand‐alone packages can be applied to conduct association analysis of imputed SNPs using the corresponding posterior probabilities. These include, for example, MACH2qtl/dat 29 , ProbABEL 30 , BEAGLE 31 , BIMBAM 32 , and SNPTEST 25 . Reviews and comprehensive comparisons of these approaches can be found in 33 , 34 . The R package snpStats also has functions to read in imputed data based on which imputation package was used (e.g., BEAGLE, IMPUTE, and MACH). For illustrations, we use the single.rhs.tests() function in R package snpStats using the imputation rules generated in step 6.

In total, we identify 22 imputed SNPs on chromosome 16 that are significant at a suggestive association threshold of 5 × 10 −6 . Next, we select only those SNPs within the region of CETP (±5 Kb) to report. Here, we use the map2gene() function we developed, which is also available in Supplementary Information B, that identifies the set of SNPs that belong to a specified gene region. This function uses gene coordinates based on Genome Reference Consortium GRCh37 (hg19), provided in the file ProCodgene_coords.csv . Further interrogation of these SNPs and the CETP region is provided in Figures 5 a and 6 as well as associated text.

UCSC Genome Browser with specified tracks open.
At this stage, we map 70 imputed SNPs to the CETP region, of which 16 are significant at the suggestive association threshold of 5 × 10 −6 .
5. Post‐analytic visualization and genomic interrogation
5.1. data integration (step 9).
At this stage, it is also common to ascribe SNPs to loci and report chromosome and base pair locations, also referred to as coordinates or positions. Notably, the SNP coordinate is dependent on the genome build, and in our data example, we use the Genome Reference Consortium GRCh37 (hg19) build. A typical presentation of results includes gene and locus name; SNP name; chromosome number; base pair location, according to a specified build; the coefficient estimate (or odds ratio) from the model fitting procedure; the corresponding standard error; and the associated p ‐value.

An additional step is required to combine the imputed data results and the typed SNP results. Notably, genotype imputation can involve imputing all SNPs, including both unobserved and typed SNPs. Thus, the analyst may chose to select from the imputation results only SNPs that were not typed or did not pass the SNP‐level filtering. In our example (step 6), we imputed non‐typed SNPs as well as SNPs that did not pass SNP‐level filtering (steps 2 and 4). GWA significant SNPs in this combined set can then be further visualized and interrogated as described in step 10.

5.2. Visualization and Quality Control (step 10)
Several plots allow us both to visualize the GWA analysis findings and to perform quality control checks at the same time. Specifically, as elaborated in each section below we are interested in identifying data inconsistencies, potential systemic biases, and consistency of our findings with previously reported results. We describe three visualization tools in the succeeding texts. In addition to these visualization approaches, association analysis using other genetic models can be a useful sensitivity analysis.
- Manhattan plots. Manhattan plots are used to visualize GWA significance level by chromosome location as shown in Figure 3 . Here, each dot corresponds to a single SNP. The x ‐axis represents gene coordinates, and the numbers shown correspond to chromosome numbers. The y ‐axis is the negative of the log p ‐value, so that large values correspond to small p ‐values. The solid horizontal line indicates the Bonferonni corrected significance threshold (− log(5 × 10 −8 )). The dotted horizontal line is a less stringent suggestive association threshold (− log(5 × 10 −6 )) that we use as an indicator of a suggestive association and requiring further validation, similar to the approach taken in 26 . Visual inspection of this plot allows for identification of SNPs with relatively small p ‐values that are in regions with relatively large and non‐significant p ‐values, suggesting potentially spurious findings. Multiple signals in the CETP region suggest that this may be a true signal. This plot is generated using the GWAS_Manhattan() that we developed and is available in Supplementary Information B.
The degree of deviation from this line is measured formally by the λ ‐statistic 35 , 36 , where a value close to 1 suggests appropriate adjustment for potential substructure. While λ is improved after adjusting for PCs (from λ = 1.014 to λ = 1.0032), a dramatic difference in values is not observed as this PennCATH sample is from a relatively homogenous population. In general, the goal is to achieve a value of λ close to one; λ > 1.2 suggests stratification, and typically, additional PCs are included in this setting, and in some cases, the study is eliminated from inclusion in subsequent meta‐analysis. Calculation of a standardized λ that accounts for sample size is particularly useful in the context of contrasting values across studies for inclusion in meta‐analysis. We apply the following code to generate standardized λ 's for the unadjusted and adjusted models, resulting in λ = 1.0108 and 1.000632 for the unadjusted and adjusted models, respectively. For binary traits, the standardization approach described in 37 can be applied.

Manhattan plot of genome‐wide association analysis results. This figure illustrates the level of statistical significance ( y ‐axis), as measured by the negative log of the corresponding p ‐value, for each single nucleotide polymorphism (SNP). Each typed SNP is indicated by a grey or black dot. SNPs are arranged by chromosomal location ( x ‐axis). Imputation was performed on chromosome 16 only using 1000 Genomes data, and imputed SNPs are indicated by blue dots. None of the SNPs reached the Bonferroni level of significance ( p < 5×10 −8 − solid horizontal line); however, two typed SNPs and 22 imputed SNPs (on chromosome 16) were suggestive of association ( p < 5×10 −6 – dashed horizontal line).

Quantile–quantile plots for quality control check and visualizing crude association. Quantile–quantile plots illustrate the relationship between observed ( y ‐axis) and expected ( x ‐axis) test statistics and are used as a tool for visualizing appropriate control of population substructure and the presence of association. The left panel (a) is based on an unadjusted model, where the deviation is below expected, while the right panel (b) is based on a model adjusted for potential confounders, which brings the tail closer to the y = x line. The extreme observed statistics are suggestive of association. Data generally falling on the y = x lines suggests no clear systemic bias. Unstandardized λ 's are reported. PCs, principal components.

Heatmap and regional association plots. Heatmap (top) illustrating linkage disequilibrium (LD) between typed (black) and imputed (red) single nucleotide polymorphisms (SNPs) in the cholesteryl ester transfer protein (CETP) region. A total of two typed SNPs and 16 imputed SNPs are significant at the less stringent 5 × 10 −6 threshold; however, the heat map only illustrates imputed SNPs with a posterior probability of 1 for the associated genotype. We observe the presence of two distinct LD blocks within the CETP gene region, with high levels of LD between SNPs within each block and lower LD between SNPs across the the two blocks. A related regional association plot (bottom) illustrates association levels and LD for a larger window surrounding CETP.
A regional association plot, provided in Figure 5 b, provides similar information for a broader region of the genome. In this case, the blue line at the top represents the SNP‐level p ‐values, the green segments indicate gene regions, and the red lines indicate LD, where we have specified to only include lines betweens SNPs with r 2 >0.8. The following code is used to generate this figure.

5.3. Additional data interrogation using external resources
Reporting SNP‐level findings from association analysis is much more meaningful when a context for the findings is also presented. For example, investigators may want to know whether a statistically significant SNP is within a protein‐coding gene, intergenic, or close to a regulatory element (e.g., a methylation mark) in specific tissues or cell types that are relevant to the disease under investigation. Possible external types of data that may be relevant are provided in Supplementary Information C, Table 1 . These fall into eight general categories, following roughly an order representing the process from DNA information to regulation to expression: (i) SNP; (ii) gene elements; (iii) chromatin state; (iv) epigenetic marks; (v) transcription factor binding; (vi) RNA expression; (vii) SNP–mRNA association; and (viii) other ‐omics data. We emphasize that this table is not intended to be comprehensive; rather, it provides a glimpse at the vast amount of external data resources available. Data associated with each of the listed categories are available from a wide range of sources (for example, column 2, Supplementary Information C, Table 1 ) and are generally based on a variety of technologies. The UCSC Genome Browser provides a well‐devised suite of integrated bioinformatic tools and databases, including many derived from the resources listed in Supplementary Information C, Table 1 , which allow for further interrogation of GWA findings.

Example data types and select resources for post‐analytic interrogation. *Listed resources are intended to provide primary examples and are not comprehensive. a National Center for Biotechnology Information (NCBI) dbSNP; b ENSEMBL Genome Browser; c NCBI RefSeq; d NCBI GenBank; e The encyclopedia of DNA elements (ENCODE) Project; f NIH Roadmap Epigenomics Project; g GTex Portal; h NCBI Sequence Read Archive (SRA); i The Universal Protein Resource Knowledgebase (UniProtKB); j The Human Metabolome Database (HMDB).
In this section, we provide a very brief introduction to the genome browser, with particular focus on how to view and interpret standard tracks, visualize data corresponding to these tracks, and create custom tracks using new data. To begin, we go to the genome browser gateway at http://genome.ucsc.edu/cgi-bin/hgGateway . We then specify assembly Feb. 2009 (GRCh37/hg19) , type the name of our most significant SNP, rs1532625 , in the field search term , and then select submit . On the next page, we select rs1532625 at chr16:57005051‐57005551 under the first heading. This choice is elaborated in the succeeding texts. The content of the next page will vary depending on the tracks remaining open at the end of the current user's last session. However, all users will see in the bottom half of this page several classes of tracks, with multiple choices within them. We make the following selections and then press refresh either after each change or after all fields have been selected: Genes and Gene Predictions: UCSC Genes select pack ; mRNA and EST: Human mRNAs select dense ; Regulation: ENCODE Regulation select show ; and Variation: common SNPs(142) select pack . All other fields should be marked hide . On the next screen, we zoom out 100x by pressing the corresponding grey button at the top of the image to acquire a better picture of the entire region.
The resulting image is illustrated in Figure 6 . Note that your tracks could be illustrated in a different order. Next is a summary of the elements of this figure. We note first that this is an image of the genetic region surrounding the rs1532625 SNP (highlighted in a black box in Figure 6 ) we entered and that the tracks illustrated (also based on our selections) are differentiated by the grey vertical rectangles on the left‐hand side of the figure.
- Variation: common SNPs(142). The bottom‐most track with the heading ‘Simple Nucleotide Polymorphisms dbSNP141) Found in >= 1% of Samples’ lists all of the common SNPs by rsNumber that are in this region, in order of their location on the genome. We see that the input SNP, rs1532625 , is highlighted with a black box. By clicking on this box, the investigator can retrieve additional information about this SNP, including the major and minor alleles and their frequencies, average heterozygosity, and the chromosome and coordinate location based on the current build. Additional information on a track can be found by pressing the grey rectangular vertical bar on the main browser window corresponding to the track. For example, for this track, we find a description of the source of the data (dbSNP build 142.)
- Genes and gene predictions: UCSC genes. The first track at the top of Figure 6 titled ‘UCSC Genes’ illustrates all protein‐coding and non‐protein coding genes that are in close proximity to the SNP we entered. We note based on this figure that our SNP rs1532625 falls in the protein‐coding gene CETP. Additional information about the display conventions and the configuration can be found by pressing the grey rectangular vertical bar on the left‐hand side of this track. Additional information about each gene, including the full gene name, coordinates, size, number of exons, and prior GWA evidence, can be retrieved by clicking on the abbreviated gene names in the browser window.
- mRNA and EST: human mRNAs. The next track entitled ‘Human mRNAs from GenBank’ provides historical information on whether there have been any reports (indicated by a vertical bar) of the presence of mRNA corresponding to sites on the genome across all tissues and cell types. By clicking on the title, an expanded view of this track is provided (not shown), allowing the user to find additional information. Consider CETP, for example, for which we expect to see mRNA expression in cells relevant to HDL production and/or regulation, such as liver tissue. By selecting mRNA {"type":"entrez-nucleotide","attrs":{"text":"M30185","term_id":"180259","term_text":"M30185"}} M30185 near the start of CETP, we learn that indeed, the mRNA was found in liver tissue.
- Regulation: ENCODE regulation. The next three tracks entitled ‘ H3K27Ac Mark ’, ‘ DNaseI Hypersensitivity Clusters in 125 cell types from ENCODE (V3) ’, and ‘ Transcription Factor ChIP‐seq (161 factors) from ENCODE with Factorbook Motifs ’ all provide information about the presence of cell and tissue‐specific regulatory elements. For example, H3K27Ac is a histone mark indicating the degree of acetylation of lysine 27 of the H3 histone protein, which in turn influences how accessible chromatin is for transcription. The color coding of the density plots in this track corresponds to different cell lines. DNA hypersensitivity is a more general measure of whether chromatin is open for transcription, while transcription factor ChIP‐seq data provides very specific information about whether given proteins can bind to the specified DNA regions.
It is possible to obtain the data corresponding to each track. As an example, consider the ‘common SNPs’ track, click on the corresponding grey vertical rectangle to the left of this track, and then select view table schema on the next page. If we scroll down, under the heading Sample Rows , we see all of the data fields associated with this track. Note also that the metadata about this table, including Database: hg19 and Primary Table: snp142Common , are available at the very top of the screen (not shown). These data can be downloaded by selecting Tools ‐> Table Browser on the top menu and then indicating the appropriate fields, including assembly: Feb. 2009 (GRCh37/hg19); group: Variation; track: Common SNPs(142); and table: snp142Common . The ‘get output’ tab at the bottom of these fields displays the data as an ascii formatted file.
We also note that it is possible to create a custom track that is displayed and linked to the information in this browser. To do this, first, we need to create what is a called a BED track file (different than the.bed file discussed in Section 2 in the preceding texts) containing all of the data contributing to this track. A BED track file must include the following five columns: chromosome number, start location, end location (one greater than the start location for individual SNPs), identifier, score, and chromosomal strand, for which the SNP is recognized on the browser. These are included as columns 2 − 6 in the table schema discussed earlier. Once we have a properly formatted BED file, we can input it directly into the genome browser as a custom track. In order to add this track to the genome browser, click ‘Add Custom Tracks’ , from the main browser window, and upload the new file. This will bring up a page with the details of our new custom track. Click ‘go to genome browser’ in order to see the new custom track in the browser.
6. Broader contemporary context and discussion
This tutorial presents fundamental analysis concepts and tools for performing a single GWA analysis and beginning the process of post‐analytic interrogation. Increasingly, GWA analysis results are being combined across a large number of studies to improve power for novel discoveries. For example, the Global Lipids Genetics Consortium recently reported the results of a meta‐analysis of 188,577 individuals across 60 studies, resulting in discovery of 62 novel loci for blood lipids 38 , 39 . Likewise, the CARDIoGRAM consortium and the CARDIoGRAMplusC4D consortium metadata (which include the PennCATH data used throughout this tutorial) include GWA study results based on 194,427 individuals and contributed to the discovery of 46 loci associated with coronary artery disease 21 , 27 . An overview of methods for GWA meta‐analysis can be found, for example, in 40 , with study‐specific details typically provided in the Supplementary Information of associated manuscripts (e.g., 21 , 38 ). Importantly, depending on consortium data harmonization procedures, we see variation in the extent and timing of SNP and sample‐level filtering, as well as the criteria for including PCs and other covariates in the final model fitting procedure. Thus, flexibility in the step‐by‐step procedure described herein may be required.
Traditionally, a two‐stage design was used, with replication of top suggestive findings ( p < 5×10 −6 ) in a large independent study sample of like design and like ethnicity 41 , 42 . A threshold for significance is set in the second stage based on the number of SNPs carried forward and typically required that the SNP met the widely held genome‐wide significance for all common SNPs ( p < 5×10 −8 ) in a combined meta‐analysis. However, often in contemporary studies, GWA data are available simultaneously in several studies, and a meta‐analysis is performed on all SNPs across all studies in the second stage, and the significance threshold of p < 5×10 −8 is applied. Typically, additional replications are sought in different ethnicities and in study designs that are not identical, for example, different age groups and with different traits that mark the same disease, in order to evaluate generalizability.
Several analytic strategies have been developed, which serve to complement the single‐SNP level testing approach described in this tutorial, including gene‐level testing strategies that require raw genotype data 16 , 43 and gene‐level testing approaches that instead leverage summary output (in the form of test statistics or p ‐values) of the GWA analysis presented herein 15 , 44 . A broad assortment of sophisticated analytic methods has also been described for gene set enrichment or biological pathway analysis 18 , 45 , 46 , 47 , 48 . Additional methods have been described to address the unique challenges inherent in rare variant analysis 8 , 9 , 10 in which the low frequencies of mutations can result in insufficient power to assess significance without regional context. Finally, linear mixed models have been described as an alternative strategy for GWA analysis, which can account for family relatedness and population substructure 49 , 50 , 51 , 52 , 53 , 54 . An additional recommended resource for more in‐depth post‐processing of GWA findings, including gene and network‐based analysis, is provided in 55 .
Defining the best practices for GWA data pre‐processing, analysis, and post‐analytic interrogation within a framework that is logical and comprehensive for statisticians is essential for standardizing methods and ensuring reproducible and comparable findings across studies. This tutorial outlines the key features that are integral to GWA studies, and provides the R code that can been applied to implement each of these features accurately. We emphasize the use of R as GWA studies are typically part of a larger data analytic investigation (e.g., gene‐based analysis as described earlier), and it is straightforward to integrate the R code provided into larger statistical coding efforts. Alternative open‐source, freely available, high‐performance programing languages, for example, Julia, which was designed specifically for parallelism and cloud computing 56 , may ultimately serve to provide additional functionalities in this big data analytic realm, particularly as post‐analytic interrogation becomes more integrated with primary GWA analysis.

Supporting information
Supporting Info Item
Reed, E. , Nunez, S. , Kulp, D. , Qian, J. , Reilly, M. P. , and Foulkes, A. S. (2015) A guide to genome‐wide association analysis and post‐analytic interrogation . Statist. Med. , 34 : 3769–3792. doi: 10.1002/sim.6605 . [ PMC free article ] [ PubMed ] [ Google Scholar ]
Support for this research is provided by NIH/NHLBI R01‐HL107196.

Integrative analysis of Transcriptome-wide and Proteome-wide association study for non-Mendelian disorders
Genome-wide association studies (GWAS) have uncovered numerous variants linked to a wide range of complex traits. However, understanding the mechanisms underlying these associations remains a challenge. To determine genetically regulated mechanisms, additional layers of gene regulation, such as transcriptome and proteome, need to be assayed. Transcriptome-wide association studies (TWAS) and Proteome-wide association studies (PWAS) offer a gene-centered approach to illuminate these mechanisms by examining how variants influence transcript expression and protein expression, thereby inferring their impact on complex traits. In the introductory chapter of this dissertation, I discuss the methodology of TWAS and PWAS, exploring the assumptions they make in estimating SNP-gene effect sizes, their applications, and their limitations. In Chapter 2, I undertake an integrative analysis of TWAS and PWAS using the largest cohort of individuals affected with Tourette’s Syndrome within the Psychiatric Genomics Consortium (PGC) – Tourette’s Syndrome working group. I identified genomic regions containing multiple TWAS and PWAS signals and integrated these results using the computational colocalization method to gain insights into genetically regulated genes implicated in the disorder. In Chapter 3, I conduct an extensive TWAS of the Myasthenia Gravis phenotype, uncovering novel genes associated with the disorder. Utilizing two distinct methodologies, I performed individual tissue-based and cross-tissue-based imputation to assess the genetic influence on transcript expression. A secondary TWAS analysis was conducted after removing SNPs from the major histocompatibility complex (MHC) region to identify significant genes outside this region. Finally, in Chapter 4, I present the conclusions drawn from both studies, offering a comprehensive understanding of the genetic architecture underlying these traits. I also discuss future directions aimed at advancing the mechanistic understanding of complex non-Mendelian disorders.
Degree Type
- Doctor of Philosophy
- Biological Sciences
Campus location
- West Lafayette
Advisor/Supervisor/Committee Chair
Additional committee member 2, additional committee member 3, additional committee member 4, usage metrics.
- Statistical and quantitative genetics
- Neurogenetics

Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- My Account Login
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Open access
- Published: 29 October 2015
Genome-wide analysis correlates Ayurveda Prakriti
- Periyasamy Govindaraj 1 na1 ,
- Sheikh Nizamuddin 1 na1 ,
- Anugula Sharath 1 na1 ,
- Vuskamalla Jyothi 1 na1 ,
- Harish Rotti 2 na1 ,
- Ritu Raval 2 na1 ,
- Jayakrishna Nayak 3 na1 ,
- Balakrishna K. Bhat 3 na1 ,
- B. V. Prasanna 3 na1 ,
- Pooja Shintre 4 na1 ,
- Mayura Sule 4 na1 ,
- Kalpana S. Joshi 4 na1 ,
- Amrish P. Dedge 4 na1 ,
- Ramachandra Bharadwaj 5 na1 ,
- G. G. Gangadharan 5 na1 ,
- Sreekumaran Nair 6 na1 ,
- Puthiya M. Gopinath 2 na1 ,
- Bhushan Patwardhan 7 na1 ,
- Paturu Kondaiah 8 na1 ,
- Kapaettu Satyamoorthy 2 na1 ,
- Marthanda Varma Sankaran Valiathan 2 na1 &
- Kumarasamy Thangaraj 1 na1
Scientific Reports volume 5 , Article number: 15786 ( 2015 ) Cite this article
86k Accesses
65 Citations
662 Altmetric
Metrics details
- Genetic association study
The practice of Ayurveda , the traditional medicine of India, is based on the concept of three major constitutional types (Vata, Pitta and Kapha) defined as “ Prakriti ”. To the best of our knowledge, no study has convincingly correlated genomic variations with the classification of Prakriti. In the present study, we performed genome-wide SNP (single nucleotide polymorphism) analysis (Affymetrix, 6.0) of 262 well-classified male individuals (after screening 3416 subjects) belonging to three Prakritis. We found 52 SNPs ( p ≤ 1 × 10 −5 ) were significantly different between Prakritis , without any confounding effect of stratification, after 10 6 permutations. Principal component analysis (PCA) of these SNPs classified 262 individuals into their respective groups (Vata, Pitta and Kapha) irrespective of their ancestry, which represent its power in categorization. We further validated our finding with 297 Indian population samples with known ancestry. Subsequently, we found that PGM1 correlates with phenotype of Pitta as described in the ancient text of Caraka Samhita, suggesting that the phenotypic classification of India’s traditional medicine has a genetic basis; and its Prakriti -based practice in vogue for many centuries resonates with personalized medicine.
Similar content being viewed by others
Genome-wide analyses disclose the distinctive HLA architecture and the pharmacogenetic landscape of the Somali population
A database of 5305 healthy Korean individuals reveals genetic and clinical implications for an East Asian population
The first insight into the genetic structure of the population of modern Serbia
Introduction.
Among the traditional systems of medicine practiced all over the world, Ayurveda of India has a documented history dating back to 1500 BCE 1 , 2 . Though contemporary medicine is currently the mainstream of medical practice in India, Ayurveda is extensively used side by side and remains highly popular, especially in South Asia. The basic concepts of Ayurveda are; 1. five elements – panchabhuta – which constitute the physical universe including the human body and; 2. three doshas (Vata, Pitta and Kapha) or constitutional types of every human. These doshas refer broadly to the functions of motion, digestion and cumulation. Though all three doshas exist in every human being one is dominant based on which an individual’s Prakriti is determined. Prakritis are discreet phenotypes and they are determined on the basis of physical, psychological, physiological and behavioural traits and independent of social, ethnic and geographical variables 1 , 3 , 4 . The etymology of these Sanskrit terms suggests that Vata originates from movement, Pitta from digestion and Kapha from cumulation. Since Prakritis underlie an individual’s predisposition to disease as well as response to treatment, it is imperative in Ayurvedic practice to identify the Prakriti of a patient before treatment 5 .
Concept of Prakriti in Ayurveda and its relationship with genomics was hypothesized over a decade ago 6 . Subsequent studies have attempted to correlate Prakriti classification with genetic information and association of single nucleotide polymorphisms (SNPs) in HLA-DRB1 7 , CYP2C19 8 , EGLN1 9 , inflammatory and oxidative stress related genes 10 , CD markers for various blood cells 11 , 12 , DNA methylation alterations 13 and risk factors of cardiovascular or inflammatory diseases have been reported 14 . While these studies have shown the association of specific genes with the phenotype of a particular Prakriti , the association of genomic variations with Prakriti classification was lacking. This is the first attempt to classify the Prakritis using genome-wide SNP markers and to provide a scientific basis for Prakriti classification.
Results and Discussions
A total of 3,416 normal healthy male subjects between 20–30 years of age were recruited by the Institute of Ayurveda and Integrative Medicine (IAIM), Bangalore, Karnataka (‘B’ in tables); Sinhgad College of Engineering (SCE) Pune, Maharashtra (‘P’ in tables); and Shri Dharmasthala Manjunatheshwara College of Ayurveda (SDMCA), Udupi, Karnataka (‘U’ in tables). Since the hormonal fluctuations during premenstrual and menstrual phases result in numerous physical and psychological disturbances, which may have confounding effect at the time of Prakriti assessment, we have excluded females from this study (detailed justification on inclusion of only males is given in the Methods section). However, several studies have included of both male and female subjects for Ayurveda -based studies 7 , 8 , 15 , 16 . The subjects belonged to diverse ethnic and linguistic groups and inhabited different geographical regions. The health status of every individual was ascertained by modern as well as Ayurvedic methods (details given in the Methods). The composition of Prakriti was determined by senior Ayurvedic physicians and confirmed independently by ‘AyuSoft’ ( http://ayusoft.cdac.in ), a software developed based on information from classical Ayurvedic literature. The subjects, whose Prakriti was in concordance between the assessment by Ayurvedic physicians and by AyuSoft were only selected for this study. Of the total 3,416 individuals evaluated, 971 had 60%–93% dominance of one Prakriti ( Table S1 ), of which 262 individuals (94 Vata-dominant, 75 Pitta-dominant and 93 Kapha-dominant) with the highest proportion of one predominant Prakriti were randomly selected and subjected to genome-wide SNP analysis (Affymetrix array, 6.0) and genotypes were fetched using Birdsuite software 17 . The proportions of each dominant and co-dominant Prakritis are given in Fig. 1 ; Figure S1 .
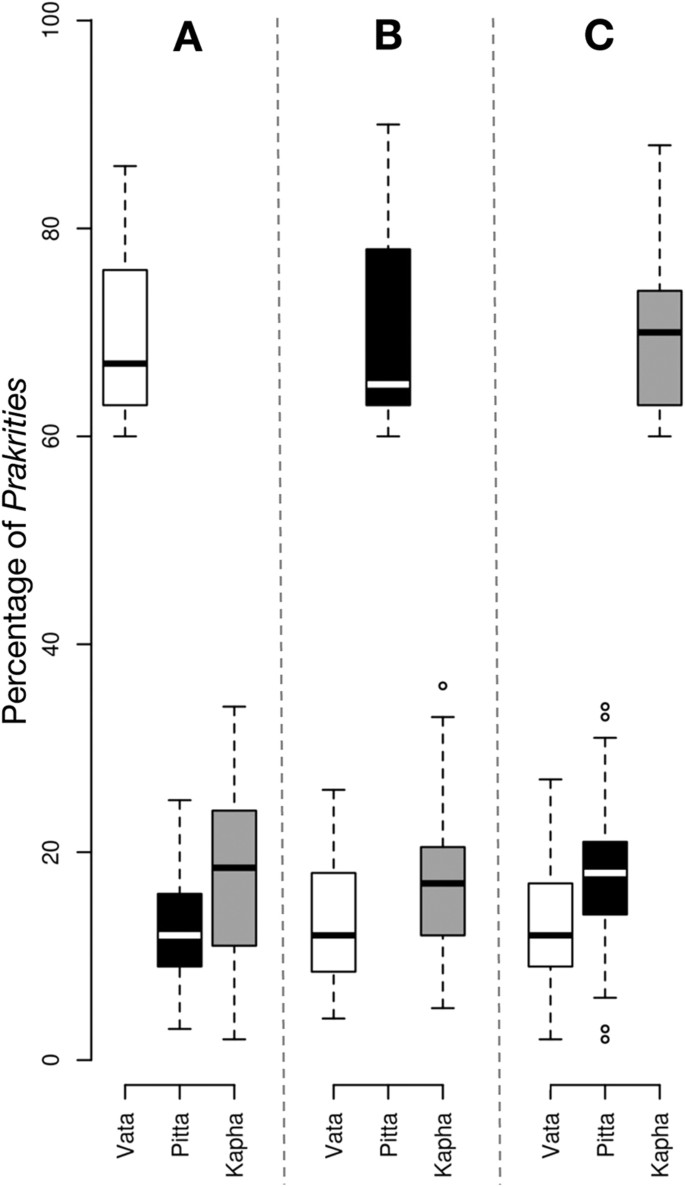
Box-plot representing the Prakriti proportion of subjects with Vata (94), Pitta (75) and Kapha (93) dominant characteristics.
( A ) Average percentage of Vata is 67%, while Pita and Kapha are 12% and 18.5%, respectively. ( B ) Average percentage of Pita is 65%, while Vata and Kapha are 12% and 17%, respectively. ( C ) Average percentage of Kapha is 70%, while Vata and Pita are 12% and 17%, respectively.
Out of 262 individuals analyzed, 245 passed the quality controls (QC) with the call rate 0.966 ± 0.0162 ( Table S2 ). In order to validate the high-throughput data set, we randomly selected 48 markers from Affymetrix array and genotyped 48 individuals using custom-designed VeraCode GoldenGate Genotyping Assay System (Illumina, San Diego, USA). The call rate of VeraCode analysis was 99.61% and the genotype matched with Affymetrix data set ( Table S3 ), suggesting that the genotypes obtained from Affymetrix array was genuine with minimum error (0.39%). Further, to increase the statistical power, we used Indian population data set as reference and imputation analysis was performed using Beagle (v3.3.1) software 18 ( Figure S1 ). As we had demonstrated earlier that Indian population has unique genetic architecture, we were skeptical of using non-Indian samples as a reference for imputation 19 . To evaluate our assumption, we masked 2%, 5% and 10% genotype of 207 unrelated Dravidian and Indo-European population samples and performed 110 simulations on chromosome 22 with four-reference populations i.e. Indian population (28 trios of Dravidian and Indo-Europeans; IN), different HapMap populations (CEU, YRI, CHB, CHS and JPT; HM), different South-Asian populations of 1000 genome project (BEB, GIH, ITU, PJL and STU; SA) and Indian along with HapMap populations (IH). As expected, imputed genotypes were more accurate with Indian samples (IN) [2% (0.9518 ± 0.0012); 5% (0.95045 ± 0.00109); 10% (0.9476 ± 0.0005)] compared to HM [2% (0.9462 ± 0.0013); 5% (0.9436 ± 0.0017); 10% (0.9396 ± 0.0005)], IH [2% (0.9463 ± 0.0014); 5% (0.9448 ± 0.0016); 10% (0.9417 ± 0.00066)] and SA [2% (0.9481 ± 0.0013); 5% (0.9471 ± 0.00098); 10% (0.9441 ± 0.00061)] samples ( Table S4 ; Figure S2 ). In all the three masked data (2%, 5% and 10%), IN showed high imputation performance compared to HM, SA and IH. Even with ~10% masked data, the imputed genotypes were more accurate with IN than other references, suggesting that it is appropriate to use Indian data set for imputation. The data set of Gujarati Indians in Houston (GIH) is the only one available in the public domain, which was admixed recently and hence does not truly represent the ANI-ASI ancestry of Indian population 19 , 20 . As the data were not suitable reference for imputation, we prepared our own reference panel of Indian population ( http://www.ccmb.res.in/bic/database_pagelink.php?page=snpdata ). To achieve this, we followed two steps (i) imputation of 15 trios of Indo-European and 15 trios of Dravidian and (ii) imputation of 229 unrelated individuals imputed with the reference genotype obtained from step-I. Further, we used this reference for imputing the Prakriti individuals. In the first step, we found 10.5% and 17.8% Mendelian inconsistency in two trios, (Kashmiri Pandit) ( Table S5 ), which were removed from the analysis. Finally, we obtained 791186 SNP markers with 0.95 ≤ R 2 ≤ 1, for further analysis.
To make sure that the Prakriti samples were collected randomly and there was no major ancestral bias while collecting samples, we performed the principal component analysis (PCA) 21 of 245 Prakriti samples ( Figure S3 ). PCA analysis revealed no significant overall differences among the Prakritis (ANOVA p-value on eigenvector 1 V vs. K-0.434; V vs. P-0.89; P vs. K-0.51; and eigenvector 2 V vs. K-0.09; V vs . P-0.06; P vs . K-0.02). In order to check the ancestry of Prakriti individuals, we used our published data set of 297 Indian population samples with known ancestry 19 , 20 . These 297 samples include; 150 Dravidians, 80 Indo-European, 35 Austro-Asiatic, 27 Tibeto-Burman and 5 Great Andamanese ( Table S6 ). We found 7,89,309 SNPs were common between Prakriti and Indian ancestral samples. In order to remove the differentiation on spurious axes 21 , we pruned 3,76,138 SNPs, which were in strong linkage disequilibrium (LD) (r 2 > 0.75) and performed PCA with 4,13,171 SNPs. Our analysis showed that most of the Prakriti samples clustered with Dravidian and Indo-European (the two major ancestral population of India) and only 3 samples seemed to be Tibeto-Burman and admixed recently ( Figure S4 ). Previous studies have shown that stratification could cause spurious association 22 , 23 , 24 , 25 , hence, PCA was performed 21 using 4,05,782 SNPs (3,85,404 SNPs were pruned with r 2 > 0.75) for 245 Prakriti samples, of which 40 were outliers and have been removed in 10 iterations with σ ≥ 6 on eigenvector 1 to 10 ( Table S7 ; Figure S3 and S5 ). ANOVA analysis revealed that the Prakriti groups were not significantly different (p-value: V vs. P - 0.40 ± 0.28; V vs. K - 0.51 ± 0.32 and P vs. K - 0.48 ± 0.29) ( Table S8 ); and 205 Prakriti samples were used for further analysis ( Figure S1 ).
Association analysis was performed using plink software 26 . Since the present study has no cases and controls (patients and healthy), we considered one Prakriti as case and the remaining two Prakritis as controls and performed association analyses in three combinations: Vata vs . Kapha and Pitta (V vs. PK); Pitta vs . Kapha and Vata (P vs. VK); Kapha vs . Pitta and Vata (K vs. VP). Prior to association analysis, 3,890; 4,153 and 4,124, respectively, markers were removed from 791186 markers, which were not in Hardy-Weinberg equilibrium (HWE) i.e. p-value < 0.001 in controls of V vs. PK, P vs. VK and K vs. VP; respectively. The three combination association results were further used to identify the SNPs that were significant. Considering the fact that none of the samples represents 100% single Prakriti , we did not expect very low p-value in the association analysis. In this scenario, truly associated loci may co-exist with false positive markers and can be identified by permutation analysis. As expected, we observed that SNPs having approximately same p-value in the extreme tail of theoretical distribution failed to achieve 10 6 permutations ( Table 1 ). For example, rs2939743 having p-value 7.61 × 10 −5 dropped at 142717 th permutation while rs10197747 having p-value 2.50 × 10 −5 achieved 10 6 permutations, which of course revealed that rs2939743 is false positive. Similarly, we found 52 true positive SNPs achieved 1 million simulations with theoretical p-value ≤ 1 × 10 −5 (details are given in Table 1 ; Figure S6 ).
It is well known that some markers differ in allele frequency more across ancestral population, compared to other set of markers. Moreover, natural selection might be the reason for this phenomenon because it acts locus-specific manner 21 . We speculate that the above so-called true positive loci might be artifacts of population stratification because of high probability of false positive results at the p-value, which observed in association analysis. Hence, we performed extensive statistical analyses to control these confounding factors and/or population stratification. Prevailing methods include genomic control and EIGENSTRAT to find such confounding effect of stratification. Genomic control uses uniform inflation factor to correct stratification, which is not sufficient for those SNPs having high frequency differences between ancestors 21 . Hence, we proceeded with EIGENSTRAT and found p-value did not change drastically ( Table S9 ). To further confirm, we used variance component model (implemented in EMMAX) 27 and mixed-linear model of association analysis (implemented in GCTA) 28 , which can correct sample structure in association, but have different statistics comparative to eigenstrat. Intriguingly, even with this analysis, we did not observe any drastic change in the p-value ( Table S9 ). This has proved that these 52 SNPs were genuine characteristics of Prakriti and not derived from ancestry. Moreover, we also explored the allele frequency differences between centers; however, we did not find any significant difference for these 52 SNPs ( Table S10 ). We further explored the power of 52 SNPs in Prakritis genetic differentiation ( Figure S1 ). In principal component analysis, 19 SNPs were excluded with r 2 > 0.75 and, as expected, we found striking separation of subjects according to their Prakriti ( Fig. 2A ). On eigenvector-1 (eigenvalue = 18.168248) Pitta significantly differentiated against Vata and Kapha (p-value = 1.11022 × 10 −16 , 4.44089 × 10 −16 , respectively); while on eigenvector 2 (eigenvalue = 15.890861) Kapha was significantly different compared to Vata and Pitta (p-value = 3.33067 × 10 −16 and ~0 respectively).
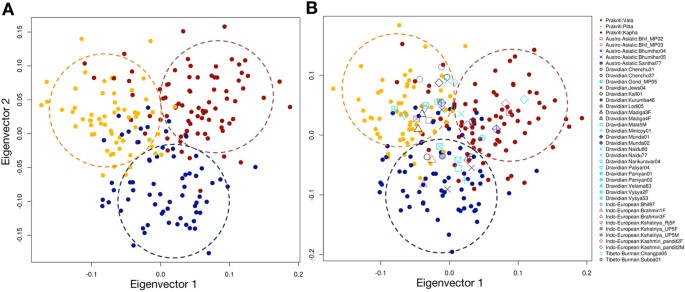
Principal component analysis (PCA) with 52 SNPs that showed p-value of <1 × 10-5 ( A ) PCA of Prakriti individuals showing three clusters (Vata, Pitta, Kapha), despite their linguistic, ethnic and geographical diversity. ( B ) PCA projection of Indian population samples with Prakriti individuals.
To examine the statistical power of these 52 markers for categorizing the samples with unknown Prakriti , we generated a statistical model (see methods). Initially, we applied it on 205 samples and found 23.9% (49 out of 205) were explained by the proposed model ( Table S11 ). Further, we applied it on 297 Indian (population) samples and found 37 individuals (5 Austro-Asiatic; 22 Dravidian; 8 Indo-European and 2 Tibeto-Burman) satisfying the model. According to the model, 7 individuals were Vata, 20 were Pitta and 11 were Kapha. Interestingly, Indian population samples, which belong to one Prakriti were from different ancestry ( Table S12 ), suggesting that these makers could separate the Prakritis , irrespective of their ancestry. To confirm the proposed model, we projected these 37 individuals on eigenvector of Prakriti samples and found that these individuals clustered with Prakriti as predicted in the model ( Fig. 2B ). It suggests that the cluster is based on Prakriti and is not due to the ancestry of samples. That would also suggest that the phenotypic variations have a genetic basis, which would be shared by Prakritis of Ayurveda .
Further, we used these 52 markers to find the genotype-phenotype correlations. We observed that 2 markers (rs10518915 and rs986846) were associated with two different Prakriti ; rs10518915 with Vata and Pitta, while rs986846 with Kapha and Vata. This observation prompted us to believe that different alleles of the same locus might be influencing different Prakriti ( Table 1 ). In order to correlate the functional relevance of these SNPs, we divided them into genic and non-genic. The SNPs, which are within 10 kb of gene, were considered genic; while others as non-genic 29 , 30 . We found 28 were genic SNPs, of which 12 were in Vata (7 genes), 11 in Pitta (7 genes) and 6 in Kapha (7 genes) ( Table 1 ). To correlate the function of these genes with respect to the characteristics of Prakritis , we searched in Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and Reactome event and found PGM1 gene associated with the Pitta phenotype. In Ayurveda , characteristics of Pitta include digestion, metabolism and energy production. Interestingly, we found PGM1 gene is in the center of many metabolic pathways i.e. glycolysis or gluconeogenesis (hsa00010); pentose phosphate pathway (hsa00030); galactose metabolism (hsa00052); purine metabolism (hsa00230) and; starch and sucrose metabolism (hsa00500) ( Figure S7 ). Our finding suggests that the function of the gene directly correlates with the role of Pitta in metabolism as described in Ayurvedic literature.
In addition, we have checked the PGM1 gene markers in Affymetrix data set and found 4 markers (rs2269241, rs2269240, rs2269239,and rs2269238) were associated with Pitta Prakriti and all are in strong Linkage Disequilibrium (LD) ( Figure S8 ). Therefore, to find the functionally relevant variants, we sequenced the whole exons and UTRs of the PGM1 gene in 78 individuals using Ion Torrent PGM (Life Technologies, USA). We found 23 variations in the gene, of which 8 were novel ( Table S13 ). Interestingly, one non-synonymous; c.1258T > C (p.Tyr420His) (rs11208257) variant was present in the LD block and found in association with Pitta Prakriti (p-value–7.049 × 10 −3 ). The frequency of the mutant allele “C” was 5.8% in Pitta and 20% in Kapha Prakriti ( Table S14 ). This result prompted us to replicate the marker (rs11208257) in additional samples. We genotyped this marker (rs11208257) for 665 Prakriti individuals (299 Vata, 164 Pitta and 202 Kapha) using Sanger sequencing method. Initially, we analyzed the distribution of the genotype among participating centres and found “U” samples (collected from Udupi centre) were not in HWE (p-value - 0.04) ( Table S15 ). Hence, we excluded 169 “U” samples from the analysis. Association analysis revealed that allelic and genotype distribution of the marker rs11208257 is significantly different in Pitta Prakriti against Vata and Kapha with p-value- 2.06 × 10 −2 ; p-value- 6.16 × 10 −3 , respectively. Further, we explored the association between P vs. V and P vs . K; and found significant p-value - 7.61 × 10 −3 and 2.35 × 10 −2 , respectively. The results would therefore suggest that Vata differs more from the Pitta Prakriti than Kapha ( Table S16 ). We further screened 1108 randomly selected Indians and 992 HapMap samples and found that the frequency of mutant allele “C” was 17.9% among Indians, 15.5–17.6% in the Europeans, 14.5–18.8% in East Asians, 42% in Mexican, 15.3% in admixed Indians (GIH) and 12.8–28.3% in Africans. Indians have comparable frequency with Europeans and GIH ( Table S17 ). Interestingly, we found Pitta has less frequency of mutant “C” allele and Vata and Kapha have comparable frequency with overall Indian population. To explore the functional relevance of the variant, we used SIFT software and found that the mutation is damaging with 0.01 score and thus substitution at this position may affect the protein function. Our data suggest that the SNP (rs11208257) in PGM1 gene is linked with one of the main features (energy production), which is more homogenous and constant in Pitta than with Vata and Kapha and a genotype correlation exists for the characteristics of Prakriti classification.
In conclusion, our preliminary study suggests that the Prakriti classification, as a foundation for the practice of Ayurveda , has a genetic basis and does provide clues for further studies.
Selection of subjects and Prakriti assessment
Selection of subjects and evaluation of the Prakriti (the human classification of Indian ancient medicine) were carried out at three centres; 1. Institute of Ayurveda and Intergrative Medicine (IAIM), Bangalore, Karnataka; 2. Sinhgad College of Engineering (SCE) Pune, Maharashtra; and 3. Shri Dharmasthala Manjunatheshwara College of Ayurveda (SDMCA) Udupi, Karnataka. This study was approved by Institutional Ethics Committees (IECs) of all the collaborative centres and the methods were carried out in accordance with the approved guidelines. We have screened normal and healthy male subjects, who were between 20–30 years. Although several Ayurveda -based studies have included both male and female subjects 7 , 8 , 15 , 16 , we have excluded female subjects from this study to minimize the confounding variations. Prakriti of an individual is determined based on defined anatomical, physiological psychological and behavioural characteristics. During actual assessment of Prakriti , the Ayurvedic physician needs to factor in these characteristics. One such aspect is the cyclical hormonal changes that occur in women, particularly the menstrual cycle. The hormonal fluctuations result in numerous physical and psychological disturbances, which occur in the premenstrual and menstrual phases. Existing evidence suggest that about 97% of young nulliparous women experience varying degrees of such disturbances 31 . These elicitable and visible features can confound or obscure the Prakriti assessment process. For example, premenstrual irritability occurring in a woman of Kapha Prakriti is confounding, since Kapha Prakriti individuals normally possess low irritability. Although the Ayurvedic physicians routinely enquire about the menstrual habits of patients while assessing the Prakriti , it would have been difficult for us to make similar enquiries to young, healthy women who volunteered to join this study. The health status of an individual was assessed based on the Ayurvedic criteria, that include; normal desire for food, easy digestion of ingested food, excretion of feces, excretion of urine, excretion of flatus, functioning of sensory organs, comfortable sleep, easy awakening and attainment of strength, bright complexion and longevity. Subjects with smoking habit, diabetes, hypertension and other chronic diseases were excluded from the study. Blood pressure (BP) was measured for each subject and BP > 130/90 mm of Hg were excluded from the study. Chronic systemic diseases such as rheumatoid arthritis, cancer, etc. and subjects having recent history of acute ailments such as fever due to infections were also excluded.
We followed three steps for the Prakriti assessment of each subjects. In the first stage, senior Ayurvedic physicians assessed the Prakriti of the subjects, applying classical Ayurveda parameters of Prakriti determination. In second stage, the same subjects were assessed using Ayusoft, a Prakriti software ( www.ayusoft.cdac.in ), which contains a comprehensive questionnaire, which had been developed based on the information from original Ayurvedic literature. In the third stage, another team of Ayurvedic physicians, who were not aware of the outcomes of assessment by senior physicians and Ayusoft, compared the Prakritis analysis. Subjects with ≥60% of single Prakriti dominance and having concordance in all the three stages were selected for the genome-wide analysis. Quantitative analysis of Prakriti was performed using Ayusoft along with traditional ayurvedic measures for the Prakriti assessment. The reason for considering ≥60% of a particular Prakriti as a dominant was mainly due to feasibility and concordance. Single dosha Prakriti with high percentage of one dosha rarely exist, hence most of the individuals possess dual-dosha Prakriti 12 . Therefore, we have considered subjects with ≥60% as single dosha dominant Prakriti . Subjects ≥60% of one dominant Prakriti were selected and blood was drawn after obtaining their informed written consent. A total of 3,416 healthy individuals were screened for their Prakriti , as per the details given above. From the total, 971 subjects who showed a predominant Prakriti of ≥60% were included in the analysis ( Figure S1 ).
High throughput genotyping, their quality control criteria and resequencing
DNA was isolated from the blood samples using standard protocol 32 . We randomly selected 262 Prakriti individuals for genotyping, using Genome-Wide Human SNP Array from Affymetrix (6.0), following manufacturer’s protocols. About 250 ng of genomic DNA was digested with Nsp I and Sty I restriction enzymes, followed by ligation of Nsp / Sty adaptors, using T4 DNA ligase. PCR was performed using the primers that are specific to these adopters. After checking the amplicons on 2% agarose gel, they were purified with deep-well plate using magnetic beads and the fragments were eluted using EB buffer, followed by quantification and fragmentation. The fragmented PCR products (<180 bp) were end-labeled using labeling kit. Labeled fragments were hybridized onto the Affymetrix (6.0) SNP arrays using hybridization cocktail. Hybridization was performed in hybridization oven for about 18 hrs at 50 °C. After hybridization, arrays were washed, stained, scanned and analyzed using Affymetrix Genotyping Console 2.0 and GeneChip® Operating Software (GCOS). The samples which passed the quality controls i.e. call rate >95% and CQC > 0.4 were considered. Affymetrix power tool (apt-geno-qc) was used for calculation of dm (dynamic model) value. The samples having dm.all_qc<0.83 were removed from further analysis and genotypes were fetched with Birdsuite software from Broad Institute 17 ( Figure S1 ).
Detection of technical artifacts
In order to validate the Affymetrix data set, we randomly selected 48 markers ( Table S3 ) from Affymetrix array and genotyped 48 individuals, who were already genotyped by Affymetrix array, using custom-designed VeraCode GoldenGate Genotyping Assay System (Illumina, San Diego, USA). Genotyping was performed according to the manufacturer’s (Illumina, San Diego, USA) instructions. The genotypes obtained by both the platforms were compared and checked for accuracy ( Figure S1 ).
Targeted resequencing
We sequenced the whole-exons and UTRs of PGM1 gene ( Figure S1 ) for randomly selected 43 Pitta and 35 Kapha individuals using Ion Torrent (Life Technologies, USA), following protocols of the manufacturer. Primer sequences were manufactured specifically for use with Ion AmpliSeq kits. The costume Ion AmpliSeq TM primer contains 35 amplicons in a single pool. For preparing amplicon libraries, about 10 ng of DNA was amplified (PCR) using AmpliSeq TM primer pools and Ion AmpliSeq TM HiFi master mix (Ion AmpliSeq kit version 2.0 Beta). The amplified products were pooled and treated with 2 μl of FuPa reagent. The amplicons were then ligated with adapters from the Ion Xpress TM barcoded adapters 17–64 kit according to the manufacturer’s instructions (Ion Torrent). After ligation, the amplicons were purified by Agencourt® AMPure® XP Reagent and additional amplification was performed to complete linkage between adapters and amplicons. In order to determine the library concentration, an Agilent 2100 Bioanalyzer high-sensitivity DNA kit (Agilent, Santa Clara, CA) was used to visualize the size range of the libraries. Equimolar concentrations of all the libraries were pooled and diluted. Using Ion One Touch TM 200 Template Kit v@ DL (Life Technologies, USA), emulsion PCR was carried out according to the manufacturer’s instructions. Ion Spheres (ISPs) were recovered according to the Ion Sphere Particles 200 recovery protocol. Sequencing was done following the Ion PGM TM 200 Sequencing Kit Protocol (version 6; Ion Torrent). The 318 sequencing chip was loaded and run on an Ion Torrent PGM (Ion Torrent). Base calling and alignment were performed using the Torrent Suite 3.0 software (Ion Torrent). In order to find the significant variation in the PGM1 whole exome data, we performed association analysis using plink software 26 and variations are annotated on EnsEMBL-BioMart.
Sanger sequencing
To validate and replicate the Pitta associated SNP (rs11208257), Sanger sequencing was carried out for 496 Prakriti samples (246 Vata, 116 Pitta and 134 Kapha) along with randomly selected 1108 Indian samples. Pair of primers (Forward primer: 5′- GCACGTTTCTTACAGCAGCT-3′ and Reverse primer: 5′-ACCTTACCTTGTACCCCAGC-3′) were designed, synthesized and PCR was performed on the GeneAmp 9700 Thermal Cycler (Applied Biosystems, Perkin-Elmer) using the following cycling conditions: 95 °C for 5 min, 35 cycles at 95 °C for 30 s, 58 °C for 30 s, 72 °C for 2 min and a final extension at 72 °C for 7 min. Amplicons were purified using with USB ExoSAP-IT (Affymetrix) according to the manufacturers instructions. The purified products were directly sequenced using the Big Dye Terminator cycle sequencing kit (Applied Biosystems, Foster City, CA, USA) and analyzed using 3730 DNA Analyzer (Applied Biosystems, Foster City, CA, USA)( Figure S1 ). The genotypes were noted and statistical analysis was performed with plink 26 and R.
Indian, HapMap and 1000 genome project sample details
For comparative analysis, we used Affymetrix (6.0) array data of 297 well-classified Indian samples with known lingustic and ethnic affiliations i.e. 150 Dravidians, 80 Indo-European, 35 Austro-Asiatic, 27 Tibeto-Burman and 5 Great Andamanese. In addition, 15 trios of Dravidian (Vysya, Madiga, Mala; 5 each) and 15 trios of Indo-European (Kshatriya, Brahmin and Kashmiri Pandit; 5 each) were used for imputation 19 , 20 . We followed the same procedure (mentioned above) for extraction of genotypes and CQC measures. The list of population and their details are given in Table S6 . We have also used 1184 HapMap ( ftp://ftp.ncbi.nlm.nih.gov/hapmap/genotypes/2009-01_phaseIII/plink_format/ ) and 1000 genome project data ( ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20130502/ ) for imputation and comparative analysis.
Imputation and their relationship with ancestry
Imputation was performed for the missing genotypes using Beagle-v3.3.2 software 18 . In order to check the power of correct imputation, we randomly masked 2%, 5% and 10% genotype of chromosome 22 in 207 unrelated Indian population samples. We imputed masked genotypes with four types of reference i.e. Indian triose (15 trios of Dravidian and 15 trios of Indo-European), HapMap samples (CEU, YRI, CHB, CHS and JPT), Indian + HapMap samples and samples of 1000 genome project with South-Asian ancestry (BEB, GIH, ITU, PJL and STU). Accuracy of imputation was calculated by comparing imputed and true genotype in 110 simulations for 2%, 5% and 10% masked data. To perform the above analysis, we used a perl script.
To impute missing genotype in Prakriti samples, we followed two steps; In the first step, imputation and phasing was done for 15 trios of Dravidian (Vysya, Madiga, Mala; 5 trios each) and 15 trios of Indo-European (Kshatriya, Brahmin and Kashmiri Pandit; 5 trios each) together without reference population. SNPs which do not follow Mendelian rule in trios were checked and masked with Beagle utility program 18 . Number of SNPs per family, which do not follow Mendelian consistency are given in Table S5 . In the second step, we performed imputation of unrelated population samples (Dravidian, Indo-European and Austro-Asiatic), with imputed familial (trios) samples as reference. Further, we imputed Prakriti samples with reference of imputed trios and unrelated Indian population samples and selected only those markers which were having R 2 > 0.95 for further analysis.
Population stratification
Principal component analysis was done with Eigensoft Package 21 . Convertf was used for converting plink ped file to Eigenstrat format. We pruned the SNPs on the basis of their Linkage Disequilibrium (r2 > 0.75) before running PCA by using Eigensoft’s killr 2 option. Ten eigenvectors were fetched. To find the ancestry of 245 Prakriti samples, we used 297 known ancestry of Indian population dataset (previously published) and performed the PCA. Stratification was checked and 40 outlier samples were excluded with cutoff sigma value ≥0.6 (default value) on 1–10 eigenvectors in 10 iterations ( Figure S1 ).
Association analysis
Plink was used for association analysis 21 . Imputed Beagle file were converted into plink ped file. Association analysis was performed for the Prakritis . Since there are no case control groups in the present study, we compared one Prakriti against the other two Prakritis (Vata vs. Pitta and Kapha, Pitta vs. Vata and Kapha and Kapha vs. Vata and Pitta) and calculated p-value from theoretical distribution. In order to exclude the markers, which could be in association by chance, we also performed adaptive permutation approach (empirical distribution) for maximum 10 6 iteration withplink and considered only those markers who achieved maximum 10 6 permutations and have p-value ≤ 1 × 10 −5 in theoretical distribution ( Figure S1 ).
Addressing issue of population stratification as possible confounder in association analysis
Even subtle stratification can cause spurious association; hence we used EIGENSTRAT software 21 for correcting association chi-square value on 10 eigenvector and to find its confounding effect. Initially we excluded 385, 404 SNPs with r2 > 0.75 and calculated eigenvector with remaining 405, 782 SNPs with SMARTPCA. Further we used these same 10 eigenvector for correction of chi-square value with EIGENSTRAT ( Figure S1 ).
To address this issue, we also used EMMAX and GCTA tools 27 , 28 . Both statistical methods consider genetic structure in association analysis. Hence, we expected major changes in p-value of 52 SNPs. First, we generated IBS matrix implemented in EMMAX and then used it with 10 eigenvector (generated with SMARTPCA) as covariate to calculate p-value with variance component model (implemented in EMMAX). To calculate mixed-model association p-value (implemented in GCTA), first, we calculated genetic relationship matrix and 10 eigenvectors with GCTA; and used it in calculation of p-value ( Figure S1 ).
Statistical determination of Prakriti in subjects
In order to prove the power of these markers in samples of unknown Prakriti percentage, we generated a statistical model ( Figure S1 ). First, this model applied to 205 Ayur samples and then replicated in Indian population data set with unknown Prakriti . For this, we calculated the weight for the genotype of each marker associated with the Prakriti . Suppose, if the frequency of genotype g in Prakriti p is f p then the weight of g (W gp ) can be calculated with equation (1)
Un-standardized total weight of the Prakriti W vs for a sample s with n number of associated markers for p Prakriti can be calculated using equation (2)
Hence, for a single sample there will be 3 weights W vs , W ps and W ks corresponding to Vata, Pitta and Kapha using equation (2) . For making weights comparable, we standardized by subtracting with mean and dividing it by standard deviation. Mean and standard deviation were calculated from total weight of each sample for each markers corresponding to each Prakriti . If total number of sample is N then standardized weight can be calculated using equation (3)
Prakriti is relative proposition (tridosha), so we calculated the differences of standardized weight for all 6 permutations; Δ VP , Δ VK , Δ PV , Δ PK , Δ KP and Δ KP for each samples and calculated representative statistics R p . For example, representative statistics for Kapha R k can be calculated using equation (4)
Since multiplication of 2 negative values is positive, the R p value could be positive for 2 negative Δ values. Hence, we considered only those R p values, which have both Δ value positive. Moreover, we consider only those samples which have R p ≥ 3 to find dominant Prakriti . We applied this model to Indian population and selected 37 samples on the basis of R p and Δ values.
Phenotype and genotype correlation
We considered markers within 10 kb flanking region of gene as genic and other as non-genic. Physical location of the genes (knownGene.txt.gz) and SNPs (snp135.txt.gz) were fetched from http://hgdownload.soe.ucsc.edu/goldenPath/hg19/database/ and; windowbed(Bedtools: https://code.google.com/p/bedtools/ ) was used to find the SNPs within 10 kb flanking region of the genes. Only genic markers were used for genotype-phenotype correlation. Genic SNPs were selected and considered for further analysis. To correlate the function of associated markers with characteristics of the individual Prakriti , we checked in KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway and Reactome event using NCBI2R R package.We used SIFT algorithm ( http://sift.jcvi.org ) for predicting the effects of non-synonomous variant (rs11208257) on protein function.
Additional Information
How to cite this article : Govindaraj, P. et al . Genome-wide analysis correlates Ayurveda Prakriti . Sci. Rep. 5 , 15786; doi: 10.1038/srep15786 (2015).
Sharma, P. V. Caraka Samhita. (Chaukhamba Orientalia, Varanasi, India,(1994).
Dwarakanath, C. The Fundamental Principles of Ayurveda. (Krishnadas Academy, Varanasi, India, 1952).
Hankey, A. Ayurvedic physiology and etiology: Ayurvedo Amritanaam. The doshas and their functioning in terms of contemporary biology and physical chemistry. J. Altern.Complement Med. 7, 567–574 (2001).
Article CAS Google Scholar
Hankey, A. A test of the systems analysis underlying the scientific theory of Ayurveda's Tridosha. J. Altern.Complement Med. 11, 385–390 (2005).
Article Google Scholar
Jayasundar, R. Ayurveda: a distinctive approach to health and disease. Curr. Sci. 98, 908–914 (2010).
Google Scholar
Patwardhan, B. AyuGenomics–Integration for customized medicine. Indian J. Nat. Prod. Resour. 19, 16–23 (2003).
Bhushan, P., Kalpana, J. & Arvind, C. Classification of human population based on HLA gene polymorphism and the concept of Prakriti in Ayurveda. J. Altern. Complement Med. 11, 349–353 (2005).
Ghodke, Y., Joshi, K. & Patwardhan, B. Traditional Medicine to Modern Pharmacogenomics: Ayurveda Prakriti Type and CYP2C19 Gene Polymorphism Associated with the Metabolic Variability. Evid. Based Complement. Alternat. Med. 2011, 249528 (2011).
Aggarwal, S. et al. EGLN1 involvement in high-altitude adaptation revealed through genetic analysis of extreme constitution types defined in Ayurveda. Proc. Natl. Acad. Sci. 107, 18961–18966 (2010).
Article CAS ADS Google Scholar
Juyal, R. C. et al. Potential of ayurgenomics approach in complex trait research: leads from a pilot study on rheumatoid arthritis. PloS one. 7, e45752 (2012).
Rotti, H. et al. Immunophenotyping of normal individuals classified on the basis of human dosha prakriti. J. Ayurveda Integr. Med. 5, 43–49 (2014).
Article ADS Google Scholar
Rotti, H. et al. Determinants of prakriti, the human constitution types of Indian traditional medicine and its correlation with contemporary science. J. Ayurveda Integr. Med. 5, 167–175 (2014).
ADS PubMed PubMed Central Google Scholar
Rotti, H. et al. DNA methylation analysis of phenotype specific stratified Indian population. J. Transl. Med. 13, 151 (2015).
Mahalle, N. P., Kulkarni, M. V., Pendse, N. M. & Naik, S. S. Association of constitutional type of Ayurveda with cardiovascular risk factors, inflammatory markers and insulin resistance. J. Ayurveda Integr. Med. 3, 150–157 (2012).
Prasher, B. et al. Whole genome expression and biochemical correlates of extreme constitutional types defined in Ayurveda. J. Transl. Med. 6, 48 (2008).
Bhalerao, S., Deshpande, T. & Thatte, U. Prakriti (Ayurvedic concept of constitution) and variations in platelet aggregation. BMC Complement. Altern. Med. 12, 248 (2012).
Korn, J. M. et al. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat. Genet. 40, 1253–1260 (2008).
Browning, B. L. & Browning, S. R. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am. J. Hum. Genet. 84, 210–223 (2009).
Reich, D., Thangaraj, K., Patterson, N., Price, A. L. & Singh, L. Reconstructing Indian population history. Nature 461, 489–494 (2009).
Moorjani, P. et al. Genetic evidence for recent population mixture in India. Am. J. Hum. Genet. 93, 422–438 (2013).
Patterson, N., Price, A. L. & Reich, D. Population structure and eigenanalysis. PLoS Genet. 2, e190 (2006).
Freedman, M. L. et al. Assessing the impact of population stratification on genetic association studies. Nat. Genet. 36, 388–393 (2004).
Marchini, J., Cardon, L. R., Phillips, M. S. & Donnelly, P. The effects of human population structure on large genetic association studies. Nat. Genet. 36, 512–517 (2004).
Helgason, A., Yngvadottir, B., Hrafnkelsson, B., Gulcher, J. & Stefansson, K. An Icelandic example of the impact of population structure on association studies. Nat. Genet. 37, 90–95 (2005).
Campbell, C. D. et al. Demonstrating stratification in a European American population. Nat. Genet. 37, 868–872 (2005).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Kang, H. M. et al. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 42, 348–354 (2010).
Yang, J., Lee, S. H., Goddard, M. E. & Visscher, P. M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88, 76–82 (2011).
Huang, R. S. et al. Identification of genetic variants contributing to cisplatin-induced cytotoxicity by use of a genomewide approach. Am. J. Hum. Genet. 81, 427–437 (2007).
Jorgenson, E. & Witte, J. S. A gene-centric approach to genome-wide association studies. Nat. Rev. Genet. 7, 885–891 (2006).
Kumar, P. & Malhotra, N. Jeffcoate's Principles of Gynecology 7th edn. (Jaypee Brothers Medical Publications, New Delhi, India, 2008).
Thangaraj, K. et al. CAG repeat expansion in the androgen receptor gene is not associated with male infertility in Indian populations. J. Androl. 23, 815–818 (2002).
CAS PubMed Google Scholar
Download references
Acknowledgements
This work was supported by the Office of the Principal Scientific Advisor to the Government of India; Department of Science and Technology (DST), Government of India (PRNSA/ADV/AYURVEDA/4/2007). KT was also supported by CSIR Network project - GENESIS (BSC0121), Government of India. We acknowledge the help of Dr. Ketaki Bapat for her constant support throughout the project tenure. We thank Dr. David Reich for his valuable suggestions.
Author information
Govindaraj Periyasamy and Nizamuddin Sheikh contributed equally to this work.
Authors and Affiliations
CSIR-Centre for Cellular and Molecular Biology, Hyderabad, Telangana, India
Periyasamy Govindaraj, Sheikh Nizamuddin, Anugula Sharath, Vuskamalla Jyothi & Kumarasamy Thangaraj
School of Life Sciences, Manipal University, Manipal, Karnataka, India
Harish Rotti, Ritu Raval, Puthiya M. Gopinath, Kapaettu Satyamoorthy & Marthanda Varma Sankaran Valiathan
Shri Dharmasthala Manjunatheshwara College of Ayurveda, Udupi, Karnataka, India
Jayakrishna Nayak, Balakrishna K. Bhat & B. V. Prasanna
Sinhgad College of Engineering, Pune, Maharashtra, India
Pooja Shintre, Mayura Sule, Kalpana S. Joshi & Amrish P. Dedge
Foundation for Revitalization of Local Health Traditions, Bangalore, Karnataka, India
Ramachandra Bharadwaj & G. G. Gangadharan
Department of Statistics, Manipal University, Manipal, Karnataka, India
Sreekumaran Nair
Interdisciplinary School of Health Sciences, University of Pune, Pune, Maharashtra, India
Bhushan Patwardhan
Department of Molecular Reproduction, Development and Genetics, Indian Institute of Science, Bangalore, Karnataka, India
Paturu Kondaiah
You can also search for this author in PubMed Google Scholar
Contributions
M.V.S.V., K.S. and K.T. conceived the idea. K.T. design the study and provided reagents. K.T., K.S., M.V.S.V., P.K., B.P. and P.M.G. supervised the study. J.N., B.K.B., B.V.P., A.P.D., R.B., G.G.G. and S.K.N. screened and selected the Prakriti samples. H.R., R.R., P.S., M.S. and K.S.J. collected blood samples and extracted DNA. P.G. performed genotyping and DNA sequencing with help of S.N., A.S. and V.J.. S.N. analyzed data under supervision of K.T.. P.G., S.N. and K.T. wrote manuscript and all the authors reviewed the manuscript.
Ethics declarations
Competing interests.
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary information, rights and permissions.
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
Reprints and permissions
About this article
Cite this article.
Govindaraj, P., Nizamuddin, S., Sharath, A. et al. Genome-wide analysis correlates Ayurveda Prakriti . Sci Rep 5 , 15786 (2015). https://doi.org/10.1038/srep15786
Download citation
Received : 17 June 2015
Accepted : 01 October 2015
Published : 29 October 2015
DOI : https://doi.org/10.1038/srep15786
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
This article is cited by
Diabetic yoga protocol improves glycemic, anthropometric and lipid levels in high risk individuals for diabetes: a randomized controlled trial from northern india.
- Navneet Kaur
- Vijaya Majumdar
- Hongasandra Ramarao Nagendra
Diabetology & Metabolic Syndrome (2021)
Exploring the signature gut and oral microbiome in individuals of specific Ayurveda prakriti
- Tirumalapura Vijayanna Shalini
- Apoorva Jnana
- G G Gangadharan
Journal of Biosciences (2021)
Predictive, Preventive and Personalized Medicine: Leads From Ayurvedic Concept of Prakriti (Human Constitution)
- Rohit Sharma
- Pradeep Kumar Prajapati
Current Pharmacology Reports (2020)
Understanding the association between the human gut, oral and skin microbiome and the Ayurvedic concept of prakriti
- Diptaraj Chaudhari
- Dhiraj Dhotre
- Yogesh Shouche
Journal of Biosciences (2019)
By submitting a comment you agree to abide by our Terms and Community Guidelines . If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing newsletter — what matters in science, free to your inbox daily.
Hybrid performance evaluation and genome-wide association analysis of root system architecture in a maize association population
Affiliations.
- 1 College of Resources and Environmental Sciences, National Academy of Agriculture Green Development, Key Laboratory of Plant-Soil Interactions of MOE, China Agricultural University, Beijing, China.
- 2 Global Institute for Food Security, University of Saskatchewan, Saskatoon, Canada.
- 3 Key Laboratory of Plant Functional Genomics of the Ministry of Education, Yangzhou University, Yangzhou, China.
- 4 College of Resources and Environment, Jilin Agricultural University, Changchun, China.
- 5 Sanya Institute of China Agricultural University, Sanya, China.
- 6 College of Resources and Environmental Sciences, National Academy of Agriculture Green Development, Key Laboratory of Plant-Soil Interactions of MOE, China Agricultural University, Beijing, China. [email protected].
- 7 Sanya Institute of China Agricultural University, Sanya, China. [email protected].
- PMID: 37606710
- DOI: 10.1007/s00122-023-04442-7
The genetic architecture of RSA traits was dissected by GWAS and coexpression networks analysis in a maize association population. Root system architecture (RSA) is a crucial determinant of water and nutrient uptake efficiency in crops. However, the maize genetic architecture of RSA is still poorly understood due to the challenges in quantifying root traits and the lack of dense molecular markers. Here, an association mapping panel including 356 inbred lines were crossed with a common tester, Zheng58, and the test crosses were phenotyped for 12 RSA traits in three locations. We observed a 1.3 ~ sixfold phenotypic variation for measured RSA in the association panel. The association panel consisted of four subpopulations, non-stiff stalk (NSS) lines, stiff stalk (SS), tropical/subtropical (TST), and mixed. Zheng58 × TST has a 2.1% higher crown root number (CRN) and 8.6% less brace root number (BRN) than Zheng58 × NSS and Zheng58 × SS, respectively. Using a genome-wide association study (GWAS) with 1.25 million SNPs and correction for population structure, 191 significant SNPs were identified for root traits. Ninety (47%) of the significant SNPs showed positive allelic effects, and 101 (53%) showed negative effects. Each locus could explain 0.39% to 11.8% of phenotypic variation. By integrating GWAS results and comparing coexpression networks, 26 high-priority candidate genes were identified. Gene GRMZM2G377215, which belongs to the COBRA-like gene family, affected root growth and development. Gene GRMZM2G468657 encodes the aspartic proteinase nepenthesin-1, related to root development and N-deficient response. Collectively, our research provides progress in the genetic dissection of root system architecture. These findings present the further possibility for the genetic improvement of root traits in maize.
© 2023. The Author(s), under exclusive licence to Springer-Verlag GmbH Germany, part of Springer Nature.
- Crops, Agricultural
- Genome-Wide Association Study*
- Zea mays* / genetics
Grants and funding
- 321CXTD443/Hainan Provincial Natural Science Foundation of China
- 31971948/National Natural Science Foundation of China
- 31972485/National Natural Science Foundation of China
Open Access Theses and Dissertations
Thursday, April 18, 8:20am (EDT): Searching is temporarily offline. We apologize for the inconvenience and are working to bring searching back up as quickly as possible.
Advanced research and scholarship. Theses and dissertations, free to find, free to use.
Advanced search options
Browse by author name (“Author name starts with…”).
Find ETDs with:
Written in any language English Portuguese French German Spanish Swedish Lithuanian Dutch Italian Chinese Finnish Greek Published in any country US or Canada Argentina Australia Austria Belgium Bolivia Brazil Canada Chile China Colombia Czech Republic Denmark Estonia Finland France Germany Greece Hong Kong Hungary Iceland India Indonesia Ireland Italy Japan Latvia Lithuania Malaysia Mexico Netherlands New Zealand Norway Peru Portugal Russia Singapore South Africa South Korea Spain Sweden Switzerland Taiwan Thailand UK US Earliest date Latest date
Sorted by Relevance Author University Date
Only ETDs with Creative Commons licenses
Results per page: 30 60 100
October 3, 2022. OATD is dealing with a number of misbehaved crawlers and robots, and is currently taking some steps to minimize their impact on the system. This may require you to click through some security screen. Our apologies for any inconvenience.
Recent Additions
See all of this week’s new additions.
About OATD.org
OATD.org aims to be the best possible resource for finding open access graduate theses and dissertations published around the world. Metadata (information about the theses) comes from over 1100 colleges, universities, and research institutions . OATD currently indexes 7,241,108 theses and dissertations.
About OATD (our FAQ) .
Visual OATD.org
We’re happy to present several data visualizations to give an overall sense of the OATD.org collection by county of publication, language, and field of study.
You may also want to consult these sites to search for other theses:
- Google Scholar
- NDLTD , the Networked Digital Library of Theses and Dissertations. NDLTD provides information and a search engine for electronic theses and dissertations (ETDs), whether they are open access or not.
- Proquest Theses and Dissertations (PQDT), a database of dissertations and theses, whether they were published electronically or in print, and mostly available for purchase. Access to PQDT may be limited; consult your local library for access information.
Literary Theory and Criticism
Home › Literature › Analysis of Jean Rhys’s Novel Wide Sargasso Sea
Analysis of Jean Rhys’s Novel Wide Sargasso Sea
By NASRULLAH MAMBROL on May 29, 2019 • ( 1 )
When Wide Sargasso Sea, her last novel, was published, Jean Rhys (24 August 1890 – 14 May 1979) was described in The New York Times as the greatest living novelist. Such praise is overstated, but Rhys’s fiction, long overlooked by academic critics, is undergoing a revival spurred by feminist studies. Rhys played a noteworthy role in the French Left Bank literary scene in the 1920’s, and between 1927 and 1939, she published four substantial novels and a number of jewel-like short stories. Although she owes her current reputation in large measure to the rising interest in female writers and feminist themes, her work belongs more properly with the masters of literary impressionism: Joseph Conrad , Ford Madox Ford, Marcel Proust, and James Joyce. She began to publish her writing under the encouragement of her intimate friend Ford Madox Ford, and she continued to write in spite of falling out of favor with his circle. As prizes and honors came to her in her old age after the publication of Wide Sargasso Sea , it must have given her grim satisfaction to realize that she had attained entirely by her own efforts a position as a writer at least equal to that of her erstwhile friends.

