If you're seeing this message, it means we're having trouble loading external resources on our website.
If you're behind a web filter, please make sure that the domains *.kastatic.org and *.kasandbox.org are unblocked.
To log in and use all the features of Khan Academy, please enable JavaScript in your browser.

High school biology
Course: high school biology > unit 1.
- Biology overview
- Preparing to study biology
- What is life?
- The scientific method
- Data to justify experimental claims examples
- Scientific method and data analysis
- Introduction to experimental design
- Controlled experiments
Biology and the scientific method review
- Experimental design and bias
The nature of biology
Properties of life.
- Organization: Living things are highly organized (meaning they contain specialized, coordinated parts) and are made up of one or more cells .
- Metabolism: Living things must use energy and consume nutrients to carry out the chemical reactions that sustain life. The sum total of the biochemical reactions occurring in an organism is called its metabolism .
- Homeostasis : Living organisms regulate their internal environment to maintain the relatively narrow range of conditions needed for cell function.
- Growth : Living organisms undergo regulated growth. Individual cells become larger in size, and multicellular organisms accumulate many cells through cell division.
- Reproduction : Living organisms can reproduce themselves to create new organisms.
- Response : Living organisms respond to stimuli or changes in their environment.
- Evolution : Populations of living organisms can undergo evolution , meaning that the genetic makeup of a population may change over time.
Scientific methodology
Scientific method example: failure to toast, experimental design, reducing errors and bias.
- Having a large sample size in the experiment: This helps to account for any small differences among the test subjects that may provide unexpected results.
- Repeating experimental trials multiple times: Errors may result from slight differences in test subjects, or mistakes in methodology or data collection. Repeating trials helps reduce those effects.
- Including all data points: Sometimes it is tempting to throw away data points that are inconsistent with the proposed hypothesis. However, this makes for an inaccurate study! All data points need to be included, whether they support the hypothesis or not.
- Using placebos , when appropriate: Placebos prevent the test subjects from knowing whether they received a real therapeutic substance. This helps researchers determine whether a substance has a true effect.
- Implementing double-blind studies , when appropriate: Double-blind studies prevent researchers from knowing the status of a particular participant. This helps eliminate observer bias.
Communicating findings
Things to remember.
- A hypothesis is not necessarily the right explanation. Instead, it is a possible explanation that can be tested to see if it is likely correct, or if a new hypothesis needs to be made.
- Not all explanations can be considered a hypothesis. A hypothesis must be testable and falsifiable in order to be valid. For example, “The universe is beautiful" is not a good hypothesis, because there is no experiment that could test this statement and show it to be false.
- In most cases, the scientific method is an iterative process. In other words, it's a cycle rather than a straight line. The result of one experiment often becomes feedback that raises questions for more experimentation.
- Scientists use the word "theory" in a very different way than non-scientists. When many people say "I have a theory," they really mean "I have a guess." Scientific theories, on the other hand, are well-tested and highly reliable scientific explanations of natural phenomena. They unify many repeated observations and data collected from lots of experiments.
Want to join the conversation?
- Upvote Button navigates to signup page
- Downvote Button navigates to signup page
- Flag Button navigates to signup page

Synthesis, Properties, and Characterization of Biological Nanomaterial
- First Online: 28 August 2023
Cite this chapter