Jean Rhys’s first novel, Quartet, reflects closely her misadventures with Ford Madox Ford. The heroine, Marya Zelli, whose husband is in prison, moves in with the rich and respectable Hugh and Lois Heidler. Hugh becomes Marya’s lover, while Lois punishes her with petty cruelties. The central figure is a woman alone, penniless, exploited, and an outsider. In her next novel, After Leaving Mr. Mackenzie, the central figure, Julia Martin, breaks off with her rich lover, Mr. Mackenzie, and finds herself financially desperate. Voyage in the Dark tells the story of Anna Morgan, who arrives in England from the West Indies as an innocent young girl, has her first affair as a chorus girl, and descends through a series of shorter and shorter affairs to working for a masseuse. In Good Morning, Midnight, the alcoholic Sasha Jensen, penniless in Paris, remembers episodes from her past which have brought her to this sorry pass. All four of these novels show a female character subject to financial, sexual, and social domination by men and “respectable” society. In all cases, the heroine is passive, but “sentimental.” The reader is interested in her feelings, rather than in her ideas and accomplishments. She is alienated economically from any opportunity to do meaningful and justly rewarding work. She is an alien socially, either from a foreign and despised colonial culture or from a marginally respectable social background. She is literally an alien or foreigner in Paris and London, which are cities of dreadful night for her. What the characters fear most is the final crushing alienation from their true identities, the reduction to some model or type imagined by a foreign man. They all face the choice of becoming someone’s gamine, garçonne , or femme fatale, or of starving to death, and they all struggle against this loss of personal identity. After a silence of more than twenty years, Rhys returned to these same concerns in her masterpiece, Wide Sargasso Sea . While the four early novels are to a large degree autobiographical, Wide Sargasso Sea has a more literary origin, although it, too, reflects details from the author’s personal life.
Wide Sargasso Sea
Wide Sargasso Sea requires a familiarity with Charlotte Brontë’s Jane Eyre (1847). In Brontë’s novel, Jane is prevented from marrying Rochester by the presence of a madwoman in the attic, his insane West Indian wife who finally perishes in the fire which she sets, burning Rochester’s house and blinding him, but clearing the way for Jane to wed him. The madwoman in Jane Eyre is depicted entirely from the exterior. It is natural that the mad West Indian wife, when seen only through the eyes of her English rival and of Rochester, appears completely hideous and depraved. Indeed, when Jane first sees the madwoman in chapter 16 of the novel, she cannot tell whether it is a beast or a human being groveling on all fours. Like a hyena with bloated features, the madwoman attacks Rochester in this episode.
Wide Sargasso Sea is a sympathetic account of the life of Rochester’s mad wife, ranging from her childhood in the West Indies, her Creole and Catholic background, and her courtship and married years with the deceitful Rochester, to her final descent into madness and captivity in England. Clearly, the predicament of the West Indian wife resembles that of Rhys herself in many ways. In order to present the alien wife’s case, she has written a “counter-text,” an extension of Brontë’s novel filling in the “missing” testimony, the issues over which Brontë glosses.
Wide Sargasso Sea consists of two parts. Part 1 is narrated by the girl growing up in Jamaica who is destined to become Rochester’s wife. The Emancipation Act has just been passed (the year of that imperial edict was 1833) and the blacks on the island are passing through a period of so-called apprenticeship which should lead to their complete freedom in 1837. This is a period of racial tension and anxiety for the privileged colonial community. Fear of black violence runs high, and no one knows exactly what will happen to the landholders once the blacks are emancipated. The girlish narrator lives in the interface between the privileged white colonists and the blacks. Although a child of landowners, she is impoverished, clinging to European notions of respectability, and in constant fear. She lives on the crumbling estate of her widowed mother. Her closest associate is Christophine, a Martinique obeah woman, or Voodoo witch. When her mother marries Mr. Mason, the family’s lot improves temporarily, until the blacks revolt, burning their country home, Coulibri, and killing her half-witted brother. She then attends a repressive Catholic school in town, where her kindly colored “cousin” Sandi protects her from more hostile blacks.
Part 2 is narrated by the young Rochester on his honeymoon with his bride to her country home. Wherever appropriate, Rhys follows the details of Brontë’s story. Rochester reveals that his marriage was merely a financial arrangement. After an uneasy period of passion, Rochester’s feelings for his bride begin to cool. He receives a letter of denunciation accusing her of misbehavior with Sandi and revealing that madness runs in the family. To counter Rochester’s growing hostility, the young bride goes to her former companion, the obeah woman Christophine, for a love potion. The nature of the potion is that it can work for one night only. Nevertheless, she administers it to her husband. His love now dead, she is torn from her native land, transported to a cruel and loveless England, and maddeningly confined. Finally, she takes candle in hand to fire Rochester’s house in suicidal destruction.
In Brontë’s novel, the character of the mad wife is strangely blank, a vacant slot in the story. Her presence is essential, and she must be fearfully hateful, so that Jane Eyre has no qualms about taking her place in Rochester’s arms, but the novel tells the reader almost nothing else about her. Rhys fills in this blank, fleshing out the character, making her live on a par with Jane herself. After all, Brontë tells the reader a great deal about Jane’s painful childhood and education; why should Rhys not supply the equivalent information about her dark rival?
It is not unprecedented for a writer to develop a fiction from another writer’s work. For example, T. H. White’s Mistress Masham’s Repose (1946) imagines that some of Jonathan Swift’s Lilliputians were transported to England, escaped captivity, and established a thriving colony in an abandoned English garden, where they are discovered by an English schoolgirl. Her intrusion into their world is a paradigm of British colonial paternalism, finally overcome by the intelligence and good feeling of the girl. This charming story depends on Swift’s fiction, but the relationship of White’s work to Swift’s is completely different from the relationship of Rhys’s work to Brontë’s. Rhys’s fiction permanently alters one’s understanding of Jane Eyre . Approaching Brontë’s work after Rhys’s, one is compelled to ask such questions as, “Why is Jane so uncritical of Rochester?” and, “How is Jane herself like the madwoman in the attic?” Rhys’s fiction reaches into the past and alters Brontë’s novel.
Rhys’s approach in Wide Sargasso Sea was also influenced by FordMadox Ford and, through Ford, Joseph Conrad. In the autumn of 1924, when Rhys first met Ford, he was writing Joseph Conrad: A Personal Remembrance . Some thirty years earlier, when Joseph Conrad was just beginning his career as a writer, his agent had introduced him to Ford in hopes that they could work in collaboration, since Conrad wrote English (a language he had adopted only as an adult) with great labor. Ford and Conrad produced The Inheritors (1901) and Romance (1903) as coauthors. During their years of association, Ford had some hand in the production of several works usually considered Conrad’s sole effort, although it has never been clear to what degree Ford participated in the creation of the fiction of Conrad’s middle period. About 1909, after Ford’s disreputable ways had become increasingly offensive to Conrad’s wife, the two men parted ways. Immediately after Conrad’s death in 1924, however, Ford rushed into print his memoir of the famous author. His memoir of Conrad is fictionalized and hardly to be trusted as an account of their association in the 1890’s, but it sheds a great deal of light on what Ford thought about writing fiction in 1924, when he was beginning his powerful Tietjens tetralogy and working for the first time with Rhys. Ford claimed that he and Conrad invented literary impressionism in English. Impressionist fiction characteristically employs limited and unreliable narration, follows a flow of associated ideas leaping freely in time and space, aims to render the impression of a scene vividly so as to make the reader see it as if it were before his eyes, and artfully selects and juxtaposes seemingly unrelated scenes and episodes so that the reader must construct the connections and relationships that make the story intelligible. These are the stylistic features of Rhys’s fiction, as well as of Ford’s The Good Soldier (1915), Conrad ’s Heart of Darkness (1902), Henry James ’s The Turn of the Screw (1898), and Joyce ’s Ulysses (1922).
An “affair”—the mainspring of the plot in an impressionist novel—is some shocking or puzzling event which has already occurred when the story begins. The reader knows what has happened, but he does not understand fully why and how it happened. The story proceeds in concentric rings of growing complication as the reader finds something he thought clear-cut becoming more and more intricate. In Conrad ’s Lord Jim (1900), the affair is the scandalous abandonment of the pilgrim ship by the English sailor. In The Good Soldier , it is the breakup of the central foursome, whose full infidelity and betrayal are revealed only gradually. Brontë’s Jane Eyre provided Rhys with an impressionist “affair” in the scene in which the mad West Indian wife burns Rochester’s house, blinding him and killing herself. Like Conrad’s Marlow, the storyteller who sits on the veranda mulling over Jim’s curious behavior, or The Good Soldier ’s narrator Dowell musing about the strange behavior of Edward Ashburnham, Rhys takes up the affair of Rochester and reworks it into ever richer complications, making the initial judgments in Jane Eyre seem childishly oversimplified. “How can Jane simply register relief that the madwoman is burned out of her way? There must be more to the affair than that,” the secondary fiction suggests.
One of the most important features of literary impressionism is the highly constructive activity which it demands of the reader. In a pointillist painting, small dots of primary colors are set side by side. At a certain distance from the canvas, these merge on the retina of the eye of the viewer into colors and shapes which are not, in fact, drawn on the canvas at all. The painting is constructed in the eyes of each viewer with greater luminosity than it would have were it drawn explicitly. In order to create such a shimmering haze in fiction, Ford advises the use of a limited point of view which gives the reader dislocated fragments of remembered experience. The reader must struggle constantly to fit these fragments into a coherent pattern. The tools for creating such a verbal collage are limited, “unreliable” narration, psychological time-shifts, and juxtaposition. Ford observes that two apparently unrelated events can be set side by side so that the reader will perceive their connection with far greater impact than if the author had stated such a connection openly. Ford advises the impressionist author to create a verbal collage by unexpected selection and juxtaposition, and Wide Sargasso Sea makes such juxtapositions on several levels. On the largest scale, Wide Sargasso Sea is juxtaposed with Jane Eyre , so that the two novels read together mean much more than when they are read independently. This increase of significance is what Ford called the “unearned increment” in impressionist art. Within Wide Sargasso Sea, part 1 (narrated by the West Indian bride) and part 2 (narrated by Rochester) likewise mean more in juxtaposition than when considered separately. Throughout the text, the flow of consciousness of the storytellers cunningly shifts in time tojuxtapose details which mean more together than they would in isolation.
Because Wide Sargasso Sea demands a highly constructive reader, it is, like The Good Soldier or Heart of Darkness, an open fiction. When the reader completes Jane Eyre , the mystery of Rochester’s house has been revealed and purged, the madwoman in the attic has been burned out, and Jane will live, the reader imagines, happily ever after. Jane Eyre taken in isolation is a closed fiction. Reading Wide Sargasso Sea in juxtaposition to Jane Eyre , however, opens the latter and poses questions which are more difficult to resolve: Is Jane likely to be the next woman in the attic? Why is a cripple a gratifying mate for Jane? At what price is her felicity purchased?
The Doppelgänger , twin, or shadow-character runs throughout Rhys’s fiction. All of her characters seem to be split personalities. There is a public role, that of the approved “good girl,” which each is expected to play, and there is the repressed, rebellious “bad girl” lurking inside. If the bad girl can be hidden, the character is rewarded with money, love, and social position. Yet the bad girl will sometimes put in an appearance, when the character drinks too much or gets excited or angry. When the dark girl appears, punishment follows, swift and sure. This is the case with Marya Zelli in Quartet, Julia Martin in After Leaving Mr. Mackenzie, Anna Morgan in Voyage in the Dark, and Sasha Jensen in Good Morning, Midnight. It is also the case in Brontë’s Jane Eyre. The education of Jane Eyre consists of repressing those dark, selfish impulses that Victorian society maintained “good little girls” should never feel. Jane succeeds in stamping out her “bad” self through a stiff British education, discipline, and self-control. She kills her repressed identity, conforms to society’s expectations, and gets her reward—a crippled husband and a burned-out house. Rhys revives the dark twin, shut up in the attic, the naughty, wild, dark, selfish, bestial female. She suggests that the struggle between repressed politeness and unrepressed self-interest is an ongoing process in which total repression means the death of a woman’s identity.

Principal long fiction Postures, 1928 (pb. in U.S. as Quartet, 1929); After Leaving Mr. Mackenzie, 1930; Voyage in the Dark, 1934; Good Morning, Midnight, 1939; Wide Sargasso Sea, 1966.
Other major works Sort Fiction: The Left Bank and Other Stories, 1927; Tigers Are Better-Looking, 1968; Sleep It Off, Lady, 1976; The Collected Short Stories, 1987. Nonfiction: Smile Please: An Unfinished Autobiography, 1979; The Letters of Jean Rhys, 1984 (also known as Jean Rhys: Letters, 1931-1966).
Bibliography Angier, Carole. Jean Rhys: Life and Work. Boston: Little, Brown, 1990. Benstock, Shari. Women of the Left Bank: Paris, 1900-1940. Austin: University of Texas Press, 1986. Harrison, Nancy R. Jean Rhys and the Novel as Women’s Text. Chapel Hill: University of North Carolina Press, 1988. Malcolm, Cheryl Alexander, and David Malcolm. Jean Rhys: A Study of the Short Fiction. New York: Twayne, 1996. Staley, Thomas. Jean Rhys: A Critical Study. London: Macmillan, 1979.
Share this:
Categories: Literature , Novel Analysis
Tags: After Leaving Mr. Mackenzie , Analysis of Jean Rhys's Novel Wide Sargasso Sea , Analysis of Jean Rhys's Novels , Analysis of Jean Rhys's Wide Sargasso Sea , Articles of Jean Rhys's Novels , Articles of Jean Rhys's Wide Sargasso Sea , Character of Jean Rhys's Novels , Character of Jean Rhys's Wide Sargasso Sea , Criticism of Jean Rhys's Novels , Criticism of Jean Rhys's Wide Sargasso Sea , Essays of Jean Rhys's Novels , Essays of Jean Rhys's Wide Sargasso Sea , Jean Rhys , Literary Criticism , Literary Theory , Notes of Jean Rhys's Novels , Notes of Jean Rhys's Wide Sargasso Sea , Plot of Jean Rhys's Novels , Plot of Jean Rhys's Wide Sargasso Sea , Postures , Quartet , Study Guide of Jean Rhys's Novels , Study Guide of Jean Rhys's Wide Sargasso Sea , Summary of Jean Rhys's Novels , Summary of Jean Rhys's Wide Sargasso Sea , Themes of Jean Rhys's Novels , Themes of Jean Rhys's Wide Sargasso Sea , Voyage in the Dark , Wide Sargasso Sea , Wide Sargasso Sea Project , Wide Sargasso Sea Themes , Wide Sargasso Sea Thesis
Related Articles

i’m afraid I find this rather sexist commentary unsatisfactory – I am a big reader, and a PhD in rhetoric having done comparative literature MA and earlier a BA in literature at university – Jean Rhys was known to me well before Ford Madox Ford, and Joseph Conrad’s effect on her may or may not be salient. Her general viewpoint is very familiar to women, and ‘Wide Sargasso Sea’ is a classic to be sure – but for its deftness and display of her observational skills of character and oppression of Mrs Rochester. You sell her short by constantly referring back to her supposed influences – it’s rather than male luminaries had to be appealed to if you were even going to get a leg up into the male-dominated world – but don’t be taken in by her independence and strong intellect – nothing to do with the male influences – more to do with Jane Austen in that particular case … I really feel this entry lets her (and you) down, Leslie