- Sarvat Zafar 7
Part of the book series: Smart Nanomaterials Technology ((SNT))
264 Accesses
1 Citations
This chapter explored the various biological methods for creating nanostructured materials, including their properties and characterization techniques. Biological approaches are gaining popularity in the synthesis of nanomaterials because of their sustainability, cost-effectiveness, speed, non-pathogenic nature, environmental friendliness, ease of scaling up for large-scale synthesis, and lack of need for high pressure, temperature, or toxic chemical components. Furthermore, these nano-sized materials are used safely and effectively for human therapeutic purposes and have distinctive dimensions with a large surface area, chemical stability, and greater binding density, making them promising for applications in diverse areas such as food processing, drug delivery, cosmetics, pharmaceuticals, chemical industry, mechanics, wastewater purification, and catalytic properties. In addition, nanomaterials are characterized by their surface morphology and compositional structure using various techniques such as energy-dispersive X-ray spectroscopy (EDS), dynamic light scattering (DLS), atomic force microscopy (AFM), Raman spectroscopy (RS), Fourier transform infrared (FT-IR) spectroscopy, and scanning and transmission electron microscopy (SEM/TEM).
This is a preview of subscription content, log in via an institution to check access.
Access this chapter
- Available as PDF
- Read on any device
- Instant download
- Own it forever
- Available as EPUB and PDF
- Durable hardcover edition
- Dispatched in 3 to 5 business days
- Free shipping worldwide - see info
Tax calculation will be finalised at checkout
Purchases are for personal use only
Institutional subscriptions
Singh P, Kim Y-J, Zhang D, Yang D-C (2016) Biological synthesis of nanoparticles from plants and microorganisms. Trends Biotechnol 34(7):588–599
Article CAS Google Scholar
Hussain I, Singh N, Singh A, Singh H, Singh S (2016) Green synthesis of nanoparticles and its potential application. Biotech Lett 38:545–560
Sanguansri P, Augustin MA (2006) Nanoscale materials development–a food industry perspective. Trends Food Sci Technol 17(10):547–556
Azzazy HM, Mansour MM, Samir TM, Franco R (2012) Gold nanoparticles in the clinical laboratory: principles of preparation and applications. Clin Chem Lab Med 50(2):193–209
Alanazi FK, Radwan AA, Alsarra IA (2010) Biopharmaceutical applications of nanogold. Saudi Pharm J 18(4):179–193
Maekawa K, Yamasaki K, Niizeki T, Mita M, Matsuba Y, Terada N, Saito H (2012) Drop-on-demand laser sintering with silver nanoparticles for electronics packaging. IEEE Trans Compon Packag Manuf Technol 2(5):868–877
Virkutyte J, Varma RS (2011) Green synthesis of metal nanoparticles: biodegradable polymers and enzymes in stabilization and surface functionalization. Chem Sci 2(5):837–846
Tsuji T, Iryo K, Watanabe N, Tsuji M (2002) Preparation of silver nanoparticles by laser ablation in solution: influence of laser wavelength on particle size. Appl Surf Sci 202(1–2):80–85
Chou K-S, Ren C-Y (2000) Synthesis of nanosized silver particles by chemical reduction method. Mater Chem Phys 64(3):241–246
Zafar S, Zafar A, Jabeen F, Siddiq MA (2021) Biological synthesis of silver nanoparticles and their biomedical activity: a review. Curr Green Chem 8(3):222–241
Bellah MM, Christensen SM, Iqbal SM (2012) Nanostructures for medical diagnostics. J Nanomater 2012:1–21
Article Google Scholar
Mukherjee P, Ahmad A, Mandal D, Senapati S, Sainkar SR, Khan MI, Parishcha R, Ajaykumar P, Alam M, Kumar R (2001) Fungus-mediated synthesis of silver nanoparticles and their immobilization in the mycelial matrix: a novel biological approach to nanoparticle synthesis. Nano Lett 1(10):515–519
Raffi M, Rumaiz AK, Hasan M, Shah SI (2007) Studies of the growth parameters for silver nanoparticle synthesis by inert gas condensation. J Mater Res 22(12):3378–3384
Meyers MA, Mishra A, Benson DJ (2006) Mechanical properties of nanocrystalline materials. Prog Mater Sci 51(4):427–556
Fang F, Kennedy J, Manikandan E, Futter J, Markwitz A (2012) Morphology and characterization of TiO 2 nanoparticles synthesized by arc discharge. Chem Phys Lett 521:86–90
Dhamale G, Mathe V, Bhoraskar S, Sahasrabudhe S, Dhole S, Ghorui S (2016) Synthesis and characterization of Nd 2 O 3 nanoparticles in a radiofrequency thermal plasma reactor. Nanotechnology 27(8):085603
Kawasaki M, Nishimura N (2006) 1064-nm laser fragmentation of thin Au and Ag flakes in acetone for highly productive pathway to stable metal nanoparticles. Appl Surf Sci 253(4):2208–2216
Tarasenko N, Butsen A, Nevar E, Savastenko N (2006) Synthesis of nanosized particles during laser ablation of gold in water. Appl Surf Sci 252(13):4439–4444
Pal S, Tak YK, Song JM (2007) Does the antibacterial activity of silver nanoparticles depend on the shape of the nanoparticle? A study of the gram-negative bacterium Escherichia coli. Appl Environ Microbiol 73(6):1712–1720
Ayano H, Miyake M, Terasawa K, Kuroda M, Soda S, Sakaguchi T, Ike M (2014) Isolation of a selenite-reducing and cadmium-resistant bacterium Pseudomonas sp. strain RB for microbial synthesis of CdSe nanoparticles. J Biosci Bioeng 117(5):576–581
Rai M, Ingle AP, Trzcińska-Wencel J, Wypij M, Bonde S, Yadav A, Kratošová G, Golińska P (2021) Biogenic silver nanoparticles: what we know and what do we need to know? Nanomaterials 11(11):2901
Xie J, Lee JY, Wang DI, Ting YP (2007) Silver nanoplates: from biological to biomimetic synthesis. ACS Nano 1(5):429–439
Gardea-Torresdey JL, Gomez E, Peralta-Videa JR, Parsons JG, Troiani H, Jose-Yacaman M (2003) Alfalfa sprouts: a natural source for the synthesis of silver nanoparticles. Langmuir 19(4):1357–1361
Shankar SS, Ahmad A, Sastry M (2003) Geranium leaf assisted biosynthesis of silver nanoparticles. Biotechnol Prog 19(6):1627–1631
Korbekandi H, Iravani S, Abbasi S (2009) Production of nanoparticles using organisms. Crit Rev Biotechnol 29(4):279–306
Sintubin L, De Windt W, Dick J, Mast J, Van Der Ha D, Verstraete W, Boon N (2009) Lactic acid bacteria as reducing and capping agent for the fast and efficient production of silver nanoparticles. Appl Microbiol Biotechnol 84:741–749
Iravani S, Korbekandi H, Mirmohammadi SV, Zolfaghari B (2014) Synthesis of silver nanoparticles: chemical, physical and biological methods. Res Pharm Sci 9(6):385
CAS Google Scholar
Iravani S (2011) Green synthesis of metal nanoparticles using plants. Green Chem 13(10):2638–2650
Mittal AK, Chisti Y, Banerjee UC (2013) Synthesis of metallic nanoparticles using plant extracts. Biotechnol Adv 31(2):346–356
Zafar S, Zafar A (2019) Biosynthesis and characterization of silver nanoparticles using fruits extract and their antimicrobial and cytotoxic effects. Open Biotechnol J 13(1)
Google Scholar
Elia P, Zach R, Hazan S, Kolusheva S, Porat ZE, Zeiri Y (2014) Green synthesis of gold nanoparticles using plant extracts as reducing agents. Int J Nanomed 9:4007
Yang X, Li Q, Wang H, Huang J, Lin L, Wang W, Sun D, Su Y, Opiyo JB, Hong L (2010) Green synthesis of palladium nanoparticles using broth of Cinnamomum camphora leaf. J Nanopart Res 12:1589–1598
Huang L, Weng X, Chen Z, Megharaj M, Naidu R (2014) Green synthesis of iron nanoparticles by various tea extracts: comparative study of the reactivity. Spectrochim Acta Part A Mol Biomol Spectrosc 130:295–301
Elumalai K, Velmurugan S, Ravi S, Kathiravan V, Ashokkumar S (2015) RETRACTED: green synthesis of zinc oxide nanoparticles using Moringa oleifera leaf extract and evaluation of its antimicrobial activity. Elsevier
Savithramma N, Rao ML, Rukmini K, Devi PS (2011) Antimicrobial activity of silver nanoparticles synthesized by using medicinal plants. Int J ChemTech Res 3(3):1394–1402
Abdel-Aziz MS, Shaheen MS, El-Nekeety AA, Abdel-Wahhab MA (2014) Antioxidant and antibacterial activity of silver nanoparticles biosynthesized using Chenopodium murale leaf extract. J Saudi Chem Soc 18(4):356–363
Edison TJI, Sethuraman M (2012) Instant green synthesis of silver nanoparticles using Terminalia chebula fruit extract and evaluation of their catalytic activity on reduction of methylene blue. Process Biochem 47(9):1351–1357
Benelli G, Lukehart CM (2017) Applications of green-synthesized nanoparticles in pharmacology, parasitology and entomology. J Cluster Sci 28:1–2
Khan A, Rashid R, Murtaza G, Zahra A (2014) Gold nanoparticles: synthesis and applications in drug delivery. Trop J Pharm Res 13(7):1169–1177
Bansod SD, Bawaskar MS, Gade AK, Rai MK (2015) Development of shampoo, soap and ointment formulated by green synthesised silver nanoparticles functionalised with antimicrobial plants oils in veterinary dermatology: treatment and prevention strategies. IET Nanobiotechnol 9(4):165–171
Velmurugan P, Anbalagan K, Manosathyadevan M, Lee K-J, Cho M, Lee S-M, Park J-H, Oh S-G, Bang K-S, Oh B-T (2014) Green synthesis of silver and gold nanoparticles using Zingiber officinale root extract and antibacterial activity of silver nanoparticles against food pathogens. Bioprocess Biosyst Eng 37:1935–1943
Vaghari H, Jafarizadeh-Malmiri H, Mohammadlou M, Berenjian A, Anarjan N, Jafari N, Nasiri S (2016) Application of magnetic nanoparticles in smart enzyme immobilization. Biotech Lett 38:223–233
Singh M, Manikandan S, Kumaraguru A (2011) Nanoparticles: a new technology with wide applications. Res J Nanosci Nanotech 1(1):1–11
Bubnova O ( 2016 ) Twists and turns. Nat Nanotechnol, pp 1–1
Wilczewska AZ, Niemirowicz K, Markiewicz KH, Car H (2012) Nanoparticles as drug delivery systems. Pharmacol Rep 64(5):1020–1037
Souza VGL, Fernando AL (2016) Nanoparticles in food packaging: biodegradability and potential migration to food—a review. Food Packag Shelf Life 8:63–70
Andra S, Balu SK, Jeevanandham J, Muthalagu M, Vidyavathy M, Chan YS, Danquah MK (2019) Phytosynthesized metal oxide nanoparticles for pharmaceutical applications. Naunyn Schmiedebergs Arch Pharmacol 392:755–771
Sorger V (2015) Nano-optics gets practical: plasmon modulators. Nat Nanotechnol 10:11–15
Stark WJ, Stoessel PR, Wohlleben W, Hafner A (2015) Industrial applications of nanoparticles. Chem Soc Rev 44(16):5793–5805
Wang X, Tabarraei A, Spearot DE (2015) Fracture mechanics of monolayer molybdenum disulfide. Nanotechnology 26(17):175703
Law M, Greene LE, Johnson JC, Saykally R, Yang P (2005) Nanowire dye-sensitized solar cells. Nat Mater 4(6):455–459
Kaegi R, Voegelin A, Sinnet B, Zuleeg S, Hagendorfer H, Burkhardt M, Siegrist H (2011) Behavior of metallic silver nanoparticles in a pilot wastewater treatment plant. Environ Sci Technol 45(9):3902–3908
Yang L, Luo W, Cheng G (2013) Graphene-supported Ag-based core–shell nanoparticles for hydrogen generation in hydrolysis of ammonia borane and methylamine borane. ACS Appl Mater Interfaces 5(16):8231–8240
Chauhan RP, Gupta C, Prakash D (2012) Methodological advancements in green nanotechnology and their applications in biological synthesis of herbal nanoparticles. Int J Bioassays (IJB)
Joerger R, Klaus T, Granqvist CG (2000) Biologically produced silver–carbon composite materials for optically functional thin-film coatings. Adv Mater 12(6):407–409
Ingale AG, Chaudhari A (2013) Biogenic synthesis of nanoparticles and potential applications: an eco-friendly approach. J Nanomed Nanotechol 4(165):1–7
Thakkar KN, Mhatre SS, Parikh RY (2010) Biological synthesis of metallic nanoparticles. Nanomed Nanotechnol Biol Med 6(2):257–262
Scaramuzza S, Agnoli S, Amendola V (2015) Metastable alloy nanoparticles, metal-oxide nanocrescents and nanoshells generated by laser ablation in liquid solution: influence of the chemical environment on structure and composition. Phys Chem Chem Phys 17(42):28076–28087
Jaworek A (2007) Micro-and nanoparticle production by electrospraying. Powder Technol 176(1):18–35
Zhang D (2004) Processing of advanced materials using high-energy mechanical milling. Prog Mater Sci 49(3–4):537–560
Bouhadoun S, Guillard C, Dapozze F, Singh S, Amans D, Bouclé J, Herlin-Boime N (2015) One step synthesis of N-doped and Au-loaded TiO 2 nanoparticles by laser pyrolysis: application in photocatalysis. Appl Catal B 174:367–375
Shou W, Mahajan BK, Ludwig B, Yu X, Staggs J, Huang X, Pan H (2017) Low-cost manufacturing of bioresorbable conductors by evaporation–condensation-mediated laser printing and sintering of Zn nanoparticles. Adv Mater 29(26):1700172
Okuyama K, Lenggoro IW (2003) Preparation of nanoparticles via spray route. Chem Eng Sci 58(3–6):537–547
Nakade S, Matsuda M, Kambe S, Saito Y, Kitamura T, Sakata T, Wada Y, Mori H, Yanagida S (2002) Dependence of TiO 2 nanoparticle preparation methods and annealing temperature on the efficiency of dye-sensitized solar cells. J Phys Chem B 106(39):10004–10010
Tsuji T, Kakita T, Tsuji M (2003) Preparation of nano-size particles of silver with femtosecond laser ablation in water. Appl Surf Sci 206(1–4):314–320
Abou El-Nour KM, Eftaiha AA, Al-Warthan A, Ammar RA (2010) Synthesis and applications of silver nanoparticles. Arab J Chem 3(3):135–140
Hornyak GL, Tibbals HF, Dutta J, Moore JJ (2008) Introduction to nanoscience and nanotechnology. CRC press
Cushing BL, Kolesnichenko VL, O’connor CJ (2004) Recent advances in the liquid-phase syntheses of inorganic nanoparticles. Chem Rev 104(9):3893–3946
Mohanpuria P, Rana NK, Yadav SK (2008) Biosynthesis of nanoparticles: technological concepts and future applications. J Nanopart Res 10:507–517
Paulkumar K, Rajeshkumar S, Gnanajobitha G, Vanaja M, Malarkodi C, Annadurai G (2013) Biosynthesis of silver chloride nanoparticles using Bacillus subtilis MTCC 3053 and assessment of its antifungal activity. Int Sch Res Not, vol 2013
Pooley F (1982) Bacteria accumulate silver during leaching of sulphide ore minerals. Nature 296(5858):642–643
Prabhu S, Poulose EK (2012) Silver nanoparticles: mechanism of antimicrobial action, synthesis, medical applications, and toxicity effects. Int Nano Lett 2:1–10
Chen X, Schluesener HJ (2008) Nanosilver: a nanoproduct in medical application. Toxicol Lett 176(1):1–12
Du L, Jiang H, Liu X, Wang E (2007) Biosynthesis of gold nanoparticles assisted by Escherichia coli DH5α and its application on direct electrochemistry of hemoglobin. Electrochem Commun 9(5):1165–1170
Suganya KU, Govindaraju K, Kumar VG, Dhas TS, Karthick V, Singaravelu G, Elanchezhiyan M (2015) Blue green alga mediated synthesis of gold nanoparticles and its antibacterial efficacy against Gram positive organisms. Mater Sci Eng C 47:351–356
Singaravelu G, Arockiamary J, Kumar VG, Govindaraju K (2007) A novel extracellular synthesis of monodisperse gold nanoparticles using marine alga, Sargassum wightii Greville. Colloids Surf B 57(1):97–101
Schröfel A, Kratošová G, Bohunická M, Dobročka E, Vávra I (2011) Biosynthesis of gold nanoparticles using diatoms—silica-gold and EPS-gold bionanocomposite formation. J Nanopart Res 13:3207–3216
Syed A, Ahmad A (2012) Extracellular biosynthesis of platinum nanoparticles using the fungus Fusarium oxysporum. Colloids Surf B 97:27–31
Ahmad A, Senapati S, Khan MI, Kumar R, Ramani R, Srinivas V, Sastry M (2003) Intracellular synthesis of gold nanoparticles by a novel alkalotolerant actinomycete, Rhodococcus species. Nanotechnology 14(7):824
Ahmad A, Senapati S, Khan MI, Kumar R, Sastry M (2003) Extracellular biosynthesis of monodisperse gold nanoparticles by a novel extremophilic actinomycete, Thermomonospora sp. Langmuir 19(8):3550–3553
Shankar SS, Ahmad A, Pasricha R, Sastry M (2003) Bioreduction of chloroaurate ions by geranium leaves and its endophytic fungus yields gold nanoparticles of different shapes. J Mater Chem 13(7):1822–1826
Mukherjee P, Senapati S, Mandal D, Ahmad A, Khan MI, Kumar R, Sastry M (2002) Extracellular synthesis of gold nanoparticles by the fungus Fusarium oxysporum. ChemBioChem 3(5):461–463
Vigneshwaran N, Kathe AA, Varadarajan P, Nachane RP, Balasubramanya R (2006) Biomimetics of silver nanoparticles by white rot fungus, Phaenerochaete chrysosporium. Colloids Surf, B 53(1):55–59
Mandal D, Bolander ME, Mukhopadhyay D, Sarkar G, Mukherjee P (2006) The use of microorganisms for the formation of metal nanoparticles and their application. Appl Microbiol Biotechnol 69:485–492
Drake PL, Hazelwood KJ (2005) Exposure-related health effects of silver and silver compounds: a review. Ann Occup Hyg 49(7):575–585
Syed A, Saraswati S, Kundu GC, Ahmad A (2013) Biological synthesis of silver nanoparticles using the fungus Humicola sp. and evaluation of their cytoxicity using normal and cancer cell lines. Spectrochim Acta Part A: Mol Biomol Spectrosc 114:144–147
Dameron C, Reese R, Mehra R, Kortan A, Carroll P, Steigerwald M, Brus L, Winge D (1989) Biosynthesis of cadmium sulphide quantum semiconductor crystallites. Nature 338(6216):596–597
Kowshik M, Deshmukh N, Vogel W, Urban J, Kulkarni SK, Paknikar K (2002) Microbial synthesis of semiconductor CdS nanoparticles, their characterization, and their use in the fabrication of an ideal diode. Biotechnol Bioeng 78(5):583–588
Kowshik M, Ashtaputre S, Kharrazi S, Vogel W, Urban J, Kulkarni SK, Paknikar KM (2002) Extracellular synthesis of silver nanoparticles by a silver-tolerant yeast strain MKY3. Nanotechnology 14(1):95
Kundu S, Maheshwari V, Saraf RF (2008) Photolytic metallization of Au nanoclusters and electrically conducting micrometer long nanostructures on a DNA scaffold. Langmuir 24(2):551–555
Cabrera FC, Mohan H, Dos Santos RJ, Agostini DL, Aroca RF, Rodríguez-Pérez MA, Job AE (2013) Green synthesis of gold nanoparticles with self-sustained natural rubber membranes. J Nanomater 2013:110–110
Pokorski JK, Steinmetz NF (2011) The art of engineering viral nanoparticles. Mol Pharm 8(1):29–43
Beatty PH, Lewis JD (2019) Cowpea mosaic virus nanoparticles for cancer imaging and therapy. Adv Drug Deliv Rev 145:130–144
Lee S-W, Mao C, Flynn CE, Belcher AM (2002) Ordering of quantum dots using genetically engineered viruses. Science 296(5569):892–895
Mao C, Flynn CE, Hayhurst A, Sweeney R, Qi J, Georgiou G, Iverson B, Belcher AM (2003) Viral assembly of oriented quantum dot nanowires. Proc Natl Acad Sci 100(12):6946–6951
Kumar B, Smita K, Cumbal L, Debut A (2014) Synthesis of silver nanoparticles using Sacha inchi (Plukenetia volubilis L.) leaf extracts. Saudi J Biol Sci 21(6):605–609
Moteriya P, Padalia H, Chanda S (2017) Characterization, synergistic antibacterial and free radical scavenging efficacy of silver nanoparticles synthesized using Cassia roxburghii leaf extract. J Genet Eng Biotechnol 15(2):505–513
Arokiyaraj S, Dinesh Kumar V, Elakya V, Kamala T, Park SK, Ragam M, Saravanan M, Bououdina M, Arasu MV, Kovendan K (2015) Biosynthesized silver nanoparticles using floral extract of Chrysanthemum indicum L.—potential for malaria vector control. Environ Sci Pollut Res 22:9759–9765
Kumar B, Angulo Y, Smita K, Cumbal L, Debut A (2016) Capuli cherry-mediated green synthesis of silver nanoparticles under white solar and blue LED light. Particuology 24:123–128
Shameli K, Ahmad MB, Zamanian A, Sangpour P, Shabanzadeh P, Abdollahi Y, Zargar M (2012) Green biosynthesis of silver nanoparticles using Curcuma longa tuber powder. Int J Nanomed, pp 5603–5610
Kumar B, Smita K, Cumbal L, Angulo Y (2015) Fabrication of silver nanoplates using Nephelium lappaceum (Rambutan) peel: a sustainable approach. J Mol Liq 211:476–480
Pallela PNVK, Ummey S, Ruddaraju LK, Gadi S, Cherukuri CS, Barla S, Pammi S (2019) Antibacterial efficacy of green synthesized α-Fe 2 O 3 nanoparticles using Sida cordifolia plant extract. Heliyon 5(11):e02765
da Silva AFV, Fagundes AP, Macuvele DLP, de Carvalho EFU, Durazzo M, Padoin N, Soares C, Riella HG (2019) Green synthesis of zirconia nanoparticles based on Euclea natalensis plant extract: optimization of reaction conditions and evaluation of adsorptive properties. Colloids Surf, A 583:123915
Pravallika PL, Mohan GK, Rao KV, Shanker K (2019) Biosynthesis, characterization and acute oral toxicity studies of synthesized iron oxide nanoparticles using ethanolic extract of Centella asiatica plant. Mater Lett 236:256–259
Mydeen SS, Kumar RR, Kottaisamy M, Vasantha V (2020) Biosynthesis of ZnO nanoparticles through extract from Prosopis juliflora plant leaf: antibacterial activities and a new approach by rust-induced photocatalysis. J Saudi Chem Soc 24(5):393–406
Dobrucka R (2019) Biofabrication of platinum nanoparticles using Fumariae herba extract and their catalytic properties. Saudi J Biol Sci 26(1):31–37
Chavan RR, Bhinge SD, Bhutkar MA, Randive DS, Wadkar GH, Todkar SS, Urade MN (2020) Characterization, antioxidant, antimicrobial and cytotoxic activities of green synthesized silver and iron nanoparticles using alcoholic Blumea eriantha DC plant extract. Mater Today Commun 24:101320
Sukri SNAM, Shameli K, Wong MM-T, Teow S-Y, Chew J, Ismail NA (2019) Cytotoxicity and antibacterial activities of plant-mediated synthesized zinc oxide (ZnO) nanoparticles using Punica granatum (pomegranate) fruit peels extract. J Mol Struct 1189:57–65
Safawo T, Sandeep B, Pola S, Tadesse A (2018) Synthesis and characterization of zinc oxide nanoparticles using tuber extract of anchote (Coccinia abyssinica (Lam.) Cong.) for antimicrobial and antioxidant activity assessment. OpenNano 3:56–63
Sadeghi B, Gholamhoseinpoor F (2015) A study on the stability and green synthesis of silver nanoparticles using Ziziphora tenuior (Zt) extract at room temperature. Spectrochim Acta Part A Mol Biomol Spectrosc 134:310–315
Gericke M, Pinches A (2006) Biological synthesis of metal nanoparticles. Hydrometallurgy 83(1–4):132–140
Sathishkumar M, Sneha K, Yun Y-S (2010) Immobilization of silver nanoparticles synthesized using Curcuma longa tuber powder and extract on cotton cloth for bactericidal activity. Biores Technol 101(20):7958–7965
Armendariz V, Herrera I, Peralta-Videa JR, Jose-Yacaman M, Troiani H, Santiago P, Gardea-Torresdey JL (2004) Size controlled gold nanoparticle formation by Avena sativa biomass: use of plants in nanobiotechnology. J Nanopart Res 6:377–382
Gauthami M, Srinivasan N, Goud NM, Boopalan K, Thirumurugan K (2015) Synthesis of silver nanoparticles using Cinnamomum zeylanicum bark extract and its antioxidant activity. Nanosci Nanotechnol-Asia 5(1):2–7
Raju D, Mehta UJ, Hazra S (2011) Synthesis of gold nanoparticles by various leaf fractions of Semecarpus anacardium L. tree. Trees 25:145–151
Patra JK, Baek K-H (2015) Green nanobiotechnology: factors affecting synthesis and characterization techniques. J Nanomater 2014:219–219
Darroudi M, Ahmad MB, Zamiri R, Zak AK, Abdullah AH, Ibrahim NA (2011) Time-dependent effect in green synthesis of silver nanoparticles. Int J Nanomed, pp 677–681
Ozel F, Kockar H (2015) Growth and characterizations of magnetic nanoparticles under hydrothermal conditions: reaction time and temperature. J Magn Magn Mater 373:213–216
Mudunkotuwa IA, Pettibone JM, Grassian VH (2012) Environmental implications of nanoparticle aging in the processing and fate of copper-based nanomaterials. Environ Sci Technol 46(13):7001–7010
Farhadi S, Ajerloo B, Mohammadi A (2017) Green biosynthesis of spherical silver nanoparticles by using date palm (Phoenix dactylifera) fruit extract and study of their antibacterial and catalytic activities. Acta Chim Slov 64(1):129–143
Baer DR (2011) Surface characterization of nanoparticles: critical needs and significant challenges. J Surf Anal 17(3):163–169
Eustis S, El-Sayed MA (2006) Why gold nanoparticles are more precious than pretty gold: noble metal surface plasmon resonance and its enhancement of the radiative and nonradiative properties of nanocrystals of different shapes. Chem Soc Rev 35(3):209–217
Khlebtsov NG, Dykman LA (2010) Optical properties and biomedical applications of plasmonic nanoparticles. J Quant Spectrosc Radiat Transf 111(1):1–35
Rong H, Jones R, Liu A, Cohen O, Hak D, Fang A, Paniccia M (2005) A continuous-wave Raman silicon laser. Nature 433(7027):725–728
Faivre D, Bennet M (2016) Magnetic nanoparticles line up. Nature 535(7611):235–236
Guo D, Xie G, Luo J (2013) Mechanical properties of nanoparticles: basics and applications. J Phys D Appl Phys 47(1):013001
Lee S, Choi S-S, Li S, Eastman J (1999) Measuring thermal conductivity of fluids containing oxide nanoparticles
Cao YC, Jin R, Mirkin CA (2002) Nanoparticles with Raman spectroscopic fingerprints for DNA nd RNA detection. Science 297(5586):1536–1540
Mughal B, Zaidi SZJ, Zhang X, Hassan SU (2021) Biogenic nanoparticles: synthesis, characterisation and applications. Appl Sci 11(6):2598
Khan I, Ali S, Mansha M, Qurashi A (2017) Sonochemical assisted hydrothermal synthesis of pseudo-flower shaped Bismuth vanadate (BiVO 4 ) and their solar-driven water splitting application. Ultrason Sonochem 36:386–392
Khan I, Yamani ZH, Qurashi A (2017) Sonochemical-driven ultrafast facile synthesis of SnO 2 nanoparticles: growth mechanism structural electrical and hydrogen gas sensing properties. Ultrason Sonochem 34:484–490
Iqbal N, Khan I, Yamani ZH, Qurashi A (2016) Sonochemical assisted solvothermal synthesis of gallium oxynitride nanosheets and their solar-driven photoelectrochemical water-splitting applications. Sci Rep 6(1):32319
Wang Y-C, Engelhard MH, Baer DR, Castner DG (2016) Quantifying the impact of nanoparticle coatings and nonuniformities on XPS analysis: gold/silver core–shell nanoparticles. Anal Chem 88(7):3917–3925
Chan GG, Koch CM, Connors LH (2017) Blood proteomic profiling in inherited (ATTRm) and acquired (ATTRwt) forms of transthyretin-associated cardiac amyloidosis. J Proteome Res 16(4):1659–1668
Ma S, Livingstone R, Zhao B, Lombardi JR (2011) Enhanced Raman spectroscopy of nanostructured semiconductor phonon modes. J Phys Chem Lett 2(6):671–674
Kestens V, Roebben G, Herrmann J, Jämting Å, Coleman V, Minelli C, Clifford C, De Temmerman P-J, Mast J, Junjie L (2016) Challenges in the size analysis of a silica nanoparticle mixture as candidate certified reference material. J Nanopart Res 18:1–22
Filipe V, Hawe A, Jiskoot W (2010) Critical evaluation of nanoparticle tracking analysis (NTA) by NanoSight for the measurement of nanoparticles and protein aggregates. Pharm Res 27:796–810
Fagerlund G (1973) Determination of specific surface by the BET method. Matériaux et Constr 6:239–245
Swinehart DF (1962) The beer-lambert law. J Chem Educ 39(7):333
Liu D, Li C, Zhou F, Zhang T, Zhang H, Li X, Duan G, Cai W, Li Y (2015) Rapid synthesis of monodisperse Au nanospheres through a laser irradiation-induced shape conversion, self-assembly and their electromagnetic coupling SERS enhancement. Sci Rep 5(1):1–9
Yu ZB, Xie YP, Liu G, Lu GQM, Ma XL, Cheng H-M (2013) Self-assembled CdS/Au/ZnO heterostructure induced by surface polar charges for efficient photocatalytic hydrogen evolution. J Mater Chem A 1(8):2773–2776
Lin G, Zhang Q, Lin X, Zhao D, Jia R, Gao N, Zuo Z, Xu X, Liu D (2015) Enhanced photoluminescence of gallium phosphide by surface plasmon resonances of metallic nanoparticles. RSC Adv 5(60):48275–48280
Gupta K, Singh R, Pandey A, Pandey A (2013) Photocatalytic antibacterial performance of TiO 2 and Ag-doped TiO 2 against S. aureus. P. aeruginosa and E. coli. Beilstein J Nanotechnol 4(1):345–351
Torchynska T, El Filali B, Ballardo Rodríguez IC, Shcherbyna L (2016) Defect related emission of ZnO and ZnO Cu nanocrystals prepared by electrochemical method. physica status solidi (c) 13(7–9):594–597
Saeed K, Khan I (2016) Preparation and characterization of single-walled carbon nanotube/nylon 6, 6 nanocomposites. Instrum Sci Technol 44(4):435–444
Download references
Author information
Authors and affiliations.
Department of Chemistry, Faculty of Science, Jazan University, P.O. Box 114, Jazan, 45142, Saudi Arabia
Sarvat Zafar
You can also search for this author in PubMed Google Scholar
Corresponding author
Correspondence to Sarvat Zafar .
Editor information
Editors and affiliations.
Department of Engineering Management, College of Engineering, Prince Sultan University, Riyadh, Saudi Arabia
Tabrej Khan
Laboratory of Biocomposites Technology, Institute of Tropical Forestry and Forest Products (INTROP), Universiti Putra Malaysia, Serdang, Malaysia
Mohammad Jawaid
Department of Aerospace Engineering, Faculty of Engineering, Universiti Putra Malaysia, Serdang, Malaysia
Kamarul Arifin Ahmad
Department of Aeronautical and Automobile Engineering, Manipal Institute of Technology, Manipal Academy of Higher Education (MAHE), Manipal, Karnataka, India
Balbir Singh
Rights and permissions
Reprints and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this chapter
Zafar, S. (2023). Synthesis, Properties, and Characterization of Biological Nanomaterial. In: Khan, T., Jawaid, M., Ahmad, K.A., Singh, B. (eds) Nanomaterials: The Building Blocks of Modern Technology. Smart Nanomaterials Technology. Springer, Singapore. https://doi.org/10.1007/978-981-99-4149-0_5
Download citation
DOI : https://doi.org/10.1007/978-981-99-4149-0_5
Published : 28 August 2023
Publisher Name : Springer, Singapore
Print ISBN : 978-981-99-4148-3
Online ISBN : 978-981-99-4149-0
eBook Packages : Chemistry and Materials Science Chemistry and Material Science (R0)
Share this chapter
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Publish with us
Policies and ethics
- Find a journal
- Track your research
Got any suggestions?
We want to hear from you! Send us a message and help improve Slidesgo
Top searches
Trending searches