Leave a Reply Cancel reply
You must be logged in to post a comment.
- Open access
- Published: 16 May 2024
Genomic analysis of severe COVID-19 considering or not asthma comorbidity: GWAS insights from the BQC19 cohort
- Omayma Amri 1 , 2 ,
- Anne-Marie Madore 1 , 2 ,
- Anne-Marie Boucher-Lafleur 1 , 2 &
- Catherine Laprise 1 , 2 , 3
BMC Genomics volume 25 , Article number: 482 ( 2024 ) Cite this article
219 Accesses
1 Altmetric
Metrics details
The severity of COVID-19 is influenced by various factors including the presence of respiratory diseases. Studies have indicated a potential relationship between asthma and COVID-19 severity.
This study aimed to conduct a genome-wide association study (GWAS) to identify genetic and clinical variants associated with the severity of COVID-19, both among patients with and without asthma.
We analyzed data from 2131 samples sourced from the Biobanque québécoise de la COVID-19 (BQC19), with 1499 samples from patients who tested positive for COVID-19. Among these, 1110 exhibited mild-to-moderate symptoms, 389 had severe symptoms, and 58 had asthma. We conducted a comparative analysis of clinical data from individuals in these three groups and GWAS using a logistic regression model. Phenotypic data analysis resulted in the refined covariates integrated into logistic models for genetic studies.
Considering a significance threshold of 1 × 10 −6 , seven genetic variants were associated with severe COVID-19. These variants were located proximal to five genes: sodium voltage-gated channel alpha subunit 1 ( SCN10A) , desmoplakin ( DSP) , RP1 axonemal microtubule associated ( RP1 ), IGF like family member 1 ( IGFL1 ), and docking protein 5 ( DOK5 ). The GWAS comparing individuals with severe COVID-19 with asthma to those without asthma revealed four genetic variants in transmembrane protein with EGF like and two follistatin like domains 2 ( TMEFF2 ) and huntingtin interacting protein-1 ( HIP1 ) genes.
This study provides significant insights into the genetic profiles of patients with severe forms of the disease, whether accompanied by asthma or not. These findings enhance our comprehension of the genetic factors that affect COVID-19 severity.
Key messages
Seven genetic variants were associated with the severe form of COVID-19;
Four genetic variants were associated with the severe form of COVID-19 in individuals with comorbid asthma;
These findings help define the genetic component of the severe form of COVID-19 in relation to asthma as a comorbidity.
Peer Review reports
Introduction
In March 2020, the World Health Organization (WHO) officially declared the outbreak of coronavirus disease 2019 (COVID-19), caused by infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), as a global pandemic [ 1 ]. By May 2023, this disease engendered a staggering 796 million infections worldwide, resulting in approximately 6.9 million deaths, equating to a mortality rate of 0.9% [ 2 ]. The range of COVID-19 symptoms varies from asymptomatic to fatality in severe cases. The majority of those infected with the virus experience mild symptoms such as cough, fever, headache, asthenia, anosmia, and ageusia [ 3 ]. However, certain cases require hospitalization and mechanical ventilation to prevent severe respiratory failure [ 4 ]. Hospitalized patients, advanced age, male sex [ 5 , 6 ], and underlying medical conditions such as hypertension, obesity, and diabetes, exhibit strong correlations with mortality [ 5 , 6 , 7 , 8 , 9 ]. The severity of COVID-19 may be affected by other factors, such as autoimmune diseases and genetic variations, which either enhance the susceptibility to severe outcomes or protect against them [ 10 ].
Various studies have work elucidating the genetic mechanisms that influence the severity of COVID-19 and associated different loci as 3p21.31 and 9q34.2 to respiratory failure and severe complications [ 11 , 12 , 13 ]. Concerning the 9q34.2 locus, it harbors the ABO, alpha 1-3-N-acetylgalactosaminyltransferase and alpha 1-3-galactosyltransferase ( ABO ) gene, which may modulate COVID-19 susceptibility and symptom severity through immunological interactions and inflammatory responses [ 11 , 14 ]. Earlier research has posited that the ABO gene, jointly associated with asthma and severe COVID-19, may partly explain the association between these conditions [ 15 ]. Additionally, studies have reported a lower prevalence of asthma among COVID-19 patients compared to the general population [ 16 , 17 ], suggesting potential resistance conferred by asthma against viral infection [ 18 ]. Furthermore, it has been proposed that allergic asthma may enhance immunity by inducing eosinophilia and a type 2 helper T cell (Th2) inflammatory response via the interleukin (IL)-13 pathway [ 19 ]. Further genetic investigations have suggested the involvement of the 12q24.13 locus, encoding oligo-adenylate synthetases ( OAS ) family, in asthma’s protective mechanisms via airway remodeling [ 20 ] and in COVID-19 [ 21 ], through mechanisms aiding in viral ribonucleic acids (RNAs) degradation and viral replication inhibition by activating latent ribonuclease [ 22 ].
The objective of this study is, firstly, to identify a genetic profile distinguishing patients with severe COVID-19 from those experiencing mild-to-moderate manifestations within the Quebecois population, and secondly, to establish a genetic profile for severe COVID-19 patients afflicted with asthma compared to those without asthma.
A schematic view of the study design is presented in Fig. 1 , with a brief description of the study population and the analyses performed, including the main objectives.
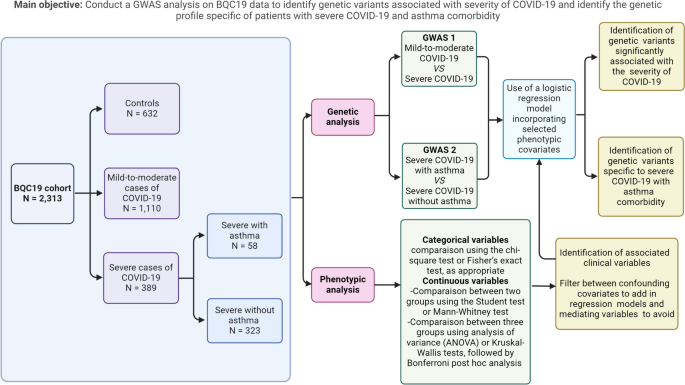
Explanatory diagram of the study design and objectives (figure created with BioRender.com)
Study population
The study participants were sourced from the Biobanque québécoise de la COVID-19 (BQC19) ( https://www.quebeccovidbiobank.ca/ ) established in Quebec, Canada. The primary objective of this biobank is to enable the scientists to access biological materials and data to facilitate COVID-19 research. Ethical approval was granted by the Research Ethics Board of the Centre intégré universitaire de santé et de services sociaux du Saguenay–Lac-Saint-Jean (IDs: 2022–388, 2021–026). Informed consent was obtained from all participants or their legal guardians in cases where the individual was unable to provide consent or was below 18 years of age [ 23 ].
This study involved 2131 patients aged from 2 months to 102.7 years old (Table 1 ). The samples and clinical data were sourced from both non-hospitalized and hospitalized individuals. All participants agreed to participate in the local clinical COVID-19 testing using SARS-CoV-2 RNA reverse transcriptase polymerase chain reaction (RT-PCR). Among these participants, 1499 tested positive for SARS-CoV-2, whereas 632 tested negative. SARS-CoV-2 PCR-negative patients were recruited as controls while patients with COVID-19 were categorized into two groups based on severity: 389 severe cases and 1110 mild-to-moderate cases. The severity of COVID-19 was classified based on WHO established criteria (Fig. 2 ) [ 24 ].
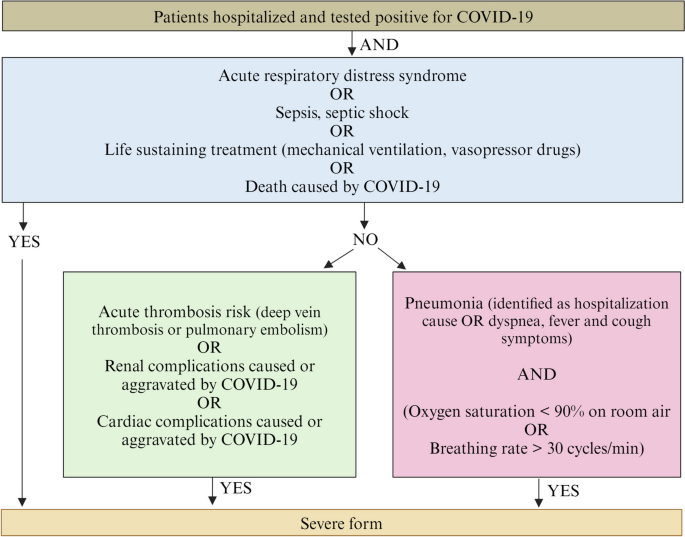
Flowchart for COVID-19 severity criteria. Participants are categorized as experiencing a severe stage of COVID-19 based on two primary criteria: a positive COVID-19 test and the necessity for hospitalization. Additionally, in conjunction with these two criteria, participants were required to satisfy the specified conditions outlined in one of the three other sections to be considered as having a severe manifestation. The figure is generated using BioRender.com
To identify the genetic profile of patients with severe COVID-19 and asthma, we divided patients with severe COVID-19 into two subgroups: 58 patients with asthma and 323 patients without asthma. Asthma was diagnosed based on patient interviews and medical history. However, no information regarding disease severity or sub phenotypes (e.g., atopy and airway hyperresponsiveness) was available.
Clinical data for all patients included individual characteristics (age, height, weight, sex) and medical history, with common medications such as systemic corticosteroids and angiotensin converting enzyme (ACE) inhibitors. Additionally, a physician conducted a physical examination to document COVID-19 symptoms (cough, headache, sore throat, ageusia, anosmia, rhinorrhea, dyspnea, fever, diarrhea, myalgia, and fatigue), asthma or respiratory conditions, comorbidities, and persistent COVID-19 symptoms. Blood cell counts (eosinophils, neutrophils, and lymphocytes), D-dimer and C-reactive protein (CRP) levels were measured.
Whole genome sequencing
DNA extraction and whole-genome sequencing (WGS) were performed at McGill Genome Center. DNA extraction involved treating samples with a lysis buffer, followed by extraction using the CMG-1091 DNA extraction kit (Perkin Elmer on a Chemagic MSM-I instrument). DNA concentration was determined using the Quant-iTPicoGreendsDNAAssay kit (ThermoFisher Scientific, P11495). For library preparation, a 25 μl aliquot from each sample at a concentration of 16 ng/μl was used with the DNA PCR-FreePrep Tagmentation kit (Illumina, 20,041,794). Libraries quality was validated through quantitative PCR using a DNA High Sensitivity Reagent Kit (Perkin Elmer Lab Chip GX, CLS760672). Twenty-seven libraries were combined in equimolar proportions, loaded into an Illumina S4 flow cell, and sequenced on the Illumina NovaSeq 6000 [ 25 ], using the NovaSeq 6000 S4 Reagent Kit v1.5 (Illumina,20,028,312). Data from WGS were analyzed for variant detection using the GenPipesDnaSeq pipeline [ 26 ]. The reads were aligned to the human reference genome (build GRCh38) using BWA-mem aligner [ 27 ]. Then, mapping accuracy was enhanced in proximal insertion and deletion regions using GATK IndelRealigner through the GATK [ 28 , 29 ] and Picard programs ( http://broadinstitute.github.io/picard/ ). Duplicate reads were labeled using Picard Mark Duplicates and quality scores were enhanced using the GATKBaseRecalibrator. Single nucleotide variants (SNVs) were detected using GATK Haplotype Caller in GVCF mode, which enabled efficient merging of multiple samples into a single variant file downstream. Samples within each cohort were merged using GATK-combined GVCFs and genotyped using Genotype GVCFs.
Quality control measures were performed during the alignment and genotype calling phases. Samples with a mean coverage below 30x were initially enhanced through top-up procedures and contamination estimation was performed using verifyBAMid2 [ 30 ]. Concordance assessments of genotypes and sexes were conducted to address potential sample mix-up, by comparing next-generation sequencing (NGS) data and SNP array information using NGS checkmate [ 31 ] and GATK cross-check fingerprints, as necessary. Moreover, variant counts in the samples were compared. Subsequently, quality filtering was applied to both individuals and genotypes using PLINK v2.0 (www.cog-genomics.org/plink/2.0/), which was guided by data completeness levels, and aimed to eliminate individuals with high coefficients of relationship [ 32 ]. The criteria for fulfillment included: a genotype call rate > 95%, an individual call rate surpassing 90%, a Hardy–Weinberg equilibrium (HWE) P -value > 10 −4 , and a minor allele frequency (MAF) of at least 0.5%. Additionally, a kinship value threshold of 0.177 (KING kinship coefficients scaled to 0.5 for duplicates) was used to detect duplicate samples and first-degree relationships between samples (including parent–child and sibling–sibling pairs). In these cases, only one individual from each pair was analyzed. Following the implementation of these primary filters, 13,185,383 variants and 2131 individuals were retained for further analyses.
Genetic analyses
Participants’ phenotypic data were compared using both group-wise analyses and comparative investigations for sex-based differences. Categorical variables were compared using the chi-square test or Fisher’s exact test, as appropriate. Continuous variables were assessed using analysis of variance (ANOVA) or Kruskal-Wallis tests, followed by Bonferroni post hoc analysis to determine specific group differences. SPSS v28.0.1 was used for the analyses and statistical significance was set at P < 0.05. For subsequent genome-wide association study (GWAS) analyses, covariates were selected based on test results and a comprehensive literature review to select confounding variables and avoid mediating variables. Principal components reflecting genotypic diversity among participants were computed and incorporated as covariates into the analysis models. This step aims to effectively address the population stratification.
The first logistic regression analysis was conducted to compare individuals with severe COVID-19 to those with mild-to-moderate forms. This model incorporated the first 10 principal components along with age and sex as covariates. Subsequently, a logistic regression analysis was conducted to compare individuals with severe COVID-19 and asthma to those without asthma. This model incorporated as covariates the first 10 principal components, the lowest values of eosinophil counts [ 16 , 33 , 34 , 35 ], and the highest values of neutrophil counts of each individuals [ 36 , 37 , 38 ]. These counts were incorporated into the model due to the frequent association of pre-existing eosinophilia with allergic asthma in individuals with asthma, and the association of non-allergic type 2 asthma with neutrophil activation [ 39 ]. Eosinopenia and neutrophilia are recognized biomarkers for severe COVID-19. Systemic corticosteroids are also included as covariate because of their frequent usage in the management of severe asthma and for treating severe COVID-19 cases as well [ 18 ].
Both models utilized PLINK v2.0 (www.cog-genomics.org/plink/2.0/) on the Digital Research Alliance of Canada’s supercomputer (alliancecan.ca). To address convergence issues, both models employed the firth-fallback option, enabling the Firth regression when logistic regression failed to converge [ 32 ]. Moreover, continuous covariates were standardized for variance normalization. A significance threshold of 1 × 10 −6 was considered [ 40 ].
Clinical analyses
The study involved 2131 participants, with a mean age of 60.34 years (± 20.26) and an average body mass index (BMI) of 27.66 kg/m 2 (± 6.50). Sex distribution was almost equal, with 49.50% females and 50.49% males. Of these participants, 80.85% ( n = 1723) were hospitalized and 19.14% ( n = 408) were treated as outpatients. Among the 381 patients with severe COVID-19 and known asthma status, 15% ( n = 58) had asthma (Table 1 ).
Table 1 highlights the significant differences between patients with severe COVID-19 and those with mild-to-moderate disease manifestations. The average age exhibited by the severe group (63.93 ± 15.98 years) is higher than that in mild-to-moderate group (58.82 ± 20.66 years). Moreover, the average BMI in the severe group (28.89 ± 7.05 kg/m 2 ) was higher than that in mild-to-moderate group (27.83 ± 6.44 kg/m 2 ). The severe category was predominantly male, whereas the mild-to-moderate category had a higher number of females. When assessing immune cell types, individuals in the severe COVID-19 group had an elevated neutrophil count (86.36% ± 45.66%) and lower eosinophil (1.11% ± 2.06%) and lymphocyte (13.62% ± 10.42%) counts. They also exhibited elevated CRP (146.49 ± 102.01 mg/L) and D-dimer levels (3157.68 ± 5082.00 μg/L). Moreover, the two groups experienced dyspnea during hospitalization: 90% ( n = 323) of patients in the severe group and 63% ( n = 518) of patients in the mild-to-moderate group. The use of ACE inhibitors was significantly higher in the severe group (37%) than that in the mild-to-moderate group (23%).
When comparing patients with severe COVID-19 with and without asthma, we observed certain differences. Specifically, the BMI was significantly higher in patients with severe COVID-19 and asthma (33.70 ± 8.49 kg/m 2 ) in comparison to those without asthma (28.17 ± 6.37 kg/m 2 ). Moreover, in the severe group with asthma, CRP levels were lower (113.26 ± 76.33 mg/L) in comparison to the group without asthma (154.08 ± 104.90 mg/L).
Table 2 highlights the significant sex-based differences within the mild-to-moderate and severe groups and delineates the clinical characteristics based on sex. In the mild-to-moderate COVID-19 group, male patients had a higher hospitalization rate (74%, n = 379) than female patients (63%, n = 380). The male patients had a significantly higher average BMI (28.35 ± 6.18 kg/m 2 ) in comparison to female patients (27.40 ± 6.64 kg/m 2 ). Moreover, 27% of male patients in the same disease group received ACE inhibitor treatment, in contrast to 20% of female patients. The biological test results demonstrated that severe COVID-19 in male patients had higher neutrophilia (87.69% ± 44.38%) compared to mild-to-moderate cases (76.92% ± 14.67%). Similarly, male patients with mild-to-moderate COVID-19 exhibited a higher incidence of lymphopenia (16.79% ± 10.42%) and elevated CRP levels (93.32 ± 73.33 mg/L) compared to females (21.08% ± 12.61% and 64.09 ± 65.22 mg/L), respectively.
Genetic analysis
First, individuals with mild-to-moderate COVID-19 and those with severe symptoms were compared (Fig. 3 ). To counteract technical biases and address population stratification, 10 principal components were included as covariates in the analyses, along with age and sex. The results indicated a significant association between seven genetic variants and severe COVID-19 (Table 3 ).
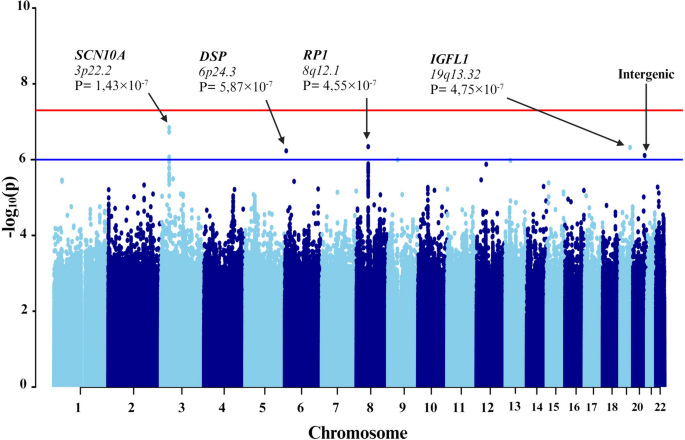
Manhattan plot of the genome-wide association study (GWAS) between mild-to-moderate and severe COVID-19. The GWAS results are shown on the y-axis as -log10 ( P -value), and on the x-axis is the chromosomal location. The red horizontal line illustrates the genome-wide association threshold ( P < 5 × 10 −8 ) and the blue line denotes the suggestive genome-wide association threshold ( P < 1 × 10 −6 ). The Manhattan plot is generated using the qqman package in R (v4.2.1) [ 41 ]
Among these variants, three (rs6599261, rs9815891, and rs62244113) were located within sodium voltage-gated channel alpha subunit 1 ( SCN10A ) at locus 3p22.2 , with P -values ranging from 8.595 × 10 −7 to 1.431 × 10 −7 (Fig. 4 ). Another variant was located within the RP1 axonemal microtubule-associated ( RP1 ) at locus 8q12.1 ( P -value = 4.547 × 10 −7 ). Additionally, rs1019213 is positioned 3747 base pairs (bp) upstream of IGF like family member 1 ( IGFL1 ) at locus 19q13.32 . Another intergenic variant, rs4809972, was positioned 161,552 bp downstream of docking protein 5 ( DOK5 ) at locus 20q13.2 ( P -value = 7.774 × 10 −7 ). Within desmoplakin ( DSP ) located at locus 6p24.3 , the variant rs4960330 was identified ( P -value =5.868 × 10 −7 ).
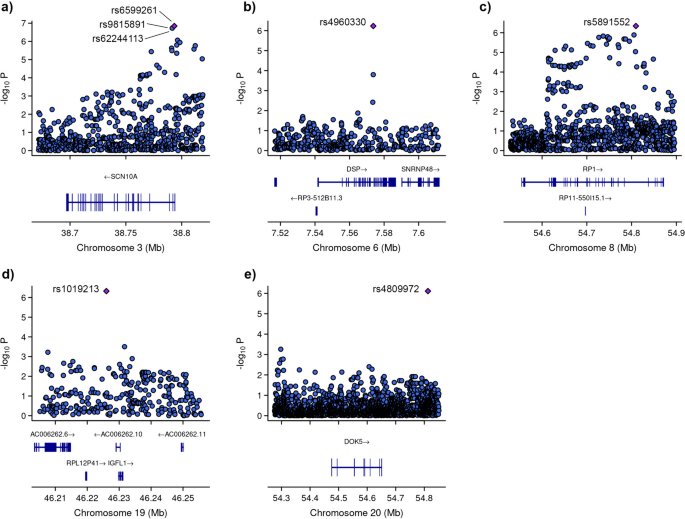
Zoom at associated loci with a severe form of COVID-19. Figure shows 25 kb regions for ( a ) sodium voltage-gated channel alpha subunit 1 ( SCN10A ), ( b ) desmoplakin ( DSP ), ( c ) RP1 axonemal microtubule-associated ( RP1 ) and ( d ) IGF like family member 1 ( IGFL1 ) genes as well as 200 kb region for e) docking protein 5 ( DOK5 ) gene. The genome-wide association study (GWAS) results are shown on the y-axis as -log10 ( P -value), and on the x-axis is the chromosomal location in Mb. At the bottom of each are the genes found in corresponding locus according to Ensemble Database library for homo sapiens v86. The plots are generated using the locuszoomr package in R (v4.3.0)
Subsequent GWAS was conducted between the groups with severe COVID-19 and asthma and those without asthma (Fig. 5 ). In addition to the 10 principal components, additional covariates included the lowest eosinophil count, highest neutrophil count, and systemic corticosteroid medication. Four genetic variants were associated. Specifically, one of the variant rs74684048 was located within the transmembrane protein with EGF like and two follistatin like domains 2 ( TMEFF2 ) at locus 2q32.3 ( P -value = 2.807 × 10 −7 ). Three additional variants (rs807875, rs807874, and rs62478485) were detected within the huntingtin interacting protein 1 ( HIP1 ) at locus 7q11.23 , with P -values ranging from 8.953 × 10 −7 to 5.860 × 10 −7 (Table 4 and Fig. 6 ).
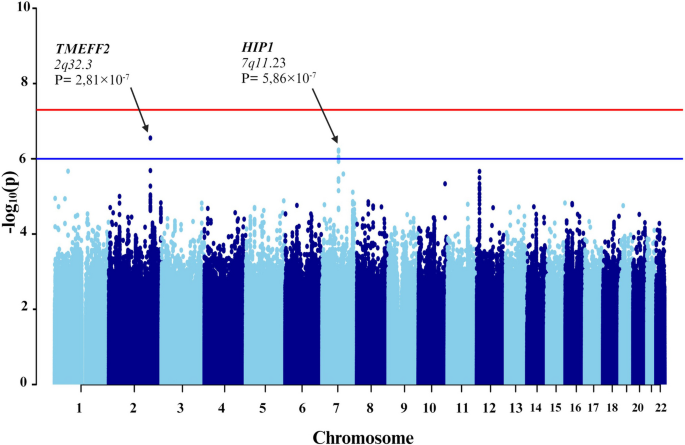
Manhattan plot of the genome-wide association study (GWAS) between patients with severe COVID-19 plus asthma and those without asthma. The GWAS results are shown on the y-axis as -log10 ( P -value), and on the x-axis is the chromosomal location. The red horizontal line illustrates the genome-wide association threshold ( P < 5 × 10 −8 ) and the blue line indicates the suggestive genome-wide association threshold ( P < 1 × 10 −6 ). The Manhattan plot is generated using the qqman package in R (v4.2.1) [ 41 ]
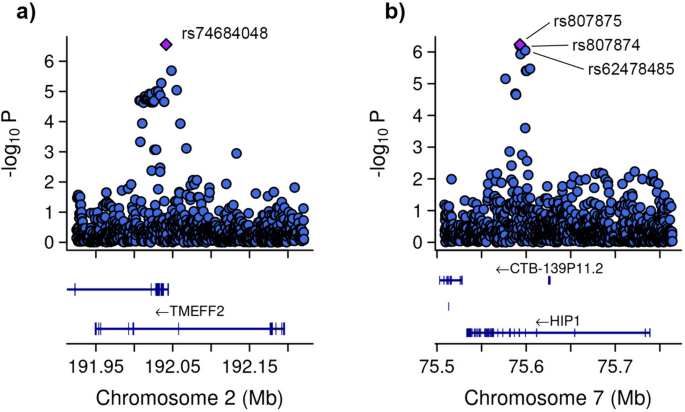
Zoom at associated loci with a severe form of COVID-19 with asthma. Figure shows 25 kb regions for ( a ) transmembrane protein with EGF like and two follistatin like domains 2 ( TMEFF2 ) and ( b ) huntingtin interacting protein 1 ( HIP1 ) genes. The genome-wide association study (GWAS) results are shown on the y-axis as -log10 ( P -value), and on the x-axis is the chromosomal location in Mb. At the bottom of each are the genes found in corresponding locus according to Ensemble Database library for homo sapiens v86. The plots are generated using the locuszoomr package in R (v4.3.0)
The primary objective of this study was to conduct a comprehensive pan-genomic analysis of individuals from BQC19, a representative sample of the Quebec population. The study aimed to acquire deeper insights into the genetic and clinical aspects of severe COVID-19 with and without asthma comorbidity. To reach this goal, it is one of the very few studies to compare genomes of patients with mild-to-moderate COVID-19 to the ones with severe COVID-19, allowing to better document the genomic profile specific to the severe form of COVID-19.
Two distinct genetic profiles were identified: one for individuals with severe COVID-19, and another for those with severe COVID-19 alongside asthma. The robust findings of this study were supported by the representative population, rendering results potentially applicable to the Quebec population. This analysis revealed multiple genomic loci associated with COVID-19 severity with or without asthma comorbidity, including DSP , HIP1 and RP1 genes. These genes have been associated through genomic or proteomic analyses in previous studies [ 42 , 43 , 44 ].
Precise asthma phenotyping (based on the Global Initiative for Asthma, 2023) [ 45 ] could distinguish the protective effect of allergic asthma from the potential risk associated with severe asthma in severe COVID-19 cases. Moreover, the genetic profiles identified in this study did not encompass new COVID-19 variants, as the samples were collected and analyzed prior to this emergence. Although these findings are significant, assessing them in an independent cohort is fundamental to enhance the validity of results. Additionally, increasing the sample size of patients with asthma could enhance the statistical robustness of the study, enabling a precise analysis.
When comparing mild-to-moderate and severe COVID-19 groups, seven significant variants were identified. The most prominent signal was observed at locus 3p22.2 , in which three specific variants (rs6599261, rs9815891, and rs62244113) were identified. SCN10A codes for the voltage-gated sodium channel Nav1.8. Shiers et al. showed that the ACE2 receptor, responsible for SARS-CoV-2 viral entry into host cells, is predominantly expressed in neuronal nociceptors labeled by this sodium channel [ 46 ]. This implies a potential route for the infection of nociceptors through the respiratory airways due to ACE2 expression. An elevated ACE2 expression was observed in the thoracic dorsal root ganglia, which house nociceptors responsible for lung innervation [ 47 , 48 ]. This is significant because of lungs being a prime site for SARS-CoV-2 viral replication [ 49 ]. The phenotypic data complement this observation with the elevated use of ACE inhibitors in severe COVID-19 cases. This could be attributed to the potential of ACE inhibitors to increase ACE2 receptor expression, enhancing viral entry [ 50 ]. Recent studies suggested an association between SCN10A and chronic obstructive pulmonary disease (COPD) [ 51 ]. Dyspnea prominently characterizes COPD, which aligns with previous clinical observations. SCN10A is also associated with cardiovascular diseases. Previous GWAS highlighted the significance of genetic variations in SCN10A on cardiac conduction [ 52 ], a factor associated with unanticipated cardiac arrest [ 53 ]. This trait is associated with a higher susceptibility to COVID-19 [ 54 , 55 , 56 ]. Moreover, research has established a correlation between cardiac conduction aberrations and SARS-CoV-2-related complications. The systemic inflammatory response to COVID-19, referred to as the “cytokine storm”, can adversely affect cardiac function and disrupt cardiac conduction [ 57 , 58 ]. The significance of SCN10A has thus been assessed in both pulmonary and cardiac disorders, in accordance with recognized risk factors for severe COVID-19.