26 templates
49 templates
11 templates

71 templates

15 templates

first day of school
68 templates
Biology Presentation templates
Give interesting presentations about biology with these free ppt templates and google slides themes. all life forms should enjoy a nice slide deck, related collections.

20 templates

31 templates

Middle School
34 templates

High School
74 templates

75 templates

It seems that you like this template!
The stomach and its relationship to emotions thesis defense.
Download the The Stomach and its Relationship to Emotions Thesis Defense presentation for PowerPoint or Google Slides. Congratulations, you have finally finished your research and made it to the end of your thesis! But now comes the big moment: the thesis defense. You want to make sure you showcase your...

Premium template
Unlock this template and gain unlimited access
Science Subject for Middle School - 9th Grade: Biology
If you encourage your students to get interested in science since middle school, who knows, some of them could even become renowned scientists! Let's go step by step: first, download this template and customize it so that you have a slideshow for your biology class. Define concepts in its slides,...
Biology Infographics
Download the "Biology Infographics" template for PowerPoint or Google Slides and discover this set of editable infographics for education presentations. These resources, from graphs to tables, can be combined with other presentations or used independently. The most important thing is what you will achieve: transmit information in an orderly and...

Biology Major for College: Inmunology
Download the Biology Major for College: Inmunology presentation for PowerPoint or Google Slides. As university curricula increasingly incorporate digital tools and platforms, this template has been designed to integrate with presentation software, online learning management systems, or referencing software, enhancing the overall efficiency and effectiveness of student work. Edit this...

Microbiology Breakthrough
Advances in medicine are always important events, and it’s great if the new findings can be presented in a clear manner. If you want to make a contribution to the medical community and share what you found out in your research, this new breakthrough presentation template is for you.

Marine Biology Science Lesson for College
Here’s a beautiful marine biology template if there ever was one! True to form, it’s all dreamy watercolors in ocean shades and an abundance of marine animal and plant life. From freshmen to grad students, everyone will be happy to listen to a lesson on this template! Download it today...

Anatomy and Physiology Major for College: Cell Biology
One of the most intriguing subjects of a major in Anatomy and Physiology is Cell Biology, and this template provides an introduction to its fascinating universe. From the basics of cellular structure and function, to specialized topics such as organelles and DNA replication, this template will provide an overview that...

Science Subject for High School - 9th Grade: The Building Blocks of Life
When you stack lots of building blocks, you end up having a nice figure or building, but when you have lots of "building blocks of life", you get organisms! Use this template for educational purposes and teach your students what are cells. We have added illustrations that are funny so...

Biochemistry Lesson for High School
How does the world around us work? That is a question too broad to be answered in a single class, but you can explain to your students in your next class a tiny and important part of its functioning and composition: what is a molecule and how they are made...

Cell Behavior Thesis Defense
You're going to need a good number of zeros in order to write how many cells there are in the human body. How do they work and what's their function? People like you, who've studied cells, can help us know a bit more about them! We've got something in return:...

Biological Roles of Micro-ARN Thesis Defense
Download the Biological Roles of Micro-ARN Thesis Defense presentation for PowerPoint or Google Slides and start impressing your audience with a creative and original design. Slidesgo templates like this one here offer the possibility to convey a concept, idea or topic in a clear, concise and visual way, by using...

AI Breakthrough in Premature Baby Care
Download the AI Breakthrough in Premature Baby Care presentation for PowerPoint or Google Slides.Treating diseases involves a lot of prior research and clinical trials. But whenever there’s a new discovery, a revolutionary finding that opens the door to new treatments, vaccines or ways to prevent illnesses, it’s great news. Should...

Sense Organs
Download the Sense Organs presentation for PowerPoint or Google Slides. If you are planning your next workshop and looking for ways to make it memorable for your audience, don’t go anywhere. Because this creative template is just what you need! With its visually stunning design, you can provide your participants...

Science Subject for Elementary: Reproductive System
This presentation template is perfect for teaching elementary school students about the reproductive system. It includes engaging and informative illustrations to help reinforce understanding of the topic. Students will learn about the anatomy of reproduction, such as components like sperm and ovum, and they will understand how these components work...

Biology Thesis
Aristotle said that “by ‘life,’ we mean a thing that can nourish itself and grow and decay.” Biology is, indeed, the study of life as such. If you have just finished your thesis on biology and your viva is approaching, use our Biology Thesis template!

Protected Areas: Biological Reserve
Download the Protected Areas: Biological Reserve presentation for PowerPoint or Google Slides. Are you looking for a way to make your school or academy stand out among the competition? This template is designed to showcase all the fantastic aspects of your center. With perfect slides that allow you to easily...

Science Subject for High School - 9th Grade: Molecular Genetics
DNA is fascinating! The Slidesgo team marvels at how within something so microscopic, there can be so much information about not only whether we have green or blue eyes, but even about our personality. So, genetics defines us and, therefore, studying it is very important. In any high school science...

Cellular Morphology
Download the "Cellular Morphology" presentation for PowerPoint or Google Slides. The education sector constantly demands dynamic and effective ways to present information. This template is created with that very purpose in mind. Offering the best resources, it allows educators or students to efficiently manage their presentations and engage audiences. With...
- Page 1 of 25

Register for free and start editing online
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List

Biological Control of Mosquito Vectors: Past, Present, and Future
Giovanni benelli.
1 Insect Behaviour Group, Department of Agriculture, Food and Environment, University of Pisa, Pisa 56124, Italy; [email protected]
Claire L. Jeffries
2 Department of Disease Control, London School of Hygiene and Tropical Medicine, London WC1E 7HT, UK; [email protected]
Thomas Walker
Mosquitoes represent the major arthropod vectors of human disease worldwide transmitting malaria, lymphatic filariasis, and arboviruses such as dengue virus and Zika virus. Unfortunately, no treatment (in the form of vaccines or drugs) is available for most of these diseases and vector control is still the main form of prevention. The limitations of traditional insecticide-based strategies, particularly the development of insecticide resistance, have resulted in significant efforts to develop alternative eco-friendly methods. Biocontrol strategies aim to be sustainable and target a range of different mosquito species to reduce the current reliance on insecticide-based mosquito control. In this review, we outline non-insecticide based strategies that have been implemented or are currently being tested. We also highlight the use of mosquito behavioural knowledge that can be exploited for control strategies.
1. Introduction
Vector control strategies have traditionally focused on killing mosquitoes using a variety of insecticides. Environmental management (through reduction or removal of mosquito breeding sites) has often been used alongside chemical or microbiological ovicides, larvicides, and pupicides [ 1 , 2 , 3 , 4 ] in areas where endemic mosquito-borne diseases occur. The use of synthetic insecticides has to be regulated given that the development of insecticide resistance is widespread [ 5 , 6 , 7 , 8 , 9 ] and that there is concern regarding the damage to the environment and effects on non-target organisms. The use of insecticides for mosquito control, including organophosphates, carbamates, and pyrethroids, can also have negative effects on human health. Personal protection against mosquito-borne diseases can involve the use of mosquito repellents such as N,N -diethyl-meta-toluamide (DEET), dimethyl phthalate (DMP), N,N -diethyl mendelic acid amide (DEM), as well as plant-borne molecules (reviewed by [ 10 ]), light-coloured clothes covering as much of the body as possible, and sleeping under mosquito nets. Insecticide-treated bednets have played a very important role in the reduction of Plasmodium falciparum infection prevalence in malaria endemic Sub-Saharan Africa, which has seen the incidence of clinical disease fall by 40% between 2000 and 2015 [ 11 ]. However, bednets are only effective against mosquitoes that bite during the night and concern is growing that insecticide resistance, particularly due to the most commonly used class of pyrethroids, could reverse this trend and lead to rising incidence of malaria and increased fatalities [ 12 ]. As insecticide resistance is now widespread in a number of mosquito species [ 6 , 8 , 9 ], there is a growing need for novel, cheap, and reliable mosquito control strategies [ 13 , 14 , 15 ]. In many countries where mosquito-borne diseases are endemic, the financial burden of insecticide-based vector control programs is also prohibitive to widespread use. Environmentally friendly alternatives have been explored to help reduce the selection pressure for insecticide resistance. These various biocontrol strategies target different stages of the mosquito lifecycle ( Figure 1 ) with the aim of being safe for the environment and sustainable. These diverse biocontrol strategies include natural organisms that kill mosquitoes, exploiting mosquito behaviour to improve mosquito mortality, and releasing mosquitoes that are either sterile or unable to transmit disease.