A new variant was identified at locus 8q12.1 in the RP1 gene. A recent study highlighted the significance of RP1 in association with SARS-CoV-2 and Middle East respiratory syndrome (MERS) viruses, indicating its role in facilitating viral infections and severe disease complications [ 42 ]. However, its precise contribution to disease pathophysiology remains uncertain.
The variant rs4960330, at locus 6p24.3 within DSP , is associated with severe COVID-19. A recent investigation demonstrated elevated DSP levels in acute COVID-19 cases [ 43 ]. Another study revealed 23 DSP variants associated with idiopathic pulmonary fibrosis (IPF). Among these, rs2076295 and rs2744371, were associated with increasing DSP expression in the respiratory epithelium of IPF-affected lungs [ 59 , 60 ]. Recent findings indicate that up to 11% of patients develop IPF [ 61 ] after recovery from COVID-19 acute phase. rs2076295 is also associated with interstitial lung abnormalities [ 60 ], a condition frequently observed in patients with COPD [ 62 ].
The variant rs1019213 at locus 19q13.32, is positioned 3747 bp upstream of IGFL1 . Elevated IGFL1 expression is associated with poor prognosis in lung adenocarcinoma [ 63 ]. However, the correlation between COVID-19 and lung cancer remains uncertain.
We identified the final intergenic variant rs4809972 at locus 20q13.2 , which was 161,552 bp downstream of DOK5 . Another variant near the DOK5 gene, rs60684837, was previously associated with COVID-19 mortality in the western Indian population [ 64 ]. This gene is also associated with obesity [ 65 ] and diabetes, [ 66 ] two comorbidities recurrently identified as COVID-19 risk factors in numerous investigations [ 67 ]. Additionally, another study indicated that overexpression of DOK5 in fibroblasts contributes to the progression of IPF [ 68 ]. However, it is important to interpret these results with caution due to the distance between the variant and the nearest gene.
The second GWAS compared the genetic profiles of individuals with severe COVID-19 and asthma to those without asthma. Correlation between COVID-19 and asthma is an ongoing research subject. The relationship between asthma and COVID-19 varies across asthma phenotypes. For instance, allergic asthma appears to offer protection through the IL-13 pathway [ 69 ], whereas severe asthma appears to be associated with severe COVID-19 outcomes through the ACE2 receptor pathway [ 70 ]. Genomic investigations can help elucidate the biological nature of these relationships. This study identified two genomic regions containing four variants significantly associated with the combined phenotypes of severe COVID-19 and asthma.
The variant rs74684048, located at locus 2q32.3 within TMEFF2 , is associated with a specific phenotype. TMEFF2 was genetically associated with submucosal eosinophils in bronchial brushing samples of patients with severe asthma [ 71 ]. An epigenome-wide association study revealed an association between DNA methylation of TMEFF2 and lung function [ 72 ]. Additionally, other studies indicated that methylation in the TMEFF2 promoter regions reduces its activity, potentially contributing to lung tumor development [ 73 ]. There is no direct association between TMEFF2 and COVID-19. Further research is required to understand the function of this gene in both asthma and COVID-19.
Three additional variants (rs807875, rs807874, and rs62478485) within HIP1 at locus 7q11.23 were identified. This finding corroborates results of a study by Pairo et al., which associated HIP1 with severe COVID-19 [ 44 ]. It is possible that HIP1 is involved in the endocytosis process of SARS-CoV-2, as the virus enters host cells through clathrin-induced endocytosis [ 74 ], a pathway involving HIP1 [ 75 , 76 ]. Additionally, other studies have revealed elevated HIP1 expression in lung cancer, with HIP1 identified as a novel fusion partner of anaplastic lymphoma kinase [ 77 , 78 ]. This indicates that HIP1 may be implicated in COVID-19 through its interaction with clathrin. However, there is no distinct association between HIP1 and asthma.
Conclusions
This study enhances our understanding of the risk factors for severe COVID-19 and highlights the significant role of genetics in determining susceptibility to this form of the disease. It delineates a specific genetic profile of severe COVID-19 compared to mild-to-moderate form and severe COVID-19 with asthma compared to severe COVID-19 without asthma. These findings have the potential to enhance preventive strategies in patients with severe COVID-19. By combining the GWAS data from this study with forthcoming data, a potential polygenetic risk score can be developed to identify individuals with a high risk of developing severe COVID-19 in relation with their asthma status. Further investigations with precise asthma phenotyping are needed to refine and fortify these findings.
Availability of data and materials
All clinical and genetic data used and results generated are available from the BQC19 via data request at https://www.bqc19.ca/en/access-data .
Abbreviations
ABO, Alpha 1-3-N-acetylgalactosaminyltransferase and alpha 1-3-galactosyltransferase
Angiotensin converting enzyme
Analysis of variance
Body mass index
Biobanque québécoise de la COVID-19
C-reactive protein
Chronic obstructive pulmonary disease
Coronavirus disease 2019
Docking protein 5
Desmoplakin
Genome-wide association study
Huntingtin interacting protein 1
Hardy-Weinberg equilibrium
IGF like family member 1
Interleukin
Idiopathic pulmonary fibrosis
Minor allele frequency
Middle East respiratory syndrome
Next-generation sequencing
Oligoadenylate synthetase
Ribonucleic acid
RP1 axonemal microtubule-associated
Reverse transcriptase polymerase chain reaction
Severe acute respiratory syndrome coronavirus 2
Sodium voltage-gated channel alpha subunit 1
Single nucleotide variant
Type 2 helper T cell
Transmembrane protein with EGF like and two follistatin like domains 2
Whole-genome sequencing
World health organization
Pandémie de maladie à coronavirus (COVID-19). https://www.who.int/emergencies/diseases/novel-coronavirus-2019 . Accessed 1 Apr 2023.
WHO Coronavirus (COVID-19) Dashboard. https://covid19.who.int/ . Accessed 30 May 2023.
Clinical Care Considerations: Clinical considerations for care of children and adults with confirmed COVID-19. https://www.cdc.gov/coronavirus/2019-ncov/hcp/clinical-guidance-management-patients.html . Accessed 30 May 2023.
Marini JJ, Gattinoni L. Management of COVID-19 respiratory distress. Jama. 2020;323(22):2329–30.
Article PubMed Google Scholar
Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, Xiang J, Wang Y, Song B, Gu X. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–62.
Article CAS PubMed PubMed Central Google Scholar
Li X, Xu S, Yu M, Wang K, Tao Y, Zhou Y, Shi J, Zhou M, Wu B, Yang Z. Risk factors for severity and mortality in adult COVID-19 inpatients in Wuhan. J Allergy Clin Immunol. 2020;146(1):110–8.
Chen R, Liang W, Jiang M, Guan W, Zhan C, Wang T, Tang C, Sang L, Liu J, Ni Z. Risk factors of fatal outcome in hospitalized subjects with coronavirus disease 2019 from a nationwide analysis in China. Chest. 2020;158(1):97–105.
Article CAS PubMed Google Scholar
Docherty AB, Harrison EM, Green CA, Hardwick HE, Pius R, Norman L, et al. Features of 20 133 UK patients in hospital with covid-19 using the ISARIC WHO clinical characterisation protocol: prospective observational cohort study. BMJ. 2020;369.
Richardson S, Hirsch JS, Narasimhan M, Crawford JM, McGinn T, Davidson KW, Barnaby DP, Becker LB, Chelico JD, Cohen SL. Presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID-19 in the new York City area. Jama. 2020;323(20):2052–9.
Taz TA, Ahmed K, Paul BK, Kawsar M, Aktar N, Mahmud SH, Moni MA. Network-based identification genetic effect of SARS-CoV-2 infections to idiopathic pulmonary fibrosis (IPF) patients. Brief Bioinform. 2021;22(2):1254–66.
Group SC-G. Genomewide association study of severe Covid-19 with respiratory failure. N Engl J Med. 2020;383(16):1522–34.
Article Google Scholar
Zeberg H, Pääbo S. The major genetic risk factor for severe COVID-19 is inherited from Neanderthals. Nature. 2020;587(7835):610–2.
Kousathanas A, Pairo-Castineira E, Rawlik K, Stuckey A, Odhams CA, Walker S, Russell CD, Malinauskas T, Wu Y, Millar J. Whole-genome sequencing reveals host factors underlying critical COVID-19. Nature. 2022;607(7917):97–103.
Zhao J, Yang Y, Huang H, Li D, Gu D, Lu X, Zhang Z, Liu L, Liu T, Liu Y. Relationship between the ABO blood group and the coronavirus disease 2019 (COVID-19) susceptibility. Clin Infect Dis. 2021;73(2):328–31.
Baranova A, Cao H, Chen J, Zhang F. Causal association and shared genetics between asthma and COVID-19. Front Immunol. 2022;13:705379.
Ferastraoaru D, Hudes G, Jerschow E, Jariwala S, Karagic M, de Vos G, Rosenstreich D, Ramesh M. Eosinophilia in asthma patients is protective against severe COVID-19 illness. J Allergy Clin Immunol: Pract. 2021;9(3):1152–62 e1153.
CAS PubMed Google Scholar
Gaspar-Marques J, van Zeller M, Carreiro-Martins P, Loureiro CC. Severe asthma in the era of COVID-19: a narrative review. Pulmonology. 2022;28(1):34–43.
Hughes-Visentin A, Paul ABM. Asthma and COVID-19: what do we know now. Clin Med Insights: Circ Respir Pulm Med. 2020;14:1179548420966242.
PubMed Google Scholar
Ramakrishnan RK, Al Heialy S, Hamid Q. Implications of preexisting asthma on COVID-19 pathogenesis. Am J Phys Lung Cell Mol Phys. 2021;320(5):L880–91.
CAS Google Scholar
Almoguera B, Vazquez L, Mentch F, Connolly J, Pacheco JA, Sundaresan AS, Peissig PL, Linneman JG, McCarty CA, Crosslin D. Identification of four novel loci in asthma in European American and African American populations. Am J Respir Crit Care Med. 2017;195(4):456–63.
Article PubMed PubMed Central Google Scholar
Prasad K, Khatoon F, Rashid S, Ali N, AlAsmari AF, Ahmed MZ, Alqahtani AS, Alqahtani MS, Kumar V. Targeting hub genes and pathways of innate immune response in COVID-19: a network biology perspective. Int J Biol Macromol. 2020;163:1–8.
Choi UY, Kang J-S, Hwang YS, Kim Y-J. Oligoadenylate synthase-like (OASL) proteins: dual functions and associations with diseases. Exp Mol Med. 2015;47(3):e144–e144.
Tremblay K, Rousseau S, Ma’n HZ, Auld D, Chassé M, Coderre D, Falcone EL, Gauthier N, Grandvaux N, Gros-Louis F. The Biobanque québécoise de la COVID-19 (BQC19)—a cohort to prospectively study the clinical and biological determinants of COVID-19 clinical trajectories. PLoS One. 2021;16(5):e0245031.
Clinical management of COVID-19: Living guideline, 13 January 2023. https://www.who.int/publications/i/item/WHO-2019-nCoV-clinical-2023.1 . Accessed 1 Apr 2023.
Modi A, Vai S, Caramelli D, Lari M. The Illumina sequencing protocol and the NovaSeq 6000 system. In: Bacterial Pangenomics: Methods and Protocols. Springer; 2021. p. 15–42.
Chapter Google Scholar
Bourgey M, Dali R, Eveleigh R, Chen KC, Letourneau L, Fillon J, Michaud M, Caron M, Sandoval J, Lefebvre F. GenPipes: an open-source framework for distributed and scalable genomic analyses. Gigascience. 2019;8(6):giz037.
Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997v2, 2013.
Van der Auwera GA, O’Connor BD. Genomics in the cloud: using Docker, GATK, and WDL in Terra. O’Reilly Media; 2020.
Google Scholar
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–303.
Zhang F, Flickinger M, SAG T, Abecasis GR, Scott LJ, SA MC, Pato CN, Boehnke M, Kang HM, Consortium IPG. Ancestry-agnostic estimation of DNA sample contamination from sequence reads. Genome Res. 2020;30(2):185–94.
Lee S, Lee S, Ouellette S, Park W-Y, Lee EA, Park PJ. NGSCheckMate: software for validating sample identity in next-generation sequencing studies within and across data types. Nucleic Acids Res. 2017;45(11):e103–e103.
Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4(1):s13742-13015-10047–3748.
Roca E, Ventura L, Zattra CM, Lombardi C. EOSINOPENIA: an early, effective and relevant COVID-19 biomarker? QJM: Int J Med. 2021;114(1):68–9.
Article CAS Google Scholar
Lombardi C, Bagnasco D, Passalacqua G. COVID-19, eosinophils, and biologicals for severe asthma. Front Allergy. 2022;3.
Saidani A, Abid S, Hamza Z, Bougherriou A, Msaad S, Bahloul N. L’éosinopénie est-elle un facteur de mauvais pronostic au cours d’une infection au COVID-19? Rev Mal Respir Actual. 2022;14(1):145.
Reusch N, De Domenico E, Bonaguro L, Schulte-Schrepping J, Baßler K, Schultze JL, Aschenbrenner AC. Neutrophils in COVID-19. Front Immunol. 2021;12:652470.
Wang J, Li Q, Yin Y, Zhang Y, Cao Y, Lin X, Huang L, Hoffmann D, Lu M, Qiu Y. Excessive neutrophils and neutrophil extracellular traps in COVID-19. Front Immunol. 2020;11:2063.
Zhang B, Zhou X, Zhu C, Song Y, Feng F, Qiu Y, Feng J, Jia Q, Song Q, Zhu B. Immune phenotyping based on the neutrophil-to-lymphocyte ratio and IgG level predicts disease severity and outcome for patients with COVID-19. Front Mol Biosci. 2020;7:157.
Ordonez CL, Shaughnessy TE, Matthay MA, Fahy JV. Increased neutrophil numbers and IL-8 levels in airway secretions in acute severe asthma: clinical and biologic significance. Am J Respir Crit Care Med. 2000;161(4):1185–90.
Margaritte-Jeannin P, Budu-Aggrey A, Ege M, Madore AM, Linhard C, Mohamdi H, von Mutius E, Granell R, Demenais F, Laprise C. Identification of OCA2 as a novel locus for the co-morbidity of asthma-plus-eczema. Clin Exp Allergy. 2022;52(1):70–81.
Turner S. Qqman: an R package for visualizing GWAS results using QQ and Manhattan plots. J Open Source Softw. 2018;3(25):731.
Maldonado LL, Bertelli AM, Kamenetzky L. Molecular features similarities between SARS-CoV-2, SARS, MERS and key human genes could favour the viral infections and trigger collateral effects. Sci Rep. 2021;11(1):4108.
Ward KE, Steadman L, Karim AR, Reynolds GM, Pugh M, Chua W, Faustini SE, Veenith T, Thwaites RS, Openshaw PJ. SARS-CoV-2 infection is associated with anti-desmoglein 2 autoantibody detection. Clin Exp Immunol. 2023;213(2):243–51.
Pairo-Castineira E, Rawlik K, Bretherick AD, Qi T, Wu Y, Nassiri I, et al. GWAS and meta-analysis identifies 49 genetic variants underlying critical COVID-19. Nature. 2023:1–15.
Global Initiative for Asthma: 2023 GINA Report, Global Strategy for Asthma Management and Prevention. https://ginasthma.org/wp-content/uploads/2023/07/GINA-2023-Full-report-23_07_06-WMS.pdf . Accessed 1 Sept 2023.
Shiers S, Ray PR, Wangzhou A, Sankaranarayanan I, Tatsui CE, Rhines LD, Li Y, Uhelski ML, Dougherty PM, Price TJ. ACE2 and SCARF expression in human DRG nociceptors: implications for SARS-CoV-2 virus neurological effects. Pain. 2020;161(11):2494.
Kummer W, Fischer A, Kurkowski R, Heym C. The sensory and sympathetic innervation of guinea-pig lung and trachea as studied by retrograde neuronal tracing and double-labelling immunohistochemistry. Neuroscience. 1992;49(3):715–37.
Springall DR, Cadieux A, Oliveira H, Su H, Royston D, Polak JM. Retrograde tracing shows that CGRP-immunoreactive nerves of rat trachea and lung originate from vagal and dorsal root ganglia. J Auton Nerv Syst. 1987;20(2):155–66.
Tay MZ, Poh CM, Rénia L, MacAry PA, Ng LF. The trinity of COVID-19: immunity, inflammation and intervention. Nat Rev Immunol. 2020;20(6):363–74.
Vaduganathan M, Vardeny O, Michel T, McMurray JJ, Pfeffer MA, Solomon SD. Renin–angiotensin–aldosterone system inhibitors in patients with Covid-19. N Engl J Med. 2020;382(17):1653–9.
Kang J, Kim KT, Lee J-H, Kim EK, Kim T-H, Yoo KH, Lee JS, Kim WJ, Kim JH, Oh Y-M. Predicting treatable traits for long-acting bronchodilators in patients with stable COPD. Int J Chron Obstruct Pulmon Dis. 2017;114:3557–65.
Chambers JC, Zhao J, Terracciano CM, Bezzina CR, Zhang W, Kaba R, Navaratnarajah M, Lotlikar A, Sehmi JS, Kooner MK. Genetic variation in SCN10A influences cardiac conduction. Nat Genet. 2010;42(2):149–52.
Bezzina CR, Barc J, Mizusawa Y, Remme CA, Gourraud J-B, Simonet F, Verkerk AO, Schwartz PJ, Crotti L, Dagradi F. Common variants at SCN5A-SCN10A and HEY2 are associated with Brugada syndrome, a rare disease with high risk of sudden cardiac death. Nat Genet. 2013;45(9):1044–9.
Bansal A, Kumar A, Patel D, Puri R, Kalra A, Kapadia SR, Reed GW. Meta-analysis comparing outcomes in patients with and without cardiac injury and coronavirus disease 2019 (COVID 19). Am J Cardiol. 2021;141:140–6.
Li J-W, Han T-W, Woodward M, Anderson CS, Zhou H, Chen Y-D, Neal B. The impact of 2019 novel coronavirus on heart injury: a systematic review and meta-analysis. Prog Cardiovasc Dis. 2020;63(4):518–24.
Vasudeva R, Challa A, Al Rifai M, Polana T, Duran B, Vindhyal M, et al. Prevalence of cardiovascular diseases in COVID-19 related mortality in the United States. Prog Cardiovasc Dis. 2022;74.
Zimmermann P, Aberer F, Braun M, Sourij H, Moser O. The arrhythmogenic face of COVID-19: Brugada ECG pattern in SARS-CoV-2 infection. J Cardiovas Dev Dis. 2022;9(4):96.
Burhan E, Mubarak F, Adilah SASU, Sari CYI, Ismail E, Astuti P, et al. Association between cardiovascular diseases and COVID-19 pneumonia outcome in Indonesia: a multi-center cohort study. Front Med. 2023;10.
Mathai SK, Pedersen BS, Smith K, Russell P, Schwarz MI, Brown KK, Steele MP, Loyd JE, Crapo JD, Silverman EK. Desmoplakin variants are associated with idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2016;193(10):1151–60.
Hobbs BD, Putman RK, Araki T, Nishino M, Gudmundsson G, Gudnason V, Eiriksdottir G, Zilhao Nogueira NR, Dupuis J, Xu H. Overlap of genetic risk between interstitial lung abnormalities and idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2019;200(11):1402–13.
Stewart I, Jacob J, George PM, Molyneaux PL, Porter JC, Allen RJ, Aslani S, Baillie JK, Barratt SL, Beirne P. Residual lung abnormalities after COVID-19 hospitalization: interim analysis of the UKILD post–COVID-19 study. Am J Respir Crit Care Med. 2023;207(6):693–703.
Liu Y, Tang J, Sun Y. Impact of interstitial lung abnormalities on disease expression and outcomes in COPD or emphysema: a systematic review. International Journal of Chronic Obstructive Pulmonary Disease; 2023. p. 189–206.
Wang Z, Liang X, Wang X, Yu Q. Elevated expression of IGFL1 indicates unfavorable prognosis in lung adenocarcinoma through promotion of cell proliferation and inhibition of apoptosis. Pol J Pathol. 2021;72(4):283–95.
Pandit R, Singh I, Ansari A, Raval J, Patel Z, Dixit R, Shah P, Upadhyay K, Chauhan N, Desai K, et al. First report on genome wide association study in western Indian population reveals host genetic factors for COVID-19 severity and outcome. Genomics. 2022;114(4):110399.
Saini S, Walia GK, Sachdeva MP, Gupta V. Genetics of obesity and its measures in India. J Genet. 2018;97(4):1047–71.
Tabassum R, Mahajan A, Chauhan G, Dwivedi OP, Ghosh S, Tandon N, Bharadwaj D. Evaluation of DOK5 as a susceptibility gene for type 2 diabetes and obesity in north Indian population. BMC Med Genet. 2010;11(1):1–7.
Escobedo-de la Peña J, Rascón-Pacheco RA, de Jesús A-MI, González-Figueroa E, Fernández-Gárate JE, Medina-Gómez OS, Borja-Bustamante P, Santillán-Oropeza JA. Borja-Aburto VH: hypertension, diabetes and obesity, major risk factors for death in patients with COVID-19 in Mexico. Arch Med Res. 2021;52(4):443–9.
Ghandikota S, Sharma M, Ediga HH, Madala SK, Jegga AG. Consensus gene co-expression network analysis identifies novel genes associated with severity of fibrotic lung disease. Int J Mol Sci. 2022;23(10):5447.
Morrison CB, Edwards CE, Shaffer KM, Araba KC, Wykoff JA, Williams DR, Asakura T, Dang H, Morton LC, Gilmore RC. SARS-CoV-2 infection of airway cells causes intense viral and cell shedding, two spreading mechanisms affected by IL-13. Proc Natl Acad Sci. 2022;119(16):e2119680119.
López-Tiro JJ, Contreras-Contreras EA, Cruz-Arellanes NN, Camargo-Pirrón MA, Cabrera-Buendía EO, Ramírez-Pérez GI, Vega-Acevedo G. Asthma and COVID-19. Rev Alerg Mex. 2022;69:15–23.
Wilson SJ, Ward JA, Sousa AR, Corfield J, Bansal AT, De Meulder B, Lefaudeux D, Auffray C, Loza MJ, Baribaud F. Severe asthma exists despite suppressed tissue inflammation: findings of the U-BIOPRED study. Eur Respir J. 2016;48(5):1307–19.
Bolund A, Starnawska A, Miller MR, Schlünssen V, Backer V, Børglum AD, Christensen K, Tan Q, Christiansen L, Sigsgaard T. Lung function discordance in monozygotic twins and associated differences in blood DNA methylation. Clin Epigenetics. 2017;9(1):1–13.
Lee SM, Park JY, Kim DS. Methylation of TMEFF2 gene in tissue and serum DNA from patients with non-small cell lung cancer. Mol Cells. 2012;34:171–6.
Bayati A, Kumar R, Francis V, McPherson PS. SARS-CoV-2 infects cells after viral entry via clathrin-mediated endocytosis. J Biol Chem. 2021;296.
Legendre-Guillemin V, Metzler M, Charbonneau M, Gan L, Chopra V, Philie J, Hayden MR, McPherson PS. HIP1 and HIP12 display differential binding to F-actin, AP2, and clathrin: identification of a novel interaction with clathrin light chain. J Biol Chem. 2002;277(22):19897–904.
Metzler M, Legendre-Guillemin V, Gan L, Chopra V, Kwok A, McPherson PS, Hayden MR. HIP1 functions in clathrin-mediated endocytosis through binding to clathrin and adaptor protein 2. J Biol Chem. 2001;276(42):39271–6.
Ou S-HI, Klempner SJ, Greenbowe JR, Azada M, Schrock AB, Ali SM, Ross JS, Stephens PJ, Miller VA. Identification of a novel HIP1-ALK fusion variant in non–small-cell lung cancer (NSCLC) and discovery of ALK I1171 (I1171N/S) mutations in two ALK-rearranged NSCLC patients with resistance to alectinib. J Thorac Oncol. 2014;9(12):1821–5.
Hong M, Kim RN, Song J-Y, Choi S-J, Oh E, Lira ME, Mao M, Takeuchi K, Han J, Kim J. HIP1–ALK, a novel fusion protein identified in lung adenocarcinoma. J Thorac Oncol. 2014;9(3):419–22.
Download references
Acknowledgements
Authors are grateful to all participants for their essential and valuable contribution.
The BQC19 received funding from the Public Health Agency of Canada, Génome Québec and Fonds de recherche du Québec – santé (FRQS) (grant number: 2021-HQ-000051).
Author information
Authors and affiliations.
Centre intersectoriel en santé durable, Université du Québec à Chicoutimi, Saguenay, Québec, G7H 2B1, Canada
Omayma Amri, Anne-Marie Madore, Anne-Marie Boucher-Lafleur & Catherine Laprise
Département des sciences fondamentales, Université du Québec à Chicoutimi, Saguenay, Québec, G7H 2B1, Canada
Centre de recherche du Centre intégré universitaire de santé et de services sociaux du Saguenay–Lac-Saint-Jean, Saguenay, Québec, G7H 7K9, Canada
Catherine Laprise
You can also search for this author in PubMed Google Scholar
Contributions
OA: Data curation, formal analysis, visualisation and writing of the original draft. AMM: Methodology, review and editing of the manuscript. AMBL: Review and editing of the manuscript. CL: Conceptualization, investigation (Saguenay–Lac-Saint-Jean site), funding acquisition, project administration, ressources, supervision, writing, reviewing and edition.
Authors’ information
Catherine Laprise is part of the Quebec Respiratory Health Network (RHN; https://rsr-qc.ca/en/ ), investigator of the CHILD Study, director of the Centre intersectoriel en santé durable of the Université du Québec à Chicoutimi (UQAC) and chairholder of the Canada Research Chair in the Genomics of Asthma and Allergic Diseases ( https://www.chairs-chaires.gc.ca/ ), and co-chairholder of the Chaire de recherche du Québec en santé durable ( https://chairesantedurable.ca/ ) .
Corresponding author
Correspondence to Catherine Laprise .
Ethics declarations
Ethic approval and consent to participate.
Ethical approval was granted by the Research Ethics Board of the Centre intégré universitaire de santé et de services sociaux du Saguenay–Lac-Saint-Jean (IDs: 2022–388, 2021–026). Informed consent was obtained from all participants or their legal guardians in cases where the individual was unable to provide consent or was below 18 years of age.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ . The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
Reprints and permissions
About this article
Cite this article.
Amri, O., Madore, AM., Boucher-Lafleur, AM. et al. Genomic analysis of severe COVID-19 considering or not asthma comorbidity: GWAS insights from the BQC19 cohort. BMC Genomics 25 , 482 (2024). https://doi.org/10.1186/s12864-024-10342-x
Download citation
Received : 04 March 2024
Accepted : 23 April 2024
Published : 16 May 2024
DOI : https://doi.org/10.1186/s12864-024-10342-x
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Severe COVID-19
BMC Genomics
ISSN: 1471-2164
- Submission enquiries: [email protected]
- General enquiries: [email protected]
Advertisement
How devante parker retiring impacts the eagles' wr position, share this article.
According to ESPN’s Adam Schefter, the Eagles suffered a loss in personnel on Monday after veteran pass catcher DeVante Parker announced that he was retiring.
A 31-year-old former 1st-rd pick entering his 10th NFL season, Parker had 33 catches for 394 yards in 13 games last season for the Patriots.
After nine NFL seasons, Eagles WR DeVante Parker has decided to retire, he said Monday night. As much as he looked forward to his time with the Eagles, Parker decided the time had come to spend more time with his family that includes his four children. “I want to see my kids,… pic.twitter.com/enakIQc4LT — Adam Schefter (@AdamSchefter) May 20, 2024
Parker played nine NFL seasons for the Dolphins and Patriots. He has 402 catches for 5,660 yards and 27 touchdowns.
Parker was a 2015 first-round pick of Miami, and his best season came in 2019 when he logged 72 tackles for 1,202 yards and nine touchdowns.
With the news and reaction still trickling in, here’s an instant analysis of the decision and how it impacts Parker.
Biggest winner