Mosquito biocontrol strategies targeting different stages of the mosquito lifecycle.
2. Using Biocontrol to Kill Mosquitoes
2.1. plant-borne mosquitocides, repellents, and oviposition deterrents.
The discovery of the plant-based drug artemisinin for malaria treatment [ 16 ] and the subsequent awarding of the Nobel prize in 2015 [ 17 ] highlights the importance of screening plants and fungi as sources of metabolites for parasitological and mosquitocidal properties. Notably, plant-borne molecules are often effective at a few parts per million (ppm) against Aedes (Ae.) , Anopheles (An.) and Culex (Cx.) young larval instars (see [ 4 ] and [ 18 ] for dedicated reviews on ovicides and larvicides, respectively). Currently, more than 80 plant species have been employed for the successful synthesis of nanomosquitocides, with particular reference to larvicidal purposes. On the other hand, studies on ovicidal and ovideterrent nanoformulates are limited [ 19 ]. Furthermore, botanicals can also be used as reducing and capping agents for the rapid synthesis of mosquitocidal nanoformulations [ 20 ], and can even be employed to prepare cheap repellents with low human toxicity [ 3 ]. Notably, much remains to be discovered about this fast-growing research area, with special reference to the following topics: (i) the chemical characterization and standardization of plant-borne botanicals used for nanobiosynthesis [ 13 ], (ii) the potential of plant-synthesized nanoparticles as mosquito ovicides and ovideterrents [ 21 ], (iii) the utility of industrial by-products of plant origin for biofabrication of nanomosquitocides (e.g., neem cake) [ 4 ], (iv) field evaluation of mosquitocidal properties of green nanoparticles against Culicidae [ 22 , 23 ], (v) the non-target effects and environmental fate of plant-synthesized nanoparticles used against mosquito vectors [ 20 ].
2.2. Mosquito Predators
Natural enemies feeding on mosquito larvae and pupae in aquatic environments can play an important role in reducing Culicidae populations [ 24 , 25 , 26 ]. Indeed, mosquito young instars are preyed upon by a large number of aquatic organisms including fish [ 21 , 27 , 28 , 29 ], amphibians [ 30 , 31 ], copepods [ 32 , 33 ], odonate young instars [ 34 ], water bugs [ 35 , 36 , 37 , 38 ], and even larvae of other mosquito species [ 39 , 40 ]. Biological control of mosquitoes using vertebrates has mostly focused on the role of larvivorous fish that consume the aquatic larval stage of mosquitoes [ 26 ]. Fish predation of mosquito larvae has been recorded in many habitats, from small plastic containers [ 41 ] to complex natural ecosystems, including coastal wetland environments [ 42 ]. Larvivorous fish have been demonstrated to be very effective at reducing mosquito larval populations in many parts of the world, and in a variety of habitats [ 25 , 27 , 43 ]. In particular, larvivorous fish belonging to the genus Gambusia and Poecilia (Poeciliidae) have been introduced in more than 60 countries for mosquito control purposes [ 27 , 28 , 44 , 45 , 46 , 47 , 48 ]. However, introduced larvivorous fish are often considered a threat to native aquatic fauna, including amphibians [ 49 , 50 ], highlighting the need to carefully consider the ecological cost of introducing predatory species intended to contribute to mosquito control.
A number of omnivorous copepods (small aquatic cyclopoid crustaceans) can prey on young mosquito larval stages [ 51 ]. Several species of copepods, such as Cyclops vernalis , Megacyclops formosanus , Mesocyclops (M.) aspericornis , M. edax , M. guangxiensis , M. longisetus and M. thermocyclopoides , have been reported as active predators of mosquito young instars [ 32 , 52 , 53 , 54 , 55 , 56 , 57 , 58 ]. Operationally, the use of copepod predators against mosquitoes in urban and semi-urban habitats is not expensive and requires minimal labour for colony maintenance, highlighting their easy and cheap potential as mass-reared biocontrol agents [ 59 , 60 ]. The largest and most successful application of copepods for mosquito control was carried out in Vietnam to target the principal vector of dengue virus (DENV), Ae. aegypti [ 61 ]. From an initial introduction of copepods into a village in northern Vietnam in 1993, Ae. aegypti was eradicated from large surrounding areas by 2000, and dengue transmission could not be detected. Copepod biocontrol for Ae. aegypti was still being actively undertaken by communities in Vietnam even after the official intervention had ceased [ 62 , 63 ]. However, there are limitations in terms of the specific mosquito species to which copepods can be efficiently applied, since the larval habitats of many mosquito species are not suitable for copepods [ 64 ].
The larvae of some Culicidae species prey on other mosquito species that are vectors of public health importance. Toxorhynchites (T.) , also known as the “elephant mosquito” or “mosquito eater”, is a large, cosmopolitan genus of mosquitoes that does not consume blood [ 40 , 65 , 66 , 67 ]. While the adults feed on sugar-rich materials such as honeydew, fruit, and nectar, the larvae prey on the larvae of other mosquitoes as well as other nektonic (free swimming) organisms. As Toxorhynchites live on a protein- and fat-rich diet of aquatic organisms such as larvae, there is no requirement for blood-feeding at the adult stage, having already accumulated the necessary nutrients for oogenesis and vitellogenesis. Most species of Toxorhynchites live in forests, with one jungle species, T. splendens , consuming mosquito larvae in tree crevices (particularly those belonging to the genus Aedes) . Toxorhynchites adults are larger than Aedes and are considered to be harmless to humans given that they do not blood feed [ 39 , 40 , 67 ]. Taken together, these findings highlight the promising role of Toxorhynchites larvae as potential biocontrol agents against mosquito vectors. However, further research on the potential threat to native aquatic fauna due to the introduction of these mosquito predators is needed.
The potential of anurans (particularly frogs and toads) for mosquito control has been barely investigated [ 31 , 68 , 69 , 70 ]. For instance, tadpoles, with various life-history characteristics, actively prey upon the eggs of Ae. aegypti . It has been shown that this mosquito species has a preference to lay eggs in tadpole water and that tadpoles of Polypedates cruciger , as well as those of the Bufo, Ramanella, Euphlyctis, and Hoplobatrachus genera, predate on the eggs [ 31 ]. Other studies, however, have shown minimal effects with three common Thai anuran species ( Bufo melanostictus , Kaloula pulchra and Hylarana raniceps ), showing no evidence of Cx. quinquefasciatus larvae predation [ 71 ]. Most importantly, the biological control programs based on the release of larvivorous organisms, with special reference to amphibians and fish, are frequently not suitable in the majority of urban environments exploited by the larvae of some Aedes species, and require further research [ 13 ].
From an integrated vector management perspective, it has been recently observed that the employment of ultra-low quantities of botanicals or green-synthesized nanomosquitocides boosts the predation rates of a range of mosquito larvae predators. This has been demonstrated for various species of copepods (e.g., M. edax [ 58 ], M. thermocyclopoides [ 54 ], Megacyclops formosanus [ 72 ], M. aspericornis [ 56 ]), tadpoles (e.g., Hoplobatrachus tigerinus [ 70 ]), fish (e.g., Gambusia affinis [ 29 ], Poecilia reticulata [ 73 ], Carassius auratus [ 74 ], Aplocheilus lineolatus [ 21 ]), odonate young instars (e.g., Anax immaculifrons nymphs [ 75 ], Brachydiplax sobrina nymphs [ 76 ]), and water bugs (e.g., Diplonychus indicus [ 77 ]). This opportunity should be explored further, since the exploitation of synergies between ultra-low doses of plant-fabricated mosquitocides and biological control agents may represent a further control option readily available in tropical and sub-tropical developing countries worldwide [ 13 ].
2.3. Bti and Entomopathogenic Fungi
Naturally occurring organisms that are pathogenic to mosquitoes can also be considered for biocontrol strategies. Bacillus thuringiensis var. israelensis (Bti) is currently the most common mosquito larvicide employed in European countries. Bti is a gram-positive, spore-forming bacterium that releases insecticidal toxins and virulence factors that selectively target the larval stages of insects [ 78 , 79 ]. Application of Bti has been used to reduce the number of Ae. aegypti [ 80 , 81 , 82 ] and Ae. albopictus [ 83 ] larvae, but longer term use is subject to the development of resistance to Bti toxins [ 84 ], and the use of Bti in large mosquito breeding sites in urban environments is logistically demanding [ 85 ]. Entomopathogenic fungi produce infective spores (conidia) that attach to and penetrate the cuticle of mosquitoes, releasing toxins that result in mosquito death [ 86 ]. Several studies have shown the pathogenic effect on malaria mosquito vectors [ 87 , 88 ] and on Ae. aegypti [ 89 , 90 , 91 ]. As entomopathogenic fungi are mostly targeted towards adult mosquitoes, and because several different toxins produced during fungal infection are lethal to mosquitoes [ 92 ], selection pressure for resistance is likely to be less intense when compared to rapid-killing insecticides. Therefore, the evolution of fungus resistance is predicted to be much slower than the evolution of insecticide resistance [ 87 ]. The paucity of studies describing the effects of fungi on mosquito populations indicates further research is needed to determine the viability, infectivity, and persistence of fungal spores in mosquito field populations [ 93 ]. Clearly to deliver large-scale application of fungal spores into wild mosquito populations, optimal methods need to be determined [ 94 ].
3. Releasing Mosquitoes for Disease Control
3.1. wolbachia endosymbiotic bacteria.
Wolbachia are endosymbiotic bacteria that naturally infect approximately 40% of insect species [ 95 ] and induce a reproductive phenotype in mosquitoes known as cytoplasmic incompatibility (CI). This phenotype results in the generation of inviable offspring when an uninfected female mates with a Wolbachia -infected male, but Wolbachia -infected females can produce viable progeny when they mate with both infected and uninfected males. The overall result is a reproductive advantage for infected females, allowing this maternally transmitted bacterium to invade host populations. Natural Wolbachia infections are present in some major mosquito disease vectors such as Cx. quinquefasciatus and Ae. albopictus , but no natural infections are present in Ae. aegypti . A recent study in Burkina Faso [ 96 ] also found a novel Wolbachia strain in An. gambiae s.s. and An. coluzzii (major vectors of malaria in Sub-Saharan Africa).
The first experiments to successfully use Wolbachia for mosquito-borne disease control utilized CI to eradicate Cx. quinquefasciatus mosquito populations from Myanmar in the late 1960s [ 97 ]. This incompatible insect technique (IIT) depends on releasing large numbers of Wolbachia -infected male mosquitoes that compete with wild type males to induce sterility and suppress the mosquito population [ 98 ]. Current targets for IIT include Ae. albopictus through the generation of a triple Wolbachia -infected strain ( w AlbA, w AlbB, and w Pip infected) [ 99 ] and Ae. polynesiensis , a vector of lymphatic filariasis in the South Pacific [ 100 ]. The biotech company MosquitoMate ( http://mosquitomate.com ) is pioneering the use of IIT using Ae. albopictus , and releases of male mosquitoes are ongoing. The application of IIT is dependent on the ecology and environment which the target mosquito population inhabits. Physically isolated populations (e.g., oceanic islands) represent the optimal conditions for IIT given that large scale releases are problematic due to the need for mosquito sex separation at the pupal stage. Irradiating at the pupal stage can overcome the potential risk of unintentional fertile female release. A Wolbachia -infected Ae. polynesiensis strain that is bi-directionally incompatible with naturally infected wild type mosquitoes was irradiated at the pupal stage and this resulted in decreased fecundity and fertility in females [ 101 ]. This dose of radiation did not negatively impact male mosquito fitness parameters, mating competitiveness, or the ability to induce CI. For Ae. albopictus , several studies have been undertaken to determine the minimum pupal irradiation dose required to induce complete sterility in Wolbachia triple-infected (HC), double-infected (GUA), and uninfected (GT) female Ae. albopictus [ 102 ]. Irradiated Ae. albopictus HC, GUA, and GT strain females had decreased fecundity and fertility when irradiated and this was inversely proportional to the dose. In addition, the fitness of three Ae. albopictus strains (triple-infected, double-infected, and uninfected) of the same genetic background revealed that the presence of Wolbachia had only minimal effects on host fitness [ 99 ]. Irradiation with a female-sterilizing dose had no negative impact on the longevity of triple infected males, while a reduced lifespan was seen in wild type males ( w AlbA and w AlbB) irradiated with a higher male-sterilizing dose [ 103 ]. These studies indicate that irradiation could be used to reduce the risk of unintentional release of Wolbachia triple-infected Ae. albopictus HC strain females during male release for population suppression.
The discovery of a virulent Wolbachia strain in Drosophila melanogaster fruit flies (named w MelPop), which significantly lowered the lifespan of its host [ 104 ], led to further work to see if this strain could shorten the lifespan of mosquitoes. Additional Wolbachia strains, including the closely related avirulent w Mel strain, were subsequently found to protect their native hosts, Drosophila fruit flies, against infection by pathogenic RNA viruses [ 105 , 106 ]. This alternative approach for mosquito vector control relies on the use of Wolbachia to prevent pathogens from replicating within the mosquito [ 107 ]. The “eliminate dengue” project ( www.eliminatedengue.com ) based in Australia has been able to demonstrate that Wolbachia bacteria can prevent DENV transmission in mosquitoes without significant fitness costs. Stable Wolbachia -infected Ae. aegypti lines have now been successfully generated using embryo microinjection [ 108 , 109 , 110 , 111 ]. All transinfected Wolbachia strains significantly reduced the vector competence of Ae. aegypti for DENV under laboratory conditions [ 110 , 112 , 113 ]. High levels of Wolbachia bacteria in salivary glands was thought to be crucial to the ability to completely block DENV transmission (shown through the absence of infectious virus in the saliva) under laboratory conditions [ 110 ]. All Wolbachia strains showed maternal transmission rates close to 100% and induced high levels of CI in Ae. aegypti [ 108 , 109 , 110 ]. Semi-field cage experiments were undertaken to assess fitness costs and the ability of two Wolbachia strains to invade mosquito populations. The fecundity of w MelPop-infected female mosquitoes was reduced by ~60% relative to uninfected wildtype and w Mel-infected mosquitoes, and this strain invaded at a slower rate when compared to w Mel [ 110 ]. Mosquitoes infected with the w Mel strain were introduced into the wild through open releases at two locations near Cairns in north Queensland, Australia, and reached near fixation within a few months [ 114 ].
The success of these preliminary field releases has led to subsequent releases in Australia and now countries that experience high dengue cases such as Indonesia, Vietnam, Colombia, and Brazil ( www.eliminatedengue.com ). One potential concern for a Wolbachia -replacement approach is the future development of resistance to Wolbachia ’s inhibitory effects. Although no studies to date have demonstrated that this is likely to happen, a Wolbachia -superinfected line was recently established in Ae. aegypti containing stable infections of the w Mel and w AlbB strains that could help mitigate potential resistance. This combination of strains resulted in greater inhibitory effects on DENV replication than the single w Mel strain when challenged with blood meals from viraemic dengue patients [ 111 ]. Wolbachia superinfections could be utilised to replace single infections in wild populations and could help overcome any resistance by DENV to singly infected strains that are present in wild mosquito populations.
As only preliminary trials are underway for this promising strategy, a number of questions remain regarding implementation in the field. The applied use of Wolbachia for dengue control needs further research to determine the best individual or combination of Wolbachia strains. This has to take into account both the effects on DENV transmission and any resulting mosquito fitness costs. To predict the impact of the w Mel strain would have on dengue transmission, mathematical models were produced to show that a 66% – 75% reduction in the basic reproductive number, R 0 , could be achieved [ 115 ]. Ultimately, further experiments are needed to determine the overall effect Wolbachia will have on DENV transmission and dengue epidemiology in the field, particularly in endemic areas. Finally, Wolbachia -infected Ae. aegypti could also play a role in reducing transmission of other mosquito-borne diseases, as Wolbachia inhibits the transmission of chikungunya virus (CHIKV) [ 112 , 116 ], yellow fever virus (YFV) [ 117 ], malaria parasites [ 118 , 119 ], and Zika virus (ZIKV) [ 120 ]. Given the recent outbreaks of ZIKV in South America, novel control strategies including Wolbachia should be considered if Ae. aegypti is responsible for outbreaks in the Americas [ 14 ]. Another arbovirus, Japanese encephalitis virus (JEV), is transmitted mostly by Cx. tritaeniorhynchus mosquitoes and the epidemiology of this zoonotic disease would suggest Wolbachia could also reduce transmission provided stable transinfection is achieved [ 15 ]. Additional mosquito species, such as Cx. quinquefasciatus and Ae. albopictus that contain resident Wolbachia strains, are also potential targets for introducing “transinfected” strains that are likely to grow to higher densities and therefore impact pathogen transmission [ 121 ].