Dec 25, 2023; Philadelphia, Pennsylvania, USA; Philadelphia Eagles wide receiver Britain Covey (18) returns a punt against the New York Giants during the first quarter at Lincoln Financial Field. Mandatory Credit: Eric Hartline-USA TODAY Sports
Parker is retiring with money in the bank and his health intact, so there are no losers, but Britain Covey and John Wilson must be the biggest winners. A.J. Brown, DeVonta Smith, and Parris Campbell look like locks to make the roster, but Parker’s decision allows Covey to make the roster outright. Covey is one of the NFL’s top punt returners, but he didn’t make the roster outright in 2023 and had competition in the return game from Ainias Smith, Isaiah Rodgers, and Cooper DeJean. For the rookie out of FSU, Parker’s retirement leaves a void, and the 6-foot-6 wide receiver could carve out a role with more training camp snaps.
Salary cap ramifications

New England Patriots wide receiver DeVante Parker (1) tries to avoid New York Giants cornerback Deonte Banks (25) at MetLife Stadium, Sunday, November 26, 2023.
Parker signed a one-year, $1.21 million contract to be the potential third wide receiver behind A.J. Brown and DeVonta Smith.
What's left

New York Giants wide receiver Parris Campbell (0) is tackled by Seattle Seahawks cornerback Devon Witherspoon (21) in the first half at MetLife Stadium on Monday, Oct. 2, 2023, in East Rutherford.
Philadelphia also signed Parris Campbell before spending two late-round picks on Ainias Smith and Johnny Wilson in the 2024 NFL Draft. The retirement will allow those younger players a few more developmental snaps during OTAs, training camp, and the mandatory minicamp.
Best of the rest

PHILADELPHIA, PENNSYLVANIA – AUGUST 17: Joseph Ngata #86 of the Philadelphia Eagles catches a pass against Gavin Heslop #41 of the Cleveland Browns in the second half of the preseason game at Lincoln Financial Field on August 17, 2023 in Philadelphia, Pennsylvania. The Browns tied the Eagles 18-18. (Photo by Mitchell Leff/Getty Images)
After Parker’s retirement, the Eagles still have ten wide receivers, second-year guys like Joseph Ngata and Austin Watkins Jr., while rookie Shaquan Davis and third-year Jacob Harris could also be players to watch.
Potential free agent targets

Nov 14, 2021; Paradise, Nevada, USA; Las Vegas Raiders wide receiver Hunter Renfrow (13) runs against Kansas City Chiefs cornerback L’Jarius Sneed (38) in the fourth quarter at Allegiant Stadium. Mandatory Credit: Kirby Lee-USA TODAY Sports
Hunter Renfro, Michael Thomas, Russell Gage, Mecole Hardman, Julio Jones, and Richie James are just a few of the names that could draw interest. Devon Allen could be a player to watch once the Olympics have been completed.
Read all the best Eagles coverage at Delaware Online and Eagles Wire.
Want the latest news and insights on your favorite team?
Sign up for our newsletter to get updates to your inbox, and also receive offers from us, our affiliates and partners. By signing up you agree to our Privacy Policy
An error has occured
Please re-enter your email address.
Thanks for signing up!
You'll now receive the top Eagles Wire stories each day directly in your inbox.
Most Popular
Eagles' updated 88-man roster ahead of otas, mandatory minicamp, 7 biggest storylines to watch during eagles otas, nfl experts predict the eagles' 2024 record after schedule release, eagles' projected offensive depth chart ahead of otas, mandatory minicamp, where do the eagles rank in spending at each position ahead of otas, eagles have one matchup make a list of the top 10 games of the 2024 nfl season, eagles 53-man roster prediction following 2024 rookie minicamp.
Please enter an email address.
Thanks for signing up.
Please check your email for a confirmation.
Something went wrong.
Hybrid performance evaluation and genome-wide association analysis of root system architecture in a maize association population
- Original Article
- Published: 22 August 2023
- Volume 136 , article number 194 , ( 2023 )
Cite this article

- Zhigang Liu 1 , 4 ,
- Pengcheng Li 2 ,
- Wei Ren 1 ,
- Zhe Chen 3 ,
- Toluwase Olukayode 4 ,
- Guohua Mi 1 ,
- Lixing Yuan 1 ,
- Fanjun Chen 1 , 5 &
- Qingchun Pan ORCID: orcid.org/0000-0001-8462-3165 1 , 5
618 Accesses
7 Altmetric
Explore all metrics
Key Message
The genetic architecture of RSA traits was dissected by GWAS and coexpression networks analysis in a maize association population.
Root system architecture (RSA) is a crucial determinant of water and nutrient uptake efficiency in crops. However, the maize genetic architecture of RSA is still poorly understood due to the challenges in quantifying root traits and the lack of dense molecular markers. Here, an association mapping panel including 356 inbred lines were crossed with a common tester, Zheng58, and the test crosses were phenotyped for 12 RSA traits in three locations. We observed a 1.3 ~ sixfold phenotypic variation for measured RSA in the association panel. The association panel consisted of four subpopulations, non-stiff stalk (NSS) lines, stiff stalk (SS), tropical/subtropical (TST), and mixed. Zheng58 × TST has a 2.1% higher crown root number (CRN) and 8.6% less brace root number (BRN) than Zheng58 × NSS and Zheng58 × SS, respectively. Using a genome-wide association study (GWAS) with 1.25 million SNPs and correction for population structure, 191 significant SNPs were identified for root traits. Ninety (47%) of the significant SNPs showed positive allelic effects, and 101 (53%) showed negative effects. Each locus could explain 0.39% to 11.8% of phenotypic variation. By integrating GWAS results and comparing coexpression networks, 26 high-priority candidate genes were identified. Gene GRMZM2G377215, which belongs to the COBRA-like gene family, affected root growth and development. Gene GRMZM2G468657 encodes the aspartic proteinase nepenthesin-1, related to root development and N-deficient response. Collectively, our research provides progress in the genetic dissection of root system architecture. These findings present the further possibility for the genetic improvement of root traits in maize.
This is a preview of subscription content, log in via an institution to check access.
Access this article
Price includes VAT (Russian Federation)
Instant access to the full article PDF.
Rent this article via DeepDyve
Institutional subscriptions
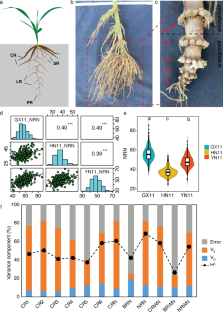
Similar content being viewed by others

Deep genotyping reveals specific adaptation footprints of conventional and organic farming in barley populations—an evolutionary plant breeding approach

Novel PHOTOPERIOD-1 gene variants associate with yield-related and root-angle traits in European bread wheat