3.2. The Sterile Insect Technique
The Sterile Insect Technique (SIT) is a genetic suppression strategy that involves rearing large numbers of males of the target species and either irradiating or treating them with chemosterilizing agents to generate chromosomal aberrations and dominant lethal mutations in sperm. These sterilized male insects are released and when they mate with wild females produce no progeny. A sustained SIT programme results in an increasing ratio of released sterile males to wild males (as the population decreases) eventually leading to population elimination. Major interventions over the past 50 years using SIT against agricultural pests have proved very successful, including the eradication of the New World screwworm, Cochliomyia hominivorax , from North and Central America, and the eradication of Glossina austeni tsetse flies from Unguja Island, Zanzibar [ 122 ]. The use of SIT for mosquitoes that transmit human disease has been limited due to the reduced performance of sterilized males caused by sterilization. An additional problem for SIT programmes (and any other mosquito suppression strategy that aims for eradication) targeted towards Aedes species is the difficulty of the initial need to reduce the wild population densities, prior to the release of sterile males [ 123 ].
In addition to the IIT approaches using Wolbachia in combination with female sterility, renewed interest in SIT for the suppression of mosquito vectors has come through experiments to combine SIT with other forms of delivering mosquito lethality [ 124 ]. SIT combined with auto-dissemination, in which adult females are contaminated with dissemination stations of juvenile hormone (e.g., pyriproxygen), could be used to treat breeding habitats of Ae. aegypti and Ae. albopictus [ 125 ]. Contaminated female mosquitoes would lay eggs in larval sites and the insect growth regulator, introduced to the aquatic environment by the female, would prevent adult mosquito eclosion. Successful suppression using juvenile hormones was achieved for both Ae. aegypti in Peru [ 123 ] and Ae. albopictus in Spain [ 125 ]. Releasing sterile males with a juvenile hormone, such as pyriproxygen, could also allow contamination of females during mating to “boost SIT” [ 126 ]. SIT could also be enhanced by using sterile males to deliver densoviruses to their wild counterparts [ 126 ]. A European project entitled “Revolutionizing insect control” has recently started to determine if dispersion of mosquito densoviruses (MDVs), species-specific natural entomopathogenic viruses, by sterile males to wild females results in detrimental effects on Ae. aegypti and Ae. albopictus larval habitats, as a result of their skipping oviposition behaviour [ 127 ]. MDVs replicate in the nuclei of mosquito cells and kill mosquito larvae in a dose-dependent manner. Larvae that survive do not pupate or eclose to adults, resulting in an overall reduction in the mosquito population. As a result, MDVs have been proposed as potential biocontrol agents as they are also highly specific to target mosquito species. Female mosquitoes infected with an MDV can transmit the virus vertically to their progeny suggesting MDVs could persist and spread through wild mosquito populations. Laboratory studies using MDVs have shown high rates (>80%) of Ae. aegypti larval mortality [ 128 ]. However, the efficacy and sustainability of MDVs as a biocontrol agent was tested in and among oviposition sites in large laboratory cages, but was not shown to significantly reduce Ae. aegypti egg densities [ 129 ]. A direct inhibitory effect of MDVs on arboviral replication in cell lines has also been shown [ 130 , 131 , 132 ] which could work synergistically with pathogenic effects on the mosquito vector. However, co-infection of MDV and CHIKV in adult Ae. aegypti mosquitoes [ 133 ] suggests that MDVs may not be effective against all medically important arboviruses.
3.3. Genetically Modified Mosquitoes
An alternative method to sterilise males for insect population suppression has been developed in which a self-limiting gene is introduced into mosquito populations through genetic engineering [ 134 ]. This approach, pioneered by the British biotech company Oxitec ( www.oxitec.com ), was named Release of Insects carrying a Dominant Lethal (RIDL). The lethal gene can be repressed using an antidote (tetracycline) so that mosquitoes can be reared to adulthood in rearing facilities prior to the release of males into wild populations, which then mate with wild females, producing offspring that die at the larval stage in the absence of tetracycline. This approach has the advantage of being species-specific (like IIT and SIT) and has no long lasting effects on the target species as the aim is to eliminate the population in the release area. Field trials in the Cayman Islands in 2009–2010 with a self-limiting strain of Ae. aegypti OX513A were shown to suppress a wild population of Ae. aegypti [ 135 ]. In Malaysia, OX513A males were shown to have similar longevity and dispersal capabilities [ 136 ] and the latest release of OX513A males in Brazil led to strong suppression of the target wild population [ 137 ]. Trials in Brazil using RIDL male releases were conducted in a small suburb of Juazeiro, Bahia, and larger trials will be required to determine if the observed level of local suppression can be scaled up to larger release areas. RIDL technology was also used to generate a strain of Ae. aegypti , LA513A, engineered to carry a dominant, repressible, non-sex-specific, late-acting lethal genetic system, resulting in death at the pupal, rather than larval stage to avoid density dependent effects on larval development in wild populations [ 138 ]. In the absence of tetracycline, larvae carrying one or more copies of the LA513A insertion develop normally but the vast majority (95%–97%) die at pupation [ 138 ]. This incomplete penetrance of the lethal phenotype, however, could potentially result in unknown environmental consequences given this strategy is reliant on a self-limiting strain of mosquitoes.
Another potential method to suppress or eliminate mosquito populations is to induce an extreme male-biased sex ratio [ 139 ]. Although naturally occurring sex ratio distorters were found in Aedes and Culex mosquitoes, population suppression was not achieved in cage experiments [ 140 ]. Genetic modification can provide a bias towards male gamete production by inducing preferential breakdown of the X chromosome during male meiosis. Breakdown of the paternal X chromosome in An. gambiae prevents it from being transmitted to the next generation, resulting in fully fertile mosquito strains that produce >95% male offspring [ 141 ]. These synthetic distorter male mosquitoes suppress caged wild type mosquito populations, providing evidence for potential new strategies for mosquito vector control. It must be noted that Culicine mosquitoes contain homomorphic sex chromosomes (containing only a small nonrecombining region) [ 142 ] which may limit this approach for major Culicine mosquito vectors.
4. Behavioural Knowledge: A Tool to Enhance Mosquito Control Programs?
4.1. behavioural quantification helps sit.
Research into understanding the basics of mosquito mating ecology (particularly sexual chemical ecology) has been limited in the context of informing vector control strategies [ 13 , 143 ]. If greater information is obtained on the mating behaviour of medically important mosquito species, it could enhance control programs. For example, a crucial factor in the success of SIT, IIT, and RIDL progams is the ability of sterile male mosquitoes to compete with wild type males when mating with females [ 124 , 144 , 145 ]. Greater knowledge of mosquito swarming and mating behaviour could be used to compare courtship and mating ethograms of different mosquito vector species. The parameters underlying male mating success can be used to inform control programs. For example, the age, body size, and density in swarms of male Ae. aegypti can influence mating success [ 146 , 147 ], in addition to the role of mosquito mating acoustics in Ae. aegypti field swarms [ 148 ]. Further information on the parameters underlying male mating success could then be used to inform control programs to generate high quality mass-released males (in the case of SIT, boosted SIT, IIT, and RIDL) and to monitor the mating performance of Wolbachia -infected males [ 13 , 149 ]. It is worth noting that quantitative analyses of mating ethograms in mosquitoes are rare, and mainly focus on the elaborate courtships found in the genera Sabethes [ 150 , 151 , 152 ] and Wyeomyia [ 153 ]. The majority of studies investigating the sexual behaviour of medically important Aedes species just compare the insemination ability in sterilised and wild type males [ 154 , 155 , 156 , 157 , 158 , 159 , 160 , 161 ]. Behavioural quantification of courtship and mating events has often been excluded in sexual behaviour studies [ 149 , 162 , 163 ]. Notably, there is also only limited information on the molecules that mediate mosquito aggregation and mating [ 164 , 165 ].
4.2. Sound Traps
Vector control stratagies incorporating sound traps were first attempted in Cuba in 1949 against An. albimanus , in which sound traps were used to collect an elevated number of male mosquitoes [ 166 ]. Further trials using sound traps resulted in the trapping of Cx. tarsalis males, leading to a reduction in insemination of females [ 167 ] and reductions in the number of Cx. tritaeniorhynchus parous females [ 168 ]. Sound traps rely on wing-beat frequencies which can overlap for different mosquito species (potentially attracting multiple vector species) [ 169 , 170 ]. However, field trials with sound traps have shown limited success for several reasons. Firstly, there are technical difficulties in designing a sound trap that has the required amplification that will attract mosquitoes from long distances. The location in which the traps are placed also needs consideration, with improved efficacy for close proximity to swarming sites. Locating swarming sites, particularly for Anopheles species, needs significant development if sound traps are to be used for mosquito control [ 169 ].
4.3. The “Lure and Kill” Technique
The “lure and kill” approach has been successful for several arthropod pest species [ 171 ] and has been proposed to have a potential role for the control of mosquitoes (particularly Anopheles species) [ 169 ]. For Anopheles mosquitoes, visual stimuli are thought to be important in the convergence of individuals to a swarming site [ 172 , 173 ]. Consequently, recent research has revealed the potential to disrupt or enhance swarms, through manipulation of artificial swarm markers (or landmarks). This could lead to the development of “kill zones” that kill large numbers of attracted mosquitoes. In order for this to be applicable in field settings, rapid and economical methods to locate swarming sites need to be developed [ 143 , 169 ].
5. Conclusions and Future Perspectives
Biocontrol strategies for mosquito-borne diseases are needed to help reduce the prolonged application of insecticides that are currently used as the primary method for mosquito control. Eco-friendly, safe, and sustainable methods should be developed that can target a range of different mosquito species. Mosquito predators can be very effective in certain conditions, as demonstrated by the elimination of Ae. aegypti populations in rural Vietnam. The pathogenic bacterium Bti has been extensively used due to its ability to selectively kill mosquito larvae, and additional pathogens, such as entomopathogenic fungi, may be effective in future control programs. One of the most promising novel strategies is the use of Wolbachia endosymbiotic bacteria, which has been targeted towards reducing DENV transmission. Despite significant progress so far, larger scale trials are needed to determine if Wolbachia -based strategies can be an effective method of mosquito biocontrol. A combination of synergistic strategies may be required for effective population suppression using methods such as SIT, RIDL, and Wolbachia -induced IIT [ 144 ]. Mosquito behaviour plays a key role in vector control programs and further knowledge regarding the chemical ecology of mate searching, swarming landmarks, and mate choice in swarming sites is required to improve control strategies.
Acknowledgments
We would like to thank Andrew G. S. Cuthbertson and Eric W. Riddick for inviting this review. Giovanni Benelli is supported by PROAPI (PRAF 2015) and the University of Pisa, Department of Agriculture, Food and Environment (Grant ID: COFIN2015_22). Thomas Walker and Claire Jeffries are supported by a Wellcome Trust/Royal Society grant (101285/Z/13/Z). Funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author Contributions
All authors prepared the manuscript. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
- Skip to primary navigation
- Skip to main content
- Skip to primary sidebar
- Skip to footer
Bringing the World's Best Biology to You
Rethinking Scientific Presentations: Slide Design and Delivery
Part 1: rethinking scientific presentations: the assertion-evidence approach.
- Duration: 00:25:56
00:00:08.13 As a research scientist, 00:00:11.03 think about how little time you spend presenting 00:00:16.20 against how much time you spend 00:00:19.11 working in the lab or on your computations. 00:00:24.08 You might give one presentation at a conference 00:00:29.18 for 20 minutes, 00:00:31.22 or you might give a colloquium at your institution 00:00:34.13 for 50 minutes. 00:00:37.28 The question comes: 00:00:40.03 how do you make that time as effective as possible 00:00:44.04 to represent all the work that you've done? 00:00:47.25 This film focuses on that question. 00:00:51.08 And in particular, this film pays a lot of attention 00:00:55.09 to the slides that you create for that presentation. 00:01:00.09 Now, you might ask, why? 00:01:02.02 And one thing that I have found 00:01:05.14 in my 30 years of researching scientific presentations 00:01:10.14 is that slides make more of a difference for the success 00:01:14.20 -- and I would say more often the downfall -- 00:01:18.10 of scientific presentations 00:01:21.11 than people realize. 00:01:23.08 First, when you're creating slides, 00:01:25.26 you make important decisions: 00:01:28.02 what information you're going to include, 00:01:30.23 and equally important, 00:01:33.22 what information you're going to leave out. 00:01:36.07 And of that information that you include, 00:01:38.07 you also make decisions... 00:01:40.16 what am I going to emphasize, 00:01:42.19 perhaps by putting on the slide, 00:01:44.13 and what am I going to de-emphasize 00:01:47.25 by folding into your speech? 00:01:50.20 A second way that slides affect a presentation's success 00:01:56.04 occurs in the delivery. 00:01:58.18 Sadly, many presenters use PowerPoint's defaults 00:02:05.14 and spend so much of their delivery 00:02:09.01 turning, reading or paraphrasing a bullet on the slide, 00:02:13.24 turning back to the audience, 00:02:16.12 then turning, reading or paraphrasing, 00:02:19.06 then turning back. 00:02:21.03 And that rhythm 00:02:24.05 -- what a lot of people call a death by PowerPoint rhythm -- 00:02:26.27 pulls down a presentation. 00:02:28.20 The best presenters, however... 00:02:30.24 they speak from what they know, 00:02:34.03 and their visual aids are in fact aids for the audience 00:02:39.13 rather than notes for them. 00:02:42.15 Yet a third way that slides 00:02:45.05 affect a presentation's success 00:02:47.05 occurs with how much the audience 00:02:50.20 understands from them. 00:02:52.20 Our research has found 00:02:55.28 that challenging PowerPoint's defaults 00:02:58.08 and using a different approach, 00:03:00.14 such as what you're gonna learn in this film, 00:03:04.01 actually increases the amount of comprehension by the audience 00:03:11.05 in a statistically significant way. 00:03:16.05 So, I've had a number of people 00:03:18.20 who have used this approach. 00:03:20.15 The woman in the upper left, Katie Kirsch... 00:03:24.10 while she was getting her PhD, 00:03:28.25 she used this approach in all her conference presentations, 00:03:31.04 and she won best presentation at the conference 00:03:35.01 three times. 00:03:37.22 The gentlemen at the bottom, 00:03:40.01 professor Are Magnus Bruaset from Simula Research Laboratory 00:03:43.26 and University of Oslo in Norway... 00:03:47.04 he uses this approach, and his colleagues use this approach, 00:03:51.12 in all their presentations 00:03:54.14 that they make to industry to present their research. 00:03:57.27 And the woman... scientist in the upper right, 00:04:01.24 Dr. Barbara Bekins from the US Geological Survey... 00:04:06.06 she had to give a lecture 00:04:09.08 to 40 different places across the country, 00:04:11.27 and she decided to use this approach 00:04:16.03 for that lecture 00:04:18.01 because so many people 00:04:21.01 were going to see her work presented in that fashion. 00:04:25.24 We've had research groups 00:04:28.16 use the assertion-evidence approach, 00:04:30.10 and what you see here on the screen 00:04:32.13 is a large gas turbine research group 00:04:35.20 at Penn State. 00:04:37.02 We've had courses adopt the approach, 00:04:40.08 and here you see a law design course 00:04:43.19 at Penn State. 00:04:45.21 And this... and the approach was used by both the professors 00:04:48.11 who taught the class 00:04:50.15 and by the students who reported on their designs. 00:04:53.20 We've even had one national organization, 00:04:57.05 the Engineering Ambassadors Network, 00:04:59.27 adopt the approach for all the presentations 00:05:02.22 that they use in high school 00:05:05.03 to recruit STEM students. 