Unique genetic architecture of prolificacy in ‘Sikkim Primitive’ maize unraveled through whole-genome resequencing-based DNA polymorphism
Data availability.
The genotype data for the association panel are available at http://www.maizego.org/Resources.html . All other data supporting the findings of this study are available within the paper, and its supplementary data is published online.
Andorf CM, Cannon EK, Portwood JL, Gardiner JM, Harper LC, Schaeffer ML, Braun BL, Campbell DA, Vinnakota AG, Sribalusu VV (2016) MaizeGDB update: new tools, data and interface for the maize model organism database. Nucleic Acids Res 44:D1195–D1201
CAS PubMed Google Scholar
Bates D, Mächler M, Bolker B, Walker S (2014) Fitting linear mixed-effects models using lme4. arXiv preprint arXiv:14065823
Bayuelo-Jiménez JS, Gallardo-Valdéz M, Pérez-Decelis VA, Magdaleno-Armas L, Ochoa I, Lynch JP (2011) Genotypic variation for root traits of maize ( Zea mays L.) from the Purhepecha Plateau under contrasting phosphorus availability. Field Crop Res 121:350–362
Google Scholar
Burton AL, Brown KM, Lynch JP (2013) Phenotypic diversity of root anatomical and architectural traits in Zea species. Crop Sci 53:1042–1055
Burton AL, Johnson JM, Foerster JM, Hirsch CN, Buell C, Hanlon MT, Kaeppler SM, Brown KM, Lynch JP (2014) QTL mapping and phenotypic variation for root architectural traits in maize ( Zea mays L.). Theor Appl Genet 127:2293–2311
PubMed Google Scholar
Burton AL, Johnson J, Foerster J, Hanlon MT, Kaeppler SM, Lynch JP, Brown KM (2015) QTL mapping and phenotypic variation of root anatomical traits in maize ( Zea mays L.). Theor Appl Genet 128:93–106
Cai H, Chen F, Mi G, Zhang F, Maurer HP, Liu W, Reif JC, Yuan L (2012) Mapping QTLs for root system architecture of maize ( Zea mays L.) in the field at different developmental stages. Theor Appl Genet 125:1313–1324
Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R (2020) TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol Plant 13:1194-1202
Chen Z, Sun J, Li D, Li P, He K, Ali F, Mi G, Chen F, Yuan L, Pan Q (2022) Plasticity of root anatomy during domestication of a maize-teosinte derived population. J Exp Bot 73:139–153
Das A, Schneider H, Burridge J, Ascanio AKM, Wojciechowski T, Topp CN, Lynch JP, Weitz JS, Bucksch A (2015) Digital imaging of root traits (DIRT): a high-throughput computing and collaboration platform for field-based root phenomics. Plant Methods 11:51
PubMed Central PubMed Google Scholar
de Dorlodot S, Forster B, Pagès L, Price A, Tuberosa R, Draye X (2007) Root system architecture: opportunities and constraints for genetic improvement of crops. Trends Plant Sci 12:474–481
Feldman L (1994) The maize root. The maize handbook. Springer, pp 29–37
Fu J, Cheng Y, Linghu J, Yang X, Kang L, Zhang Z, Zhang J, He C, Du X, Peng Z (2013) RNA sequencing reveals the complex regulatory network in the maize kernel. Nat Commun 4:1–12
CAS Google Scholar
Gu R, Chen F, Long L, Cai H, Liu Z, Yang J, Wang L, Li H, Li J, Liu W (2016) Enhancing phosphorus uptake efficiency through QTL-based selection for root system architecture in maize. J Genet Genomics 43:663–672
Guo J, Li C, Zhang X, Li Y, Zhang D, Shi Y, Song Y, Li Y, Yang D, Wang T (2020) Transcriptome and GWAS analyses reveal candidate gene for seminal root length of maize seedlings under drought stress. Plant Sci 292:110380
He X, Ma H, Zhao X, Nie S, Li Y, Zhang Z, Shen Y, Chen Q, Lu Y, Lan H (2016) Comparative RNA-Seq analysis reveals that regulatory network of maize root development controls the expression of genes in response to N stress. PLoS ONE 11:e0151697
Hirsch CN, Foerster JM, Johnson JM, Sekhon RS, Muttoni G, Vaillancourt B, Peñagaricano F, Lindquist E, Pedraza MA, Barry K (2014) Insights into the maize pan-genome and pan-transcriptome. Plant Cell 26:121–135
CAS PubMed Central PubMed Google Scholar
Hochholdinger F, Woll K, Sauer M, Dembinsky D (2004) Genetic dissection of root formation in maize ( Zea mays ) reveals root-type specific developmental programmes. Ann Bot 93:359–368
Hochholdinger F, Wen TJ, Zimmermann R, Chimot-Marolle P, Da Costa e Silva O, Bruce W, Lamkey KR, Wienand U, Schnable PS (2008) The maize ( Zea mays L.) roothairless3 gene encodes a putative GPI-anchored, monocot-specific, COBRA-like protein that significantly affects grain yield. Plant J 54:888–898
Hoopes GM, Hamilton JP, Wood JC, Esteban E, Pasha A, Vaillancourt B, Provart NJ, Buell CR (2019) An updated gene atlas for maize reveals organ-specific and stress-induced genes. Plant J 97:1154–1167
Jin M, Zhang X, Zhao M, Deng M, Du Y, Zhou Y, Wang S, Tohge T, Fernie AR, Willmitzer L (2017) Integrated genomics-based mapping reveals the genetics underlying maize flavonoid biosynthesis. BMC Plant Biol 17:1–17
Kano M, Inukai Y, Kitano H, Yamauchi A (2011) Root plasticity as the key root trait for adaptation to various intensities of drought stress in rice. Plant Soil 342:117–128
Kassambara A, Mundt F (2017) Factoextra: extract and visualize the results of multivariate data analyses. R Package Version 1:337–354
Kidd BN, Edgar CI, Kumar KK, Aitken EA, Schenk PM, Manners JM, Kazan K (2009) The mediator complex subunit PFT1 is a key regulator of jasmonate-dependent defense in Arabidopsis . Plant Cell 21:2237–2252
Kim D, Langmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12:357–360
Lai J, Li R, Xu X, Jin W, Xu M, Zhao H, Xiang Z, Song W, Ying K, Zhang M (2010) Genome-wide patterns of genetic variation among elite maize inbred lines. Nat Genet 42:1027–1030
Li Q, Li L, Yang X, Warburton ML, Bai G, Dai J, Li J, Yan J (2010) Relationship, evolutionary fate and function of two maize co-orthologs of rice GW2associated with kernel size and weight. BMC Plant Biol 10:1–15
Li Q, Yang X, Xu S, Cai Y, Zhang D, Han Y, Li L, Zhang Z, Gao S, Li J (2012) Genome-wide association studies identified three independent polymorphisms associated with α-tocopherol content in maize kernels. PLoS ONE 7:e36807
Li H, Peng Z, Yang X, Wang W, Fu J, Wang J, Han Y, Chai Y, Guo T, Yang N (2013) Genome-wide association study dissects the genetic architecture of oil biosynthesis in maize kernels. Nat Genet 45:43–50
Li P, Chen F, Cai H, Liu J, Pan Q, Liu Z, Gu R, Mi G, Zhang F, Yuan L (2015) A genetic relationship between nitrogen use efficiency and seedling root traits in maize as revealed by QTL analysis. J Exp Bot 66:3175–3188
Li J, Chen F, Li Y, Li P, Wang Y, Mi G, Yuan L (2019) ZmRAP2.7, an AP2 transcription factor, is involved in maize brace roots development. Front Plant Sci 10:820
Li D, Wang H, Wang M, Li G, Chen Z, Leiser WL, Weiss TM, Lu X, Wang M, Chen S, Chen F, Yuan L, Wurschum T, Liu W (2021) Genetic dissection of phosphorus use efficiency in a maize association population under two P levels in the field. Int J Mol Sci 22(17):9311
Lipka AE, Tian F, Wang Q, Peiffer J, Li M, Bradbury PJ, Gore MA, Buckler ES, Zhang Z (2012) GAPIT: genome association and prediction integrated tool. Bioinformatics 28:2397–2399
Liu X, Huang M, Fan B, Buckler ES, Zhang Z (2016) Iterative usage of fixed and random effect models for powerful and efficient genome-wide association studies. PLoS Genet 12:e1005767
Liu Z, Gao K, Shan S, Gu R, Wang Z, Craft EJ, Mi G, Yuan L, Chen F (2017) Comparative analysis of root traits and the associated QTLs for maize seedlings grown in paper roll, hydroponics and vermiculite culture system. Front Plant Sci 8:436
Liu Z, Zhao Y, Guo S, Cheng S, Guan Y, Cai H, Mi G, Yuan L, Chen F (2019) Enhanced crown root number and length confers potential for yield improvement and fertilizer reduction in nitrogen-efficient maize cultivars. Field Crop Res 241:107562
Liu S, Barrow CS, Hanlon M, Lynch JP, Bucksch A (2021) DIRT/3D: 3D root phenotyping for field-grown maize (Zea mays). Plant Physiol 187:739–757
Lynch J (1995) Root architecture and plant productivity. Plant Physiol 109:7
Lynch JP (2018) Rightsizing root phenotypes for drought resistance. J Exp Bot 69:3279–3292
Ma L, Qing C, Frei U, Shen Y, Lübberstedt T (2020) Association mapping for root system architecture traits under two nitrogen conditions in germplasm enhancement of maize doubled haploid lines. The Crop Journal 8:213–226
Mi G, Chen F, Yuan L, Zhang F (2016) Ideotype root system architecture for maize to achieve high yield and resource use efficiency in intensive cropping systems. Adv Agron 139:73–97
Muszynski MG, Moss-Taylor L, Chudalayandi S, Cahill J, Del Valle-Echevarria AR, Alvarez-Castro I, Petefish A, Sakakibara H, Krivosheev DM, Lomin SN (2020) The maize hairy sheath frayed1 (Hsf1) mutation alters leaf patterning through increased cytokinin signaling. Plant Cell 32:1501–1518
Pace J, Gardner C, Romay C, Ganapathysubramanian B, Lübberstedt T (2015) Genome-wide association analysis of seedling root development in maize ( Zea mays L.). BMC Genomics 16:1–12
Pertea M, Pertea GM, Antonescu CM, Chang T-C, Mendell JT, Salzberg SL (2015) StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33:290–295
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, De Bakker PI, Daly MJ (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Human Genet 81:559–575
Raihan MS, Liu J, Huang J, Guo H, Pan Q, Yan J (2016) Multi-environment QTL analysis of grain morphology traits and fine mapping of a kernel-width QTL in Zheng58 × SK maize population. Theor Appl Genet 129:1465–1477
Ren W, Zhao L, Liang J, Wang L, Chen L, Li P, Liu Z, Li X, Zhang Z, Li J, He K, Zhao Z, Ali F, Mi G, Yan J, Zhang F, Chen F, Yuan L, Pan Q (2022) Genome-wide dissection of changes in maize root system architecture during modern breeding. Nat Plants 8(12):1408–1422
Saengwilai P, Tian X, Lynch JP (2014) Low crown root number enhances nitrogen acquisition from low-nitrogen soils in maize. Plant Physiol 166:581–589
Schaefer RJ, Michno J-M, Jeffers J, Hoekenga O, Dilkes B, Baxter I, Myers CL (2018) Integrating coexpression networks with GWAS to prioritize causal genes in maize. Plant Cell 30:2922–2942
Shin J-H, Blay S, Lewin-Koh N, McNeney B, Yang G, Reyers M, Yan Y, Graham J (2016) Package ‘LDheatmap’. R package
Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T (2011) Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27:431–432
Steklov MY, Lomin SN, Osolodkin DI, Romanov GA (2013) Structural basis for cytokinin receptor signaling: an evolutionary approach. Plant Cell Rep 32:781–793
Stelpflug SC, Sekhon RS, Vaillancourt B, Hirsch CN, Buell CR, de Leon N, Kaeppler SM (2016) An expanded maize gene expression atlas based on RNA sequencing and its use to explore root development. Plant Genome 9(1):plantgenome2015–plantgenome2104
Sun B, Gao Y, Lynch JP (2018) Large crown root number improves topsoil foraging and phosphorus acquisition. Plant Physiol 177:90–104
Team RC (2013) R: A language and environment for statistical computing
Thorup-Kristensen K, Halberg N, Nicolaisen M, Olesen JE, Crews TE, Hinsinger P, Kirkegaard J, Pierret A, Dresbøll DB (2020) Digging deeper for agricultural resources, the value of deep rooting. Trends Plant Sci 25:406–417
Trachsel S, Kaeppler SM, Brown KM, Lynch JP (2011) Shovelomics: high throughput phenotyping of maize ( Zea mays L.) root architecture in the field. Plant Soil 341:75–87
Tracy SR, Nagel KA, Postma JA, Fassbender H, Wasson A, Watt M (2020) Crop improvement from phenotyping roots: highlights reveal expanding opportunities. Trends Plant Sci 25:105–118
Viana WG, Scharwies JD, Dinneny JR (2022) Deconstructing the root system of grasses through an exploration of development, anatomy and function. Plant, Cell Environ 45:602–619
Walbot V (2009) 10 reasons to be tantalized by the B73 maize genome. PLoS Genet 5:e1000723–e1000723
Wang H, Lockwood SK, Hoeltzel MF, Schiefelbein JW (1997) The ROOT HAIR DEFECTIVE3 gene encodes an evolutionarily conserved protein with GTP-binding motifs and is required for regulated cell enlargement in Arabidopsis . Genes Dev 11:799–811
Wang H, Xu C, Liu X, Guo Z, Xu X, Wang S, Xie C, Li W-X, Zou C, Xu Y (2017) Development of a multiple-hybrid population for genome-wide association studies: theoretical consideration and genetic mapping of flowering traits in maize. Sci Rep 7:40239
Wang K, Zhang Z, Sha X, Yu P, Li Y, Zhang D, Liu X, He G, Li Y, Wang T, Guo J, Chen J, Li C (2023) Identification of a new QTL underlying seminal root number in a maize-teosinte population. Front Plant Sci 14:1132017
Wen W, Li D, Li X, Gao Y, Li W, Li H, Liu J, Liu H, Chen W, Luo J, Yan J (2014) Metabolome-based genome-wide association study of maize kernel leads to novel biochemical insights. Nat Commun 5:3438
Wickham H (2016) ggplot2: elegant graphics for data analysis Springer-Verlag New York; 2009. Book
Wu B, Ren W, Zhao L, Li Q, Sun J, Chen F, Pan Q (2022) Genome-wide association study of root system architecture in maize. Genes (basel) 13(2):181
Xue X-H, Guo C-Q, Du F, Lu Q-L, Zhang C-M, Ren H-Y (2011) AtFH8 is involved in root development under effect of low-dose latrunculin B in dividing cells. Mol Plant 4:264–278
Yan J, Kandianis CB, Harjes CE, Bai L, Kim E-H, Yang X, Skinner DJ, Fu Z, Mitchell S, Li Q, Fernandez MGS, Zaharieva M, Babu R, Fu Y, Palacios N, Li J, DellaPenna D, Brutnell T, Buckler ES, Warburton ML, Rocheford T (2010) Rare genetic variation at Zea mays crtRB1 increases β-carotene in maize grain. Nat Genet 42:322–327
Yan J, Warburton M, Crouch J (2011) Association mapping for enhancing maize ( Zea mays L.) genetic improvement. Crop Sci 51:433–449
Yang X, Gao S, Xu S, Zhang Z, Prasanna BM, Li L, Li J, Yan J (2011) Characterization of a global germplasm collection and its potential utilization for analysis of complex quantitative traits in maize. Mol Breeding 28:511–526
Yin L (2020) CMplot: circle manhattan plot. R Package Version 3:6
Yonekura-Sakakibara K, Kojima M, Yamaya T, Sakakibara H (2004) Molecular characterization of cytokinin-responsive histidine kinases in maize. Differential ligand preferences and response to cis-zeatin. Plant Physiol 134:1654–1661
Yu P, Hochholdinger F, Li C (2015) Root-type-specific plasticity in response to localized high nitrate supply in maize ( Zea mays ). Ann Bot 116:751–762
Yu T, Liu C, Lu X, Bai Y, Zhou L, Cai Y (2019) ZmAPRG, an uncharacterized gene, enhances acid phosphatase activity and Pi concentration in maize leaf during phosphate starvation. Theor Appl Genet 132:1035–1048
Zhang X, Warburton ML, Setter T, Liu H, Xue Y, Yang N, Yan J, Xiao Y (2016) Genome-wide association studies of drought-related metabolic changes in maize using an enlarged SNP panel. Theor Appl Genet 129:1449–1463
Zheng Z, Hey S, Jubery T, Liu H, Yang Y, Coffey L, Miao C, Sigmon B, Schnable JC, Hochholdinger F, Ganapathysubramanian B, Schnable PS (2019) Shared Genetic Control of Root System Architecture between Zea mays and Sorghum bicolor 1 [OPEN]. Plant Physiol 182:977–991
Zhu J, Kaeppler SM, Lynch JP (2005) Mapping of QTL controlling root hair length in maize ( Zea mays L.) under phosphorus deficiency. Plant Soil 270:299–310
Download references
Acknowledgements
The authors gratefully acknowledge Dr. Jianbin Yan, Huazhong Agricultural University, who provided the germplasm resources and established the genotypes for the association mapping population.We also thank Dr. Philip James Kear, from International Potato Center–China Center for Asia and the Pacific, for providing valuable feedback and editing the revised version of our manuscript.
This study was financially supported by the Hainan Provincial Natural Science Foundation of China (321CXTD443) and the National Natural Science Foundation of China (31972485, 31971948).
Author information
Authors and affiliations.
College of Resources and Environmental Sciences, National Academy of Agriculture Green Development, Key Laboratory of Plant-Soil Interactions of MOE, China Agricultural University, Beijing, China
Zhigang Liu, Wei Ren, Guohua Mi, Lixing Yuan, Fanjun Chen & Qingchun Pan
Key Laboratory of Plant Functional Genomics of the Ministry of Education, Yangzhou University, Yangzhou, China
Pengcheng Li
College of Resources and Environment, Jilin Agricultural University, Changchun, China
Global Institute for Food Security, University of Saskatchewan, Saskatoon, Canada
Zhigang Liu & Toluwase Olukayode
Sanya Institute of China Agricultural University, Sanya, China
Fanjun Chen & Qingchun Pan
You can also search for this author in PubMed Google Scholar
Contributions
FC and QP designed the experiment; ZL analyzed the data and wrote the manuscript; PL performed the experiments; WR and ZC assisted in data analysis; and OT, GM, LY, FC, and QP contributed to manuscript editing.
Corresponding author
Correspondence to Qingchun Pan .
Ethics declarations
Conflict of interest.
The authors declare that they have no conflict of interests.
Additional information
Publisher's note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
122_2023_4442_MOESM1_ESM.xlsx
Supplement Table S1: Information of the 356 representative maize panel. Supplement Table S2: Environmental information of Guangxing, Yunnan and Hainan locations for field experiments in this study. Supplement Table S3: Summary of the significant SNPs association loci for root traits. (XLSX 43 KB)
122_2023_4442_MOESM2_ESM.tif
Supplement Figure 1 Distribution of 1.25 million polymorphic SNPs in the maize genome. Heatmap of SNP density on the chromosome within a 1-Mb interval, colors were used to indicate the number of SNPs within the 1-Mb interval. The physical position of the SNPs was based on the B73 reference sequence (RefGen_V2). (TIF 9090 KB)
122_2023_4442_MOESM3_ESM.tif
Supplement Figure 2 Distribution and correlation of root system architecture traits between each pair of locations. The significance levels of pairwise t-tests were added. * indicates P ≤ 0.05, ** indicates P ≤ 0.01; *** indicates P ≤ 0.001; **** indicates P ≤ 0.0001. Different lowercase letters indicate significant differences (P < 0.05) in different locations, as determined by Tukey's HSD test. Abbreviations for root traits are as follows: BRN, brace root number; BRWN, brace root whorl number; CR1, 1 st whorl crown roots; CR2, 2 nd whorl crown roots; CR3, 3 rd whorl crown roots; CR4, 4 th whorl crown roots; CR5, 5 th whorl crown roots; CR6, 6 th -8 th whorl crown roots; CRN, crown root number; CRWN, crown root whorl number; NRWN, nodal root whorl number. GX11: Guangxi location in 2011, HN11: Hainan location in 2011, YN11: Yunnan location in 2011. (TIF 9318 KB)
122_2023_4442_MOESM4_ESM.tif
Supplement Figure 3 The phenotypic distribution of root traits in the association panel. Abbreviations for root traits are as follows: BRN, brace root number; BRWN, brace root whorl number; CR1, 1 st whorl crown roots; CR2, 2 nd whorl crown roots; CR3, 3 rd whorl crown roots; CR4, 4 th whorl crown roots; CR5, 5 th whorl crown roots; CR6, 6 th -8 th whorl crown roots; CRN, crown root number; CRWN, crown root whorl number; NRWN, nodal root whorl number. (TIF 65415 KB)
122_2023_4442_MOESM5_ESM.tif
Supplement Figure 4 Phenotypic correlations among the root traits in the association panel. Abbreviations for root traits are as follows: BRN, brace root number; BRWN, brace root whorl number; CR1, 1 st whorl crown roots; CR2, 2 nd whorl crown roots; CR3, 3 rd whorl crown roots; CR4, 4 th whorl crown roots; CR5, 5 th whorl crown roots; CR6, 6 th -8 th whorl crown roots; CRN, crown root number; CRWN, crown root whorl number; NRWN, nodal root whorl number. (TIF 61613 KB)
122_2023_4442_MOESM6_ESM.tif
Supplement Figure 5 The percentage of total variance explained by each principal component. Abbreviations for root traits are as follows: BRN, brace root number; BRWN, brace root whorl number; CR1, 1 st whorl crown roots; CR2, 2 nd whorl crown roots; CR3, 3 rd whorl crown roots; CR4, 4 th whorl crown roots; CR5, 5 th whorl crown roots; CR6, 6 th -8 th whorl crown roots; CRN, crown root number; CRWN, crown root whorl number; NRN, nodal root number; NRWN, nodal root whorl number. (TIF 11460 KB)
122_2023_4442_MOESM7_ESM.tif
Supplement Figure 6 Comparison of root traits between PA and SPT heterotic groups. Different lowercase letters indicate significant differences ( P < 0.05) in different locations, as determined by Tukey's HSD test. Abbreviations for root traits are as follows: BRN, brace root number; BRWN, brace root whorl number; CR1, 1 st whorl crown roots; CR2, 2 nd whorl crown roots; CR3, 3 rd whorl crown roots; CR4, 4 th whorl crown roots; CR5, 5 th whorl crown roots; CR6, 6 th -8 th whorl crown roots; CRN, crown root number; CRWN, crown root whorl number; NRN, nodal root number; NRWN, nodal root whorl number. (TIF 11656 KB)
122_2023_4442_MOESM8_ESM.tif
Supplement Figure 7 Quantile–quantile plots of root traits. (a) BRN, brace root number, (b) BRWN, brace root whorl number, (c) CR1, 1 st whorl crown roots, (d) CR2, 2 nd whorl crown roots, (e) CR3, 3 rd whorl crown roots, (f) CR4, 4 th whorl crown roots, (g) CR5, 5 th whorl crown roots, (h) CR6, 6 th -8 th whorl crown roots, (i) CRN, crown root number, (j) CRWN, crown root whorl number, (k) NRN, nodal root number, (l) NRWN, nodal root whorl number. (TIF 60252 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Reprints and permissions
About this article
Liu, Z., Li, P., Ren, W. et al. Hybrid performance evaluation and genome-wide association analysis of root system architecture in a maize association population. Theor Appl Genet 136 , 194 (2023). https://doi.org/10.1007/s00122-023-04442-7
Download citation
Received : 02 December 2022
Accepted : 04 August 2023
Published : 22 August 2023
DOI : https://doi.org/10.1007/s00122-023-04442-7
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Find a journal
- Publish with us
- Track your research
Advertisement
49ers only team spending more at wr than seahawks in 2024, share this article.
Nobody ever said that winning was cheap. As much as any industry it applies in the NFL, where choosing how to allocate your cap money is as critical to building a contender as who your head coach is. Under former Seahawks head coach Pete Carroll the team tended to overspend at non-premium positions like linebacker sand safety. It seems general manager John Schneider is determined to correct that based on how he approached this offseason.
One area where Carroll was correct to invest a lot of resources is the wide receiver position, and that’s carried over into the Mike Macdonald era. Heading into the 2024 season, only the 49ers are spending more money at wide receiver based on cap hits:
Most expensive WR groups in 2024 based on cap hit: #49ers – $55.01M #Seahawks – $54.65M #Cowboys – $51.22M #Dolphins – $48.82M #Bears – $45.60M Cheapest WR groups: #Bills – $15.21M #Cardinals – $13.46M #Chargers – $12.77M #Steelers – $11.71M #Packers – $11.52M — NFL Stats (@NFL_Stats) May 16, 2024
In Deebo Samuel and Brandon Aiyuk the Niners have two of the league’s best receivers – we have both of them ranked in the top 20 . They also added Florida’s Ricky Pearsall in the draft, potentially giving them their third Musketeer.
While as a general rule it makes sense to spend at this position, the value is somewhat lost in San Francisco’s case as the extremely overrated Brock Purdy can’t really take full advantage of their talents. Kyle Shanhan and the 49ers offense would probably be better served by shifting some of those resources to their offensive line – or better yet, a QB upgrade.
As for the Seahawks, they’re paying a good amount of money to WR1 DK Metcalf, as well as veteran Tyler Lockett and second-year wide out Jaxon Smith-Njigba. That cap number should drop a bit when Lockett inevitably leaves and is replaced by someone on a rookie contract.
More Seahawks Wire stories
Ranking all 32 teams by ESPN projected win totals
4 biggest takeaways from Seahawks’ 2024 schedule
Seahawks all-time record against each ’24 opponent
Week by week predictions for all 17 Seahawks games
Want the latest news and insights on your favorite team?
Sign up for our newsletter to get updates to your inbox, and also receive offers from us, our affiliates and partners. By signing up you agree to our Privacy Policy
An error has occured
Please re-enter your email address.
Thanks for signing up!
You'll now receive the top Seahawks Wire stories each day directly in your inbox.
Most Popular
Ranking all 32 nfl teams (including the seahawks) by espn's projected win totals, predicting each game on the seattle seahawks 2024 schedule, ranking the league's top 50 quarterbacks after the 2024 nfl draft, 4 biggest takeaways from the seattle seahawks 2024 schedule, seattle seahawks all-time record vs. every team on their 2024 schedule, seahawks 90-man roster by jersey number with undrafted free agents, espn: seahawks have improved most along their defensive line.
Please enter an email address.
Thanks for signing up.
Please check your email for a confirmation.
Something went wrong.

IMAGES
VIDEO
COMMENTS
Abstract. This tutorial is a learning resource that outlines the basic process and provides specific software tools for implementing a complete genome‐wide association analysis. Approaches to post‐analytic visualization and interrogation of potentially novel findings are also presented. Applications are illustrated using the free and open ...
The aim of GWAS is exceedingly simple—namely to detect association between allele or genotype frequency and trait status. The first step of such analysis is to identify the traits to be scored and select an appropriate study population considering both the size of the population and the amounts of genetic and trait variance that it possesses (Fig. 1).
Genome-wide association studies (GWAS) have uncovered numerous variants linked to a wide range of complex traits. However, understanding the mechanisms underlying these associations remains a challenge. To determine genetically regulated mechanisms, additional layers of gene regulation, such as transcriptome and proteome, need to be assayed. Transcriptome-wide association studies (TWAS) and ...
Genome-wide analysis of trimethylated lysine 4 of histone H3 (H3K4me3) in Aspergillus niger Christina Sawchyn A Thesis in The Department of ... well, in the preparation of this Thesis, Dr Tsang provided me with a great deal of feedback, which has greatly impacted my writing skills in scientific communication. Finally, Dr Tsang has supported me in
A genome-wide association study (GWAS) is a study design used to find associations between a given trait or disease and loci across the entire genome. -wide During annotation of genome association studies, false positive results can mislead and misdirect researchers, leading to wasted resources, time, and money.
Genome-wide characterization of genetic variation may have immense potential for the exploitation of natural genetic resources in fruit species as observed in grapes [].Robust and equally distributed genome-wide SNP markers linked with reference genetic linkage maps, help us to utilize new genomic-based approaches like GWAS and GS [] which are currently developing as effective tools in various ...
Our meta-analysis of genome-wide association (GWA) studies (GWAS) in plant pathosystems highlights the power of GWA mapping to characterize thoroughly the genetic architecture of plant responses to a wide range of pathogens, subsequently leading to the identification of novel defense mechanisms. GWAS in pathogens revealed fewer, but nonetheless ...
2.1 Assembly and Phenotyping of an Association Panel. The foundational step in any GWAS is the selection of the accessions that will form the association panel. This has two important components to it: the number of accessions and their degree of relationship or how wide (genetically diverse) the panel should be.
Transcriptome-wide association studies (TWAS) aim to detect relationships between gene expression and a phenotype, and are commonly used for secondary analysis of genome-wide association study (GWAS) results. Results from TWAS analyses are often interpreted as indicating a geneticrelationship between gene expression and a phenotype, but this interpretation is not consistent with the null ...
GWAS analysis performed on a genome-wide SNP matrix that was designed to cover coding as well as the regulatory regions such as promoters, alternative spliced junctions, 5′ and 3′ untranslated ...
1 Introduction. The Wnt/β-catenin pathway is an important cell-to-cell signaling mechanism conserved among animals including humans (Loh, van Amerongen, & Nusse, 2016).Wnt/β-catenin signaling function has been linked to a wide range of biological processes as diverse as embryonic axis formation and patterning, cell proliferation and differentiation, as well as tissue regeneration and cancer ...
SARS-CoV-2 mortality has been extensively studied in relation to host susceptibility. How sequence variations in the SARS-CoV-2 genome affect pathogenicity is poorly understood. Starting in October 2020, using the methodology of genome-wide association studies (GWAS), we looked at the association be …
In the present study, we performed genome-wide SNP (single nucleotide polymorphism) analysis (Affymetrix, 6.0) of 262 well-classified male individuals (after screening 3416 subjects) belonging to ...
This design analysis and interpretation of genome wide association scans will help people to enjoy a good book with a cup of coffee in the afternoon instead of juggled with some infectious bugs inside their laptop. Thank you for downloading design analysis and interpretation of genome wide association scans. As you may know, people have search numerous times for their favorite books like this ...
The C2H2 zinc finger protein (C2H2-ZFP) gene family plays important roles in response to environmental stresses and several other biological processes in plants. Ginseng is a precious medicinal herb cultivated in Asia and North America. However, little is known about the C2H2-ZFP gene family and its …
The genetic architecture of RSA traits was dissected by GWAS and coexpression networks analysis in a maize association population. ... number (CRN) and 8.6% less brace root number (BRN) than Zheng58 × NSS and Zheng58 × SS, respectively. Using a genome-wide association study (GWAS) with 1.25 million SNPs and correction for population structure ...
You may also want to consult these sites to search for other theses: Google Scholar; NDLTD, the Networked Digital Library of Theses and Dissertations.NDLTD provides information and a search engine for electronic theses and dissertations (ETDs), whether they are open access or not. Proquest Theses and Dissertations (PQDT), a database of dissertations and theses, whether they were published ...
Table of contents. Step 1: Reading the text and identifying literary devices. Step 2: Coming up with a thesis. Step 3: Writing a title and introduction. Step 4: Writing the body of the essay. Step 5: Writing a conclusion. Other interesting articles.
Genome‑wide association studies (GWAS) It was reported on 11 January 2019 that for humans 3730 GWAS studies had been published with a total of 37 730 sin-gle nucleotide variations and 52 415 unique SNV-trait asso-ciations above a genome-wide signicance threshold [1 , 2]. Analysis of the staggering increase in the number of associa-
DESIGN AND ANALYSIS OF ULTRA WIDE BAND CMOS LNA by Janmejay Adhyaru An Ultra WideBand CMOS Low Noise Amplifier (LNA) is presented. Due to really low power consumption and extremely high data rates the UWB standard is bound to be popular in the consumer market. The LNA is the outer most part of an UWB transceiver.
When Wide Sargasso Sea, her last novel, was published, Jean Rhys (24 August 1890 - 14 May 1979) was described in The New York Times as the greatest living novelist. Such praise is overstated, but Rhys's fiction, long overlooked by academic critics, is undergoing a revival spurred by feminist studies. Rhys played a noteworthy role….
Background The severity of COVID-19 is influenced by various factors including the presence of respiratory diseases. Studies have indicated a potential relationship between asthma and COVID-19 severity. Objective This study aimed to conduct a genome-wide association study (GWAS) to identify genetic and clinical variants associated with the severity of COVID-19, both among patients with and ...
Critical analysis is a process of examining a piece of work or an idea in a systematic, objective, and analytical way. It involves breaking.. About us; ... Develop your thesis statement: Based on your analysis, develop a clear and concise thesis statement that summarizes your overall evaluation of the text.
Glenn Erby. May 20, 2024 9:42 pm ET. According to ESPN's Adam Schefter, the Eagles suffered a loss in personnel on Monday after veteran pass catcher DeVante Parker announced that he was retiring ...
Download app. Subscription Support: 1-347-509-6837. With a solid dividend yield mitigating downside risks, Enbridge is poised to rally. Find out if ENB stock is a buy.
Summary. TSLX was one of my favorite BDC picks going into 2024. So far the BDC has generated flat performance, while the overall BDC market has surged higher. The deviation here is mostly ...
Key Message The genetic architecture of RSA traits was dissected by GWAS and coexpression networks analysis in a maize association population. Abstract Root system architecture (RSA) is a crucial determinant of water and nutrient uptake efficiency in crops. However, the maize genetic architecture of RSA is still poorly understood due to the challenges in quantifying root traits and the lack of ...
jiefeng jiang. Investment Thesis. 2024 is said to be the Year of the Dragon.In Chinese culture, the dragon is a highly auspicious symbol, revered, and known to be a symbol of fortune.
Rookie receivers hit the ground running in the NFL each year, with Puka Nacua of the Los Angeles Rams the standout in 2023. At least one rookie wide receiver has cracked 1,000 receiving yards in the regular season in each of the past five seasons, and it is often as much about the situation they land in as much as their talent.
Heading into the 2024 season, only the 49ers are spending more money at wide receiver based on cap hits: Most expensive WR groups in 2024 based on cap hit: #49ers - $55.01M #Seahawks - $54.65M #Cowboys - $51.22M #Dolphins - $48.82M #Bears - $45.60M.