00:05:08.18 Now, one assumption that I'm going to make 00:05:12.10 in this presentation 00:05:15.04 is that the goals for your research presentations 00:05:17.26 are that they are understood, 00:05:21.04 that your presentations are remembered, 00:05:23.10 and that your presentations are believed. 00:05:27.00 I'm also gonna have another assumption 00:05:28.28 that on a personal level 00:05:31.09 you have a goal of wanting to feel 00:05:36.09 and project more confidence. 00:05:38.03 So, where do we start? 00:05:41.12 Where I would like to start is 00:05:44.08 I would like you to think about 00:05:47.00 when you watch scientific presentations 00:05:49.18 and you see the slides, 00:05:51.24 what are the biggest problems that you see with those slides? 00:05:56.05 I've asked this question 00:05:59.00 to hundreds of scientists and engineers around the world, 00:06:03.05 and the number one answer 00:06:05.12 -- and it's not even close -- 00:06:07.15 is too many words. 00:06:09.14 And no doubt, 00:06:11.27 you see many examples of that weekly 00:06:15.27 in presentations that are given at conferences, in seminars, 00:06:20.26 in classrooms that are... 00:06:24.17 that include slides. 00:06:26.01 And you... and that particular problem 00:06:29.11 is not one that actually is so surprising. 00:06:34.20 In the mid-1980s, some research came out, 00:06:39.06 and that research points to why 00:06:43.00 having too many words is a problem. 00:06:47.09 So, let's say you have a speaker and you have an audience. 00:06:49.09 Now, we've known forever 00:06:51.25 that the speaker's spoken words... 00:06:54.19 those are gonna be taken in through the ears. 00:06:57.06 And if the speaker has any written words, 00:07:00.13 those are gonna be taken in through the eyes. 00:07:03.05 But it wasn't until the mid-1980s 00:07:07.16 that a Canadian psychology researcher 00:07:10.02 by the name of Allan Paivio 00:07:12.13 found that those written words and spoken words 00:07:17.26 are processed in the same part of the brain. 00:07:20.26 And another researcher, 00:07:24.12 this one from Australia, John Sweller... 00:07:27.14 he thought about Paivio's research 00:07:30.11 and he asked this question: 00:07:32.02 if written words and spoken words 00:07:34.02 are processed in the same part of the brain, 00:07:36.06 could that part of the brain become overloaded, 00:07:39.27 much as a central processing unit 00:07:43.00 can become overloaded 00:07:45.17 when it tries to do too many tasks? 00:07:47.14 And so Sweller... he did experiments. 00:07:49.11 He had one room where people just read, 00:07:51.23 one room where people just listened, 00:07:53.20 and another room where people read and listened. 00:07:57.29 And when there weren't too many words, 00:08:00.04 in the comprehension tests that he gave after those presentations, 00:08:04.16 the room where they read and listened, 00:08:09.01 they did the best. 00:08:10.28 But what he found is... 00:08:13.00 that when there were too many words projected, 00:08:16.12 that what happened is that room 00:08:20.04 that was both reading and listening 00:08:23.08 went from first to worst. 00:08:25.28 And so, Sweller came up with this theory 00:08:28.26 that if audiences try to process 00:08:31.08 too many words, 00:08:33.12 this cognitive overload occurs, 00:08:37.04 and that is what happens in many presentations. 00:08:41.19 Now, scientists and engineers 00:08:43.16 will also talk about a couple of other problems. 00:08:46.10 I mean, they'll actually talk about a lot of problems, 00:08:48.28 but there are three that stand out. 00:08:51.08 So, too many words is number one. 00:08:53.00 A second one is that the slides are cluttered. 00:08:57.21 And by cluttered, they mean that the... 00:09:01.00 that the audience isn't sure where to look. 00:09:03.22 And so, you have a slide, 00:09:05.28 and maybe what you want to do is you want to look at a graph on the slide 00:09:09.05 because you think the graph contains the most important things... 00:09:12.06 but there's text and arrows 00:09:16.13 and other things that are impinging on that graph, 00:09:19.00 and your eye gets pulled away. 00:09:22.05 A third problem is that many people 00:09:26.07 find that much of the text on slides is not readable. 00:09:29.17 And you can see in this graph 00:09:33.17 that even though maybe you can see the curves, 00:09:36.09 what you cannot read are the axes. 00:09:38.05 And if you can't read the axis of a... 00:09:40.23 axes of a graph, 00:09:43.08 then the value of that graph plummets. 00:09:47.03 A big takeaway is that 00:09:51.08 because so many people -- something like 95% -- 00:09:55.05 use PowerPoint, 00:09:58.19 that then PowerPoint's defaults become important. 00:10:02.00 And what we realize is PowerPoint 00:10:06.20 came out about the same time that Paivio 00:10:09.00 was coming out with his research. 00:10:11.07 And the two gentlemen who created that program, 00:10:15.26 Robert Gaskins, who was an entrepreneur, 00:10:18.00 and Dennis Austin, who was a computer scientist... 00:10:21.22 they were not aware of Paivio's research. 00:10:25.00 Now, they ended up, I think, doing the best they could 00:10:28.24 with the computer architecture of the day. 00:10:31.01 But a problem is that the defaults 00:10:35.09 were not based on any research. 00:10:38.18 Worse yet, and maybe the big tragedy, 00:10:42.08 is that PowerPoint's defaults have not changed significantly. 00:10:46.08 Yeah, in 2003 they changed from Times New Roman to Arial, 00:10:50.03 and in 2007 they changed from Arial typeface to Calibri, 00:10:53.22 and they threw in Microsoft's little artistic insert... 00:11:01.23 but... but nothing actually changed here. 00:11:04.27 So, one thing that I want you to realize 00:11:11.16 is that PowerPoint's defaults run counter to how people learn. 00:11:15.11 That text box in the body 00:11:18.12 that has all those nested bullets... 00:11:20.12 that leads people to create too many written words. 00:11:25.07 As John Sweller says, 00:11:27.02 it is a disaster how many words 00:11:31.04 people will put on slides. 00:11:33.01 That textbox also consumes valuable space 00:11:35.13 that could be used for images, 00:11:38.20 which makes the slides cluttered. 00:11:41.22 And then one last thing, 00:11:43.15 and something that I've been paying a lot of attention to, 00:11:45.15 is that that headline 00:11:50.09 leads scientists and engineers to write phrase headlines. 00:11:54.23 On the surface, that sounds like a good idea, 00:11:57.27 but a problem is that a phrase headline 00:12:00.15 does not filter noise. 00:12:03.06 And as you can remember 00:12:05.09 from when we first talked about why slides are important, 00:12:09.01 it's important for us to have a filter 00:12:13.18 on what to include and what not to include. 00:12:16.01 I'm gonna give you one quotation, 00:12:20.05 because we can do better. 00:12:21.24 I'm gonna give you one quotation... 00:12:23.19 and I could have chosen a lot, 00:12:25.17 but I'm gonna choose this one. 00:12:27.08 "The real mystery to me 00:12:29.29 is why PowerPoint's default style has been adopted so widely. 00:12:34.11 Why do medical researchers use the PowerPoint style 00:12:38.23 at academic conferences? 00:12:40.23 Why do engineers use the PowerPoint style 00:12:42.29 for technical discussions? 00:12:45.24 And the reason I like this quotation 00:12:48.28 as a criticism of PowerPoint 00:12:51.11 is that it was said by none other than Robert Gaskins, 00:12:57.15 the creator of PowerPoint. 00:12:59.18 I mean, if Gaskins himself 00:13:03.14 challenges the defaults, 00:13:05.08 then you should as well. 00:13:07.05 So, the question comes, what should we do? 00:13:09.09 But before we do that, 00:13:11.08 I have yet another assumption I want you to have. 00:13:12.26 And that is, 00:13:14.25 you should not have slides 00:13:18.01 if slides do not support the presentation. 00:13:22.24 In other words, if sl... 00:13:25.03 if slides do not help the audience 00:13:28.14 understand, remember, or believe the content. 00:13:31.28 And someone who was very astute 00:13:34.29 at that particular point, 00:13:36.28 and did not include slides if they weren't needed, 00:13:40.16 was Steve Jobs. 00:13:43.04 Steve Jobs thought about his presentations, 00:13:45.19 in a sense, as a story, 00:13:47.22 and then each slide or blank screen... 00:13:50.05 that was a scene. 00:13:52.09 And so, if you don't need a slide, 00:13:55.02 press the B on the control panel of your... 00:13:59.09 of your computer, 00:14:01.10 or use your advancer to blank the screen, 00:14:04.23 or better yet, insert a black slide 00:14:07.23 so that then the audience knows to focus on you. 00:14:13.23 And while that focus might seem frightening at first, 00:14:18.24 that focus is important 00:14:21.14 because, as Faraday said, 00:14:23.18 for your presentation to be a success, 00:14:25.21 the audience has to believe in you. 00:14:29.08 You have to show ownership of the information. 00:14:32.23 Now, maybe you won't have a blank screen 00:14:35.05 at a conference presentation 00:14:37.20 because those are so compressed. 00:14:39.12 But in a symposium that's 50 minutes, 00:14:41.20 think about having at least one scene 00:14:43.22 where there is no slide 00:14:46.04 and you move to a part of the room 00:14:48.26 where no one typically stands. 00:14:51.27 That will command attention. 00:14:53.15 Okay, now we're ready. 00:14:55.16 So then, what should we do? 00:14:57.26 And my first piece of advice to you is, 00:15:01.04 build your talk on messages, not on topics. 00:15:06.15 Most scientists and engineers in research 00:15:09.11 build their presentations on topics: 00:15:12.22 introduction, methods, results, discussion. 00:15:16.04 You can do better than that. 00:15:19.19 So, what often happens is... 00:15:21.21 let's say you're putting together a presentation, 00:15:23.29 and you're at a particular scene 00:15:26.17 and you decide... mmm... electron acceptors, 00:15:29.07 that's what I'm gonna be talking about here. 00:15:31.07 And so you write "electron acceptors" 00:15:34.00 in your biggest typeface up at the top, 00:15:36.06 and then you write down all the things you want to say... 00:15:40.02 you write those down below. 00:15:42.00 And then, if... and you put those in this bulleted list. 00:15:44.20 And then, if there's any room, 00:15:46.24 then you include an image. 00:15:50.02 What I'm telling you here is 00:15:53.02 go back to step one and stop there. 00:15:56.00 You can do better. 00:15:58.01 Build your talk on messages. 00:16:00.08 And so, Dr. Barbara Bekins, 00:16:02.01 when she was putting together her presentation 00:16:05.08 on the effect of hydrology 00:16:10.04 on the 25-year degradation of a crude oil spill... 00:16:18.01 on this particular scene, 00:16:20.23 she thought deeply about what she wanted the audience 00:16:24.01 to walk out the door with. 00:16:26.04 And then she wrote this sentence: 00:16:28.17 a succession of electron acceptors occurs 00:16:31.17 when an aquifer becomes contaminated with oil. 00:16:36.14 And then, she took that sentence, 00:16:39.28 tightened it as much as she could, 00:16:41.29 and put that at the top of a slide. 00:16:44.11 And once she had that, 00:16:46.19 then she had a filter. 00:16:48.17 And then she created visual evidence 00:16:51.00 to support that. 00:16:53.00 And so, when you've got an oil spill 00:16:56.18 that occurs in an aerobic aquifer, 00:17:01.09 what depletes first is the oxygen. 00:17:07.27 And so, what you see here is that this oxygen... 00:17:12.13 this becomes depleted in this outer band of the plume. 00:17:16.18 And once that is depleted, 00:17:19.08 then you get these bands of other types of depletions. 00:17:25.20 You get a reduction of nitrate and manganese, 00:17:29.10 and then you get a reduction of iron, 00:17:32.14 and then there's a reduction of sulfate. 00:17:36.29 So, what she has done 00:17:40.24 is she has stated her assertion up at the top 00:17:43.08 and then supported that assertion visually. 00:17:47.22 That's principle number one. 00:17:51.14 Principle number two 00:17:54.04 is to support your messages with visual evidence 00:17:57.04 -- not a bullet list. 00:18:00.00 It could be photographs, drawings, 00:18:03.05 diagrams, graphs. 00:18:05.17 It could be an equation. It could be a film. It could be a short table. 00:18:10.07 And then what you are to do is... 00:18:14.18 by creating that, you allow the audience 00:18:19.27 to focus on the visual evidence 00:18:23.14 and then what it is you're saying. 00:18:25.07 You avoid that cognitive overload. 00:18:27.29 So, let's go back to Paivio. 00:18:30.05 And if you remember, 00:18:32.27 Paivio found that written words and spoken words... 00:18:35.16 they're processed in the same part of the brain. 00:18:38.10 But his research also found 00:18:42.09 that images are processed in a different part of the brain. 00:18:46.01 And another researcher, professor Richard Mayer 00:18:49.19 from UC Santa Barbara... 00:18:52.05 what he did is he really ran with the question of, 00:18:55.27 what is the effect of using images in a presentation? 00:19:00.25 What is the effect on the learning 00:19:04.23 that the audience has? 00:19:06.20 And so, he's done a number of experiments, 00:19:08.22 and what his big takeaway 00:19:12.06 is that people learn much more deeply 00:19:14.05 from words and relevant images 00:19:17.10 than from words alone. 00:19:19.28 So, let's take a look at an example, 00:19:22.07 and I'm gonna choose a PhD graduate student, Jacob Snyder, 00:19:25.02 who's presenting a scene from 00:19:29.00 one of his conference presentations and from his PhD defense. 00:19:32.20 And so, in this scene 00:19:36.05 he's talking about the effect of using additive manufacturing, 00:19:40.18 or 3D printing, 00:19:42.25 on some really small channels. 00:19:47.09 And so, he begins the scene 00:19:51.04 by showing a channel that is built in the vertical direction, 00:19:55.10 much as a smokestack is. 00:19:57.09 And what you can see 00:19:59.18 -- and what he points out -- 00:20:01.26 is the variation that occurs in that 3D printed channel. 00:20:05.27 And then, once he has set that up, 00:20:09.14 then what he does is he shows a channel 00:20:12.24 that was built as a pipeline. 00:20:15.10 And what you see is that there's much more variation 00:20:20.12 in that particular channel, 00:20:22.19 that parts of it actually almost cave in. 00:20:26.11 And so, you might think, ohh, well, it's obvious 00:20:30.02 -- we would go with the vertical one. 00:20:32.10 Except that these channels... they are... 00:20:36.09 serve to produce heat transfer of these very hot gases. 00:20:40.22 And so, as it turns out, 00:20:43.03 having that roughness can be an asset. 00:20:47.03 And then he went on to show another channel 00:20:51.12 -- this one at 45 degrees -- 00:20:53.20 and he made the point that manufacturers 00:20:57.10 could choose different angles 00:21:01.24 to balance having the overall shape that they want 00:21:04.18 with increased roughness. 00:21:07.17 Now, we've looked at an assertion-evidence slide here. 00:21:12.01 Let's take it apart. 00:21:14.15 And the slide began with the assertion headline. 00:21:18.11 Now, some of you might say, 00:21:20.23 you know, I... I bet if I had seen Jacob Snyder give that talk, 00:21:25.28 I would understand exactly what was going on 00:21:29.05 and I would not have read that sentence. 00:21:31.15 And what I would say is, fantastic. 00:21:33.28 You were on your game as a listener 00:21:36.06 and then Jacob was on his game as a speaker. 00:21:40.09 But that sentence at a conference or in a symposium... 00:21:44.15 that sentence serves as a safety rope for the audience 00:21:50.04 in case they zone out, they get tired, 00:21:53.21 they receive a text, or whatever... 00:21:56.07 that allows them to stay in the presentation. 00:22:02.06 A couple of other things about the sentence headline: 00:22:05.10 keep it to one or two lines. 00:22:07.26 If it goes more than two lines, 00:22:09.22 our focus groups find that people won't read it... 00:22:12.28 perhaps it's just too much time away from the speaker. 00:22:16.13 Capitalize it the way you would a sentence. 00:22:19.09 It's just easier for people to read a sentence 00:22:22.08 that's capitalized as a sentence. 00:22:24.10 Also, don't center it; left justify it. 00:22:27.12 It's an easier read for the audience. 00:22:32.09 And the period... do you need a period? 00:22:36.14 Because it's a standalone sentence, 00:22:38.21 a period isn't required. 00:22:40.17 However, if you hear the voice of your fifth grade teacher 00:22:43.29 to put a period there and it haunts you at night, 00:22:47.22 put the period. 00:22:49.16 It's not that big of a deal. 00:22:52.16 So, that's the sentence. 00:22:54.12 What about the visual evidence? 00:22:56.11 With the visual evidence, I'd say the big thing is to avoid clutter. 00:22:59.07 In other words, try to have the slide breathe. 00:23:01.19 And I think Jacob Snyder did a really good job here 00:23:04.10 with the positioning of the three contour plots 00:23:08.20 and not allowing them to crowd the headline. 00:23:15.08 Leave some space there -- very nice. 00:23:17.17 And one last thing is, think about how you're gonna tell the story of the scene. 00:23:22.29 So, in this case, Jacob Snyder 00:23:25.28 discussed the one contour plot 00:23:28.13 and then animated in the second one, 00:23:30.23 when he was ready and when the audience was ready, 00:23:33.16 and then animated in the third, 00:23:36.22 again when the audience was ready. 00:23:39.13 So, we've talked about two of the principles 00:23:43.05 of the assertion-evidence approach: 00:23:45.28 build your talk on messages, not on topics, 00:23:48.01 and support those messages with visual evidence 00:23:51.01 not bullet lists. 00:23:53.15 The third principle of the assertion-evidence approach 00:23:56.15 is that when you present that visual evidence 00:24:00.04 fashion sentences on the spot. 00:24:02.18 In other words, show that you own the information. 00:24:06.20 Now, many of you might be 00:24:09.14 afraid of this particular principle. 00:24:11.25 You think, oh, I don't think I can do it. 00:24:14.01 I need those bullet lists for me to know what to say. 00:24:16.20 And what I would say is, you don't. 00:24:19.16 It is your research. 00:24:21.29 If you choose visual evidence 00:24:25.10 that is from your work, 00:24:28.05 you can present it. 00:24:29.29 You don't need those bullet lists. 00:24:32.14 As a research scientist or a research engineer, 00:24:35.04 be an experimentalist. 00:24:37.02 Try this approach. 00:24:39.19 And in trying this approach, 00:24:42.02 rather than starting with PowerPoint's defaults, 00:24:44.06 go to our website -- www.assertion-evidence.com -- 00:24:47.07 and download one of our PowerPoint templates. 00:24:50.00 They're free. It doesn't cost you anything. 00:24:53.02 But it's gonna save you a lot of time. 00:24:55.02 You'll also find some example presentations 00:24:57.13 by people such as Jacob Snyder and Katie Kirsch, 00:25:01.13 whom I had mentioned earlier. 00:25:04.08 So, what we've done here is that 00:25:10.04 I have shown you a different approach 00:25:12.23 to give a scientific research presentation. 00:25:16.05 And I hope that you'll be an experimentalist 00:25:18.23 and try this approach. 00:25:21.13 In a second film that's coming up, 00:25:23.22 we're going to walk through 00:25:27.11 a research presentation 00:25:30.03 -- title slide, mapping slide, the body slides, conclusion slide -- 00:25:33.16 to show some best examples on what it is that you can do. 00:25:38.00 Thank you.
Part 2: Assertion-Evidence Slides for a Research Talk
- Duration: 00:27:42
00:00:08.07 This film is a second part 00:00:11.24 in a three-part series for iBiology 00:00:15.18 on creating effective scientific research presentations. 00:00:22.05 Now, in the first part, 00:00:25.12 I introduced the assertion-evidence approach. 00:00:28.23 And the assertion-evidence approach has three main principles: 00:00:33.06 number one, build your talk on messages not on topics; 00:00:37.27 number two, support those messages with visual evidence not bullet lists; 00:00:42.13 and then number three is, when explaining that visual evidence, 00:00:46.18 fashion sentences on the spot. 00:00:50.14 Whereas you might have just one sentence written on the slide, 00:00:54.26 you will say six, seven, eight, or nine sentences 00:01:00.14 in your talk. 00:01:03.01 This second film does two things. 00:01:06.15 This first is this film 00:01:10.27 presents evidence showing that the assertion-evidence approach 00:01:14.14 is more effective. 00:01:16.21 After all, this approach requires quite a bit more work. 00:01:20.16 You have to think deeply about what those message headlines are. 00:01:23.19 You have to create that visual evidence. 00:01:26.13 And then you have to practice enough 00:01:29.08 that you can fashion sentences on the spot. 00:01:32.15 The second part of the talk 00:01:36.02 is to walk through a research presentation 00:01:38.07 and show you what I consider some of my greatest hits: 00:01:42.09 slides that students have created 00:01:45.14 for various research talks. 00:01:48.20 So, why the assertion-evidence approach? 00:01:52.20 Why should we use it? 00:01:54.27 At Penn State, we've done some tests 00:01:58.09 in which we've found that the assertion-evidence approach 00:02:02.14 is more effective than a typical presentation slide, 00:02:07.13 such as what you see on the screen. 00:02:10.00 So, we created a presentation... 00:02:12.16 this one happens to be on magnetic resonance imaging... 00:02:15.23 and we first created it as a typical traditional talk 00:02:22.29 that follows PowerPoint's defaults. 00:02:25.12 In fact, we looked at a lot of presentations 00:02:29.17 that were on magnetic resonance imaging, 00:02:32.06 and we patterned it after that. 00:02:34.20 Then we created a set of slides 00:02:38.01 for the exact same words that would be spoken, 00:02:42.26 but then followed the assertion-evidence approach. 00:02:45.22 So, we've got 50-60 participants in a room. 00:02:51.10 They see slides that follow the traditional approach. 00:02:56.14 Then we've got 50-60 students in another room. 00:03:00.07 They see slides that follow the assertion-evidence approach. 00:03:05.00 And after the presentation, 00:03:08.00 both sets of participants... 00:03:10.08 they essentially tried to explain, 00:03:13.26 how does magnetic resonance imaging work? 00:03:16.21 And then we had scorers who scored each of those attempts. 00:03:22.09 But the scorers, or the raters, 00:03:25.09 they didn't know from which group that they came. 00:03:29.02 But what the raters found is... 00:03:32.18 they found that the traditional approach... 00:03:35.14 they scored on average about 42% 00:03:38.05 in terms of how much they understood. 00:03:41.23 Whereas the people who viewed the assertion-evidence approach, 00:03:45.28 they found 50... 00:03:48.21 they scored that 59%. 00:03:51.01 And the statistical difference between those two 00:03:53.21 -- at these levels, 50-60 participants -- 00:03:57.16 is about... less than 0.01. 00:04:02.08 And so, what we were left to conclude 00:04:04.26 is the assertion-evidence slides 00:04:07.17 led to better comprehension and recall of information, 00:04:12.19 and that increase in comprehension and recall 00:04:16.29 is statistically significant. 00:04:19.14 One thing that we also found 00:04:23.13 is that the misconceptions that participants had 00:04:27.12 in the traditional approach, 00:04:29.27 with traditional PowerPoint slides, 00:04:32.22 is about 10 times higher 00:04:36.06 than for the assertion-evidence group -- ten times. 00:04:39.18 And what we think is happening there 00:04:42.13 is that the participants... 00:04:45.13 they're reading a bullet or something 00:04:49.23 while the speaker is talking about something else. 00:04:52.24 Those are two reasons 00:04:57.11 for you to think about using these slides 00:05:00.01 to present your research: 00:05:02.18 to increase comprehension and recall, 00:05:04.29 and to reduce number of misconceptions 00:05:08.21 that people have about your work. 00:05:11.08 So, that's a reason for doing the assertion-evidence approach. 00:05:14.14 How do you... how do you apply it 00:05:18.02 in a research presentation? 00:05:20.14 And so, what I'd like to do is walk through some common scenes, 00:05:23.18 sometimes talking about what's typically done, 00:05:27.08 and then show you what's done in the assertion-evidence approach. 00:05:30.17 Okay. So, here we go. 00:05:31.29 The first scene is typically a title slide. 00:05:34.29 And at so many research conferences, 00:05:38.10 or at symposiums and seminars, 00:05:41.00 you'll see people have a slide such as the following. 00:05:44.14 Hello. My name is Stuart Apple. 00:05:46.20 I'm workin' with Kerry Cho and Dale Gray, 00:05:49.03 and we are from the Environmental Science Department 00:05:52.14 at such and such a university. 00:05:54.19 And what we are going to present today is 00:05:58.02 Atmospheric Mercury Depletion Events 00:06:00.18 in Polar Regions During Arctic Spring. 00:06:03.25 And then that slide is gone. 00:06:06.05 And in those few nanoseconds 00:06:09.04 between that slide and the next slide, 00:06:11.27 think about how you're feeling as an audience member. 00:06:15.14 Are you... are you confident 00:06:18.09 that you're gonna learn a lot in this talk? 00:06:20.19 Do you feel as if the train has left the station, 00:06:22.27 and yet you're not really clear on what the talk is about 00:06:28.29 or what the title means? 00:06:33.27 I think that's what many people have... 00:06:35.09 or, the state that they're in in a research talk. 00:06:37.27 We can do better than that. 00:06:40.05 And now, you kind of look at this slide 00:06:42.21 and you wonder... gosh, I see a lot of slides like that. 00:06:45.17 Maybe they've got different decoration. 00:06:47.23 But why? 00:06:49.28 Why do so many slides have this title on the top, 00:06:52.05 and then just the name and the affiliation beneath, 00:06:55.05 and maybe just some decoration? 00:06:57.19 And you know why. 00:07:00.02 I mean, the reason is that people are just following PowerPoint's defaults. 00:07:03.05 PowerPoint tells them to do that. 00:07:05.10 And so, they'll put their title and then they'll put their affiliation. 00:07:08.07 And what we're saying is, 00:07:10.03 you can do better than that. 00:07:12.01 You can do better than that. 00:07:14.10 So, what can we do? 00:07:16.27 So, I'd like to look at... actually, this research 00:07:20.06 was done by a young researcher at the University of Oslo, 00:07:25.02 a chemistry student by the name of Katrine Aspmo. 00:07:27.24 And she was gonna give her first research presentation. 00:07:31.18 And she was a little nervous about that. 00:07:34.18 Moreover, the presentation was gonna occur in Portland, Oregon, 00:07:38.21 and so the people were gonna be speaking in English 00:07:43.14 -- not her native Norwegian. 00:07:45.27 But her English is good. 00:07:48.02 Still, that's just one more thing for her to consider. 00:07:50.25 But before this talk, 00:07:53.20 she went to her advisor and she asked her advisor, 00:07:57.22 how many people will be in the audience? 00:07:59.23 And her advisor said, well, it'll be about 50. 00:08:02.07 She said, whoa. 00:08:04.19 And then she said, how many of them know 00:08:05.20 what an atmospheric mercury depletion event is? 00:08:08.21 And her advisor said, hmm... I'd say five. 00:08:11.15 You're one. I'm another one. 00:08:14.20 Our collaborator, Grethe -- she's a third. 00:08:17.08 And so, that kind of gave her this idea that... 00:08:20.23 whoa... you know... 00:08:23.11 for her to orient that audience, 00:08:25.20 she's gonna have to spend some time on that initial scene. 00:08:29.05 And so, here's what she came up with. 00:08:33.12 Hello, my name is Katrine Aspmo, 00:08:36.00 and I'm working with Torunn Berg 00:08:38.21 at the Norwegian Institute of Air Research, 00:08:40.22 and I'm also working with Professor Grethe Wibetoe 00:08:42.23 from the Department of Chemistry at the University of Oslo. 00:08:46.00 And what we're trying to determine 00:08:48.20 is where atmospheric mercury goes 00:08:54.03 when it depletes or falls out of the atmosphere. 00:08:59.03 Now, one thing you may not realize 00:09:02.08 is that in this atmosphere -- in this room right here -- 00:09:03.29 there's a certain amount of mercury. 00:09:06.16 It's not much. It's about 1.5 nanograms per liter. 00:09:09.16 And here, in this room, 00:09:11.16 it remains in the atmosphere. 00:09:13.15 Even when we breathe it in, we breathe it out. 00:09:15.24 It still stays in that gaseous state. 00:09:18.19 But for some reason 00:09:21.09 -- and scientists aren't exactly sure why -- 00:09:24.14 in the polar regions of the world, 00:09:26.24 such as this beautiful Ny-Alesund region, 00:09:29.00 that you see here of Norway, 00:09:32.04 there are these events where that level of mercury 00:09:36.23 will drop to zero. 00:09:39.20 And the question comes... 00:09:41.27 where does that mercury go? 00:09:43.28 So, these events... they've got this fancy name, 00:09:47.10 atmospheric mercury depletion events. 00:09:49.16 But where does that mercury go? 00:09:51.20 Now, while you may not have realized 00:09:53.21 that mercury is in the atmosphere, 00:09:55.14 I think everyone in the room knows 00:09:58.03 that mercury is toxic. 00:09:59.29 And at high enough levels, 00:10:02.07 it can cause paralysis and even death for mammals. 00:10:04.29 And so, for the polar bears in the region, 00:10:08.01 for the arctic foxes, 00:10:10.01 for the stray Norwegians who wander through there, 00:10:14.23 how... where is that mercury? 00:10:17.03 What and... and... and how does... 00:10:20.03 how does it get to that particular spot? 00:10:22.19 That is, a... you know, a research question. 00:10:26.17 And so, what we did is... you may not be able to see, 00:10:29.02 but in this one corner here is a cabin. 00:10:32.27 And we stayed in that cabin for two months. 00:10:37.00 And what we did is 00:10:39.20 we made simultaneous measurements of the amount of mercury 00:10:42.29 that's in the atmosphere 00:10:44.24 with the amount of mercury that's in the surface snow 00:10:47.06 to test our hypothesis. 00:10:49.21 That is in fact where that mercury goes. 00:10:54.15 Wow. I remember seeing this talk at... 00:10:57.19 seeing the beginning of this talk 00:11:00.03 at the University of Oslo in 2004. 00:11:02.03 And I thought to myself, you know, 00:11:04.15 I'm not an environmental scientist, 00:11:06.17 but I am lucky. 00:11:08.18 I am lucky to be here 00:11:10.26 because I'm gonna learn something in this talk. 00:11:12.20 I have already learned something in this talk. 00:11:14.20 And that's something that I would suggest to you: 00:11:17.08 in that first scene, give the audience something. 00:11:21.02 You know at the beginning of a scientific presentation, 00:11:23.28 there are certain things 00:11:26.27 that you want to accomplish before you get to the middle. 00:11:29.05 One thing is you want to show the importance of the research. 00:11:31.15 You want to present background 00:11:33.15 that the audience needs. 00:11:35.11 You also want to present background 00:11:37.20 about yourself to build credibility. 00:11:39.09 And you want to give your research question. 00:11:41.14 Katrine did all of those... 00:11:44.13 all of those on that title slide. 00:11:47.07 Wow -- that's effective. 00:11:49.11 So, we've talked about one scene here. 00:11:54.18 Another scene that often occurs 00:11:58.24 also happens at the beginning, before you get to the middle, 00:12:00.14 and it's something people call an outline slide. 00:12:02.10 I like to call it kind of a mapping slide. 00:12:04.17 Sadly, many people's slide looks like the following. 00:12:08.18 You've got this detailed list 00:12:10.24 of every single thing they're gonna do 00:12:13.00 -- things they've already done, actually, 00:12:14.27 in the talk by the time they give it -- 00:12:16.24 and then things that you already know were gonna come. 00:12:19.06 And then they will dutifully read through that list. 00:12:21.20 You know, they'll get to... you know, 00:12:23.29 almost all the way through, 00:12:25.24 and then they'll realize they misspelled a word like acknowledgments, 00:12:28.06 and they'll apologize for it... 00:12:30.14 and then they will finish, 00:12:32.05 and then they will move on. 00:12:34.00 And when they move on, 00:12:35.25 then the audience is, to themselves, 00:12:38.08 thinking, well, he gave me all these things, 00:12:41.14 or she gave me all these things, 00:12:43.13 and I've forgotten them already. 00:12:45.11 You can do better than that. 00:12:47.20 One thing is, remember, 00:12:51.04 people feel comfortable with lists of two's, three's, and four's. 00:12:54.01 So, a list, here, of eleven is not very good. 00:12:57.19 Another thing is you don't need to give them some things: 00:13:00.01 introduction -- they know the talk has an introduction; 00:13:02.08 background -- you've already given the background 00:13:04.05 before you do this mapping; 00:13:06.04 conclusions; acknowledgments; questions. 00:13:08.02 They know that's happening. 00:13:09.28 It's the stuff in the middle. 00:13:11.24 And you don't have to give the sub-levels. 00:13:13.22 I mean, that's not what's so important. 00:13:15.13 It's the main things that you need to give. 00:13:17.16 So, let's look at what Katrine Aspmo did. 00:13:19.20 I thought she did an excellent job. 00:13:22.00 So, in her talk, she divided it into three parts. 00:13:25.22 One is this theory for mercury cycling. 00:13:28.21 And then she talked ab... 00:13:30.18 mentioned that she was gonna, you know, talk about that. 00:13:33.06 And then she had another part, 00:13:35.16 where she had those measurements from that station... 00:13:37.29 actually, on those two types of mercury measurements: 00:13:41.24 in the atmosphere and then in the surface snow. 00:13:46.03 And then, her third part of the talk 00:13:49.00 was environmental implications, 00:13:51.05 and she showed this particular picture. 00:13:53.00 And the polar bear is not there in a gratuitous fashion. 00:13:55.18 Actually, a lot of the data 00:13:58.05 on the effects of these mercury depletion events 00:14:00.08 occurs through studying polar bear carcasses, 00:14:04.02 and so that is what she went over. 00:14:06.20 But whoa... what a great mapping scene. 00:14:09.20 And I have seen people sometimes take those images 00:14:12.20 and put those as icons in the corners of all the slides 00:14:17.05 that were from that particular section. 00:14:19.05 That's just another thing, here, 00:14:21.05 that you could do that doesn't clutter a slide 00:14:24.04 or take up too much space 00:14:25.29 or too much time for the audience's attention. 00:14:28.15 Yeah. 00:14:30.07 But that's the kind of mapping that you want to do. 00:14:32.19 Let's try... let's think about literature review. 00:14:35.02 So, literature view... oh my gosh. 00:14:36.27 This is often one of the most boring parts. 00:14:38.13 People walk through all the... 00:14:40.08 all the major work that's been done. 00:14:42.15 You'll get a bullet list slide, 00:14:44.06 and they'll talk about everything that they've done. 00:14:46.07 And I'd like you to rethink that. 00:14:48.29 You know, what are you doing in the literature review? 00:14:51.11 You're trying to show that there's a gap in the work, 00:14:56.04 and that... and that your research question... 00:14:58.21 your research hypothesis... 00:15:00.06 your research fills that gap. 00:15:03.12 And so, you're trying to show that gap exists. 00:15:05.14 So, here's one that Jacob Snyder did on his. 00:15:09.23 And so, what he did is... 00:15:12.01 he wanted to make it clear 00:15:14.23 that there are a number of people 00:15:17.09 -- not a lot of people, but a number of people -- 00:15:19.09 who have tried to use additive manufacturing 00:15:22.18 to create these heat transfer devices 00:15:25.24 for these gas turbine engines. 00:15:27.28 And so, he showed a couple of images 00:15:30.21 right at the beginning: 00:15:32.06 one from the work of Kirsch and Thole, 00:15:34.03 another one from the work Collins. 00:15:35.24 Then it talked a little bit about what they had done. 00:15:38.14 Then he brought in the work of Ferster 00:15:40.26 and the work of somebody else. 00:15:42.17 Then he brought in the work of Siemens, 00:15:44.17 that they... and he talked about what they had done. 00:15:46.21 But then he made it clear that there was a gap 00:15:50.24 between what they've done and what still could be done. 00:15:53.04 Wow. 00:15:54.26 That's something that people can follow. 00:15:56.29 Really, really nice. 00:15:58.18 And it's thinking about what details are essential 00:16:00.29 and then what details are secondary. 00:16:06.20 So, we've looked at a literature review. 00:16:08.26 Let's talk a little bit about methods. 00:16:11.00 So, with methods... 00:16:13.07 I'll show you one that's more from an experimental talk. 00:16:17.19 And so, here, rather than having, again, 00:16:21.19 a bullet list with all these details, 00:16:23.09 what Nick Cardwell did in this one is 00:16:26.13 he started with an image of his experiment. 00:16:29.08 So, he's got this recirculating wind tunnel. 00:16:31.19 And then he shows this one particular part 00:16:34.28 of that wind tunnel 00:16:36.19 where they split the flow. 00:16:38.26 And so, the flow there splits. 00:16:42.02 You see some of it goes in the blue part 00:16:44.08 and some of it goes in that white part. 00:16:46.04 And then, in that... in that center part, or that white part, 00:16:48.17 what happens is they heat some of that flow. 00:16:51.00 So, you've got... now, you've got two flows: 00:16:53.14 the secondary flow in the flu... in the blue part, 00:16:58.00 and then the primary flow, but the primary flow is heated. 00:17:01.00 And then they bring that secondary flow 00:17:03.22 back in to try to cool the surfaces after... 00:17:09.20 after the heated flow has occurred. 00:17:11.12 And so, they're simulating what goes on in a jet engine, 00:17:14.08 where they try to cool the parts 00:17:16.28 downstream of the combustor. 00:17:18.23 But... really nice. 00:17:21.01 Now, Nick Cardwell... he knows a lot of the numbers 00:17:24.03 on flow rates and temperatures, 00:17:25.29 and so he was just able to say those. 00:17:27.27 What you could do, if you're... 00:17:30.21 don't have the confidence you could remember those, 00:17:33.23 you could animate them into that white space, 00:17:36.02 but then I would animate them out 00:17:38.08 when they're not needed. 00:17:40.02 Don't let those clutter the slide. 00:17:42.14 But remember, the audience will have your paper. 00:17:45.20 They can go back and refer to what things are. 00:17:49.16 You definitely want to have those details, 00:17:51.19 and you want to know them 00:17:53.12 and be able to respond to questions, 00:17:55.08 but you don't need to put everything on the slide 00:17:58.13 as so many people do. 00:18:02.07 Let's look at a second method slide. 00:18:04.06 And on this one, 00:18:07.08 the speaker didn't have images 00:18:09.19 such as the last one had. 00:18:11.26 So, let's see how he handled it. 00:18:14.19 So, this particular work was done by Jimmy Webber. 00:18:18.06 And in a sense, 00:18:20.03 what he's trying to do in this part of the methods section 00:18:22.27 is just talk about how he is going 00:18:27.14 to characterize this pollution of 53 streams 00:18:30.15 in the Northeast. 00:18:32.08 And so, he introduces these six criteria 00:18:34.21 that he will use to characterize that pollution. 00:18:39.19 And then the second thing that he does 00:18:42.15 is he says, as a reference, 00:18:44.06 he's gonna compare the pollution of those streams 00:18:47.03 versus the pollution 00:18:49.15 -- or those same criteria -- 00:18:51.09 of 12 reference streams. 00:18:54.03 And then he gives a teaser 00:18:58.16 on a couple of results from his work, 00:19:01.19 and I thought that was interesting that he did so. 00:19:03.29 And what he did is he showed 00:19:06.19 that the sulfate levels were beyond that reference point, 00:19:11.10 on average, for the 53 streams. 00:19:14.20 Not to a place where it's dangerous 00:19:16.26 for the plants or wildlife, 00:19:18.26 but certainly beyond the reference. 00:19:21.12 And then he also showed that the chloride levels... 00:19:24.28 those were in fact at the impaired level. 00:19:29.13 I love this scene. 00:19:31.02 I love how he tells this part of his methods 00:19:36.09 as a story. 00:19:38.06 And I love the imagination. 00:19:40.09 Somebody else would have just had a bulleted list, 00:19:42.27 and we would have slept through it. 00:19:44.25 But here, he has essentially told us a story 00:19:49.08 that we can remember. 00:19:52.10 And that is so important. 00:19:54.15 It's not just that people understand our work; 00:19:56.19 we want them to remember our work. 00:19:59.20 So, here... this part of the talk 00:20:01.26 is where you have to... 00:20:03.16 you're making some of your best arguments. 00:20:07.12 And I just want to show an example, here, 00:20:10.23 that comes from some work 00:20:14.11 that's being done in Norway 00:20:17.11 by this... Siri Larsen. 00:20:19.12 And so, what Siri is doing is 00:20:24.26 she's looking at satellite images 00:20:27.00 and kind of looking at them in interesting ways, 00:20:28.26 with different filters, 00:20:30.23 to extract some valuable information. 00:20:32.20 And so, these images 00:20:36.08 happen to come from parts of Norway 00:20:38.10 where there might be avalanches. 00:20:40.18 And that's important because 00:20:42.27 if you have an avalanche... 00:20:44.24 you know... or if... 00:20:46.25 if a place is susceptible to one avalanche, 00:20:49.23 then more could occur, 00:20:51.20 and you want to keep people away 00:20:54.22 from that particular area. 00:20:56.12 But the problem is when you look at the satellite images, 00:20:58.23 sometimes it's hard to distinguish 00:21:01.16 an avalanche from a sparse forest. And so, what she has done is 00:21:08.01 she's applied these filters 00:21:10.11 that kind of helped her make that distinguishing. 00:21:12.23 And so, one of the filters is this aspect direction, 00:21:15.18 and she would talk about how she applied that 00:21:18.24 and what she would find. 00:21:20.08 Another one, then, is of the vertical direction 00:21:22.27 -- how she applied that and what they have found. 00:21:25.19 But I like, again, 00:21:28.08 how she tells that scene, tells it as a story. 00:21:31.12 So, that's an example of results that has images. 00:21:35.24 Another kind of result would be a graph, 00:21:38.06 and so here's... here's a graph that Steve Weaver 00:21:40.26 put together in one of his talks. 00:21:43.24 And so, in this particular graph, 00:21:47.02 what he is doing is he's trying 00:21:51.20 to examine the roughness in this heat transfer channel. 00:21:55.20 And so, one way to determine that 00:21:59.14 is by this particular graph, 00:22:02.20 where you graph friction factor, f, 00:22:06.00 on the y axis 00:22:07.24 versus Reynolds number on the x axis. 00:22:10.08 And so, the first thing that Weaver did 00:22:15.06 is he has that straight line. 00:22:17.06 And so, that would be a perfectly smooth pipe. 00:22:19.23 That would be the theoretical explanation, 00:22:23.12 so... from Blasius in 1894. 00:22:25.10 And then, here... then, the dots... 00:22:29.23 those are his experiments showing that 00:22:34.23 within experimental error he matches the curve of Blasius. 00:22:36.28 But then he's gonna roughen up these surfaces, 00:22:40.07 and so he roughened them up in different ways, 00:22:42.09 and so... different colors... 00:22:44.08 red is not as rough as blue, 00:22:46.05 which is not as rough as green. 00:22:48.05 So, what's the effect of roughening the surface? 00:22:50.27 What effect does it have on the friction factor? 00:22:53.08 And then he animates in what that effect is, 00:22:57.18 and then he shows that it's actually... 00:22:59.12 it increases that friction factor in all of those cases. 00:23:04.03 A couple of things here that I liked... 00:23:06.18 one is I liked that he told things as a story. 00:23:08.24 Another thing I liked is that, here, 00:23:13.04 he was giving his most important result, 00:23:15.24 and he didn't start with the assertion at the beginning 00:23:18.06 because he knew there would be some people 00:23:20.20 who would be skeptical about that result, 00:23:23.12 and he knew there would be others that, 00:23:25.08 if he gave that assertion at the beginning, 00:23:27.07 it would be too much for them 00:23:29.20 -- they would be... they would be overwhelmed. 00:23:31.19 So, he started with a question that they understood. 00:23:34.23 And then, when he was ready, 00:23:36.20 then he brought in his assertion. 00:23:40.01 And so, in a way, I think of this kind of scene 00:23:43.27 as evidence-assertion. 00:23:45.09 Evidence comes first, and then, 00:23:47.08 when the audience is ready, 00:23:49.05 then you bring the assertion. 00:23:51.06 And in real persuasive types of situations, 00:23:53.06 that's a good thing to do. 00:23:55.14 So, let's move on to our last scene. 00:23:59.08 And our last scene is the scene that you show at the end. 00:24:02.09 And interestingly, the most common scene that I see people show 00:24:08.13 as their last slide is something that looks like that. 00:24:11.01 Oh my gosh. 00:24:13.04 Let's think about this. 00:24:14.19 Let's just think about this scene for just a second. 00:24:16.01 What are we trying to do here? 00:24:18.01 What we're trying to do here is... 00:24:19.11 we have presented our work to the world 00:24:21.28 and now we want the world's response. 00:24:24.04 And so, people are seeing our research, here, 00:24:28.19 for the first time. 00:24:30.18 And so that... maybe they're spending a minute and a half 00:24:34.15 or two minutes on a graph, 00:24:36.05 but now they're trying to formulate questions. 00:24:38.09 How does this slide help them formulate questions 00:24:40.19 about research that they've never seen before? 00:24:44.10 And the fact is it doesn't. 00:24:46.08 This is a terrible scene. 00:24:48.04 You can do so much better. 00:24:49.28 Don't do that -- please. 00:24:51.27 I mean think about it. 00:24:53.18 Think about it from the audience's perspective. 00:24:56.01 Your best scene to end with 00:24:58.22 is a scene that shows your conclusion, 00:25:01.28 that shows really what your main takeaways are. 00:25:05.16 And that's what Danielle Lesso 00:25:08.02 did in this particular scene. 00:25:09.22 So, she had three main points 00:25:12.09 that she had made in her talk 00:25:14.26 to show that Miscanthus 00:25:17.14 could be a promising fuel crop 00:25:20.10 for the northeastern part of the United States. 00:25:22.20 And the first one is that Miscanthus 00:25:25.06 is able to have a cold tolerance 00:25:29.00 down to 6 degrees Celsius, 00:25:30.24 which is important in the Northeast. 00:25:32.15 Another thing is that the root depth 00:25:35.10 goes down to 2 meters, 00:25:37.21 which is important because a lot of the Northeast, 00:25:39.25 as she pointed out in her talk, 00:25:41.18 is on hilly surfaces, 00:25:43.15 and so the plant could be washed away 00:25:45.19 if it doesn't have sufficient root depth. 00:25:48.14 And then in the third part, she showed this energy ratio, 00:25:52.02 which the National Academy of Sciences had found, 00:25:55.00 that for every BTU of energy that you put in to 00:26:02.08 planting, fertilizing, harvesting, and transporting Miscanthus, 00:26:07.11 you get 6 BTUs out of energy 00:26:10.00 -- a ratio was 6 to 1. 00:26:13.21 And at this point, she gave her closure to her talk. 00:26:17.04 And then, when she was ready, 00:26:19.28 then she asked for questions. 00:26:22.08 Now, the audience has something 00:26:25.06 in which they can ask. 00:26:28.01 They've got words up here to help them formulate questions. 00:26:33.25 So, what have we done today? 00:26:36.14 We've talked about why the assertion-evidence approach... 00:26:39.01 why it is worth 00:26:42.27 at least an experiment on your part, 00:26:44.27 for you to try the gains 00:26:48.09 that we have in audience comprehension. 00:26:50.23 And then we walked through various scenes 00:26:53.00 to give you ideas 00:26:55.09 on how you might design your title slide, 00:26:57.18 a mapping slide, 00:27:00.02 a slide on your literature review, 00:27:01.12 talk about your methods, your results, 00:27:03.19 and then finally your conclusion. 00:27:07.04 So, this particular talk has focused 00:27:09.26 on how we might design slides for a research talk. 00:27:14.13 The third part of the series talks 00:27:17.28 about how you would deliver those slides 00:27:19.24 and, in particular, 00:27:22.11 how you would deliver those slides with confidence.
Part 3: Attaining Confidence in Your Scientific Presentations
- Duration: 00:20:40
00:00:08.02 This film is the third of a three-part series for iBiology 00:00:14.17 on improving your scientific presentations. 00:00:17.14 The first two films introduced a different approach, 00:00:20.28 the assertion-evidence approach, 00:00:23.04 for presentations, 00:00:25.24 what it is, why to use it, 00:00:28.19 and then how to incorporate it 00:00:31.17 into a research presentation. 00:00:33.17 This third film focuses on an important aspect of presentations, 00:00:39.15 and that is delivery. 00:00:41.07 And in particular, 00:00:43.22 how do you exude confidence in your delivery? 00:00:49.10 Now, I have had the fortune of teaching scientific presentations 00:00:54.13 over the past 30 years 00:00:58.10 on 4 continents, 17 countries, 00:01:02.17 and more than 100 institutions -- 00:01:08.00 laboratories, companies, universities, and agencies. 00:01:12.28 And by far, the number one question 00:01:16.05 that young scientists and engineers asked me is, 00:01:21.09 how do I handle nervousness? 00:01:24.08 Now, they don't ask this question in front of the big crowd. 00:01:27.11 Rather, they catch me in the hallway and they ask. 00:01:31.01 And to that question, I have the same answer. 00:01:35.00 In the days before the presentation, 00:01:38.14 think positively. 00:01:40.13 Just as a professional tennis player 00:01:43.00 will imagine a shot going in a certain spot, 00:01:46.15 you too, in your presentations, 00:01:49.25 should imagine success -- 00:01:53.22 the audience understanding your results, for instance. 00:01:57.19 On that day of the presentation, 00:02:00.27 arrive early. 00:02:04.10 I cannot tell you how many times arriving early 00:02:08.08 has saved me from embarrassment. 00:02:12.03 Learning that my computer 00:02:15.09 doesn't hook up with their particular projector, 00:02:18.19 finding that chairs are not where they should be... 00:02:22.27 arrive early. 00:02:25.11 That way, you can remove one of the biggest sources of nervousness, 00:02:29.23 and that is that your presentation slides will not work. 00:02:36.11 And then, just before you speak, 00:02:40.28 think about some advice that Mark Twain 00:02:43.24 shared with a nervous presenter backstage. 00:02:47.20 Don't worry -- they're not expecting much. 00:02:51.15 So, those are the three things 00:02:54.10 that I generally tell people 00:02:57.01 on how to handle nervousness 00:02:59.12 before a scientific presentation. 00:03:01.09 And I have to say, I've chosen those three 00:03:03.28 because those three things have helped me. 00:03:06.14 But what I would also say is "how do I handle nervousness?" 00:03:11.04 is not your best question. 00:03:13.24 The better question to ask is, 00:03:16.07 how do I attain confidence when I present? 00:03:20.13 And I'm talking about the confidence of the neuroscientist Jill Bolte Taylor 00:03:25.03 or the physicist Brian Cox. 00:03:29.19 So, this question's a little bit different. 00:03:31.23 And if you think about confidence, 00:03:35.19 there are really two types of confidence. 00:03:38.09 There's that confidence that you feel inside, 00:03:41.03 the internal confidence. 00:03:44.17 And then there's that confidence 00:03:47.20 that you project to the audience. 00:03:50.02 Even though inside you might be hurting, 00:03:52.16 to the audience you can still project 00:03:55.18 that you at least seem confident. 00:03:58.28 So, I'd like to talk about those two. 00:04:01.21 And I'll start first with the internal confidence. 00:04:05.15 And I'd like to draw upon my interviews 00:04:08.29 with a number of scientists and engineers 00:04:12.21 who've given excellent presentations 00:04:15.02 to large audiences, 00:04:16.29 such as Jill Bolte Taylor or Brian Cox. 00:04:23.01 And one of the key ingredients for that internal confidence 00:04:29.09 is having strong content. 00:04:32.15 If you have tested or vetted that content of yours 00:04:37.24 before audiences whom you respect, 00:04:41.12 and that content has withstood those audiences, 00:04:45.20 and in fact received positive responses 00:04:49.16 from those audiences, 00:04:51.16 that will help you with your internal confidence. 00:04:57.00 A second ingredient comes from a short interview 00:05:02.01 I had with Jane Goodall. 00:05:04.18 I happened to meet her at an airport, 00:05:06.27 and I didn't have much time to speak with her, 00:05:10.21 but I mentioned that I have this opportunity 00:05:14.08 to talk about presentations 00:05:16.25 to many young scientists and engineers 00:05:20.00 around the world, 00:05:22.05 and what piece of advice would she give them? 00:05:25.22 And her demeanor changed. 00:05:29.22 She kind of took on a harsh demeanor, 00:05:31.26 and she said, 00:05:34.00 well, it certainly wouldn't be the same piece of advice 00:05:37.09 that someone gave me. 00:05:39.26 And I had this sense that 00:05:43.05 I had just ripped the Band-Aid off an old wound. 00:05:45.13 She said that before her first presentation 00:05:49.13 someone had told her to just go up there. 00:05:52.16 You're the expert. 00:05:54.18 And she said that was horrible advice. 00:05:57.17 And then she went on to tell me 00:05:59.29 that she was here touring the United States, 00:06:02.14 and she was giving essentially 00:06:06.12 the same talk at ten different places. 00:06:09.12 And at each of those places, 00:06:13.26 before the talk, 00:06:16.25 she insisted on having 45 minutes by herself 00:06:21.00 so that she could walk through that talk 00:06:24.02 and imagine herself giving it to the crowd. 00:06:28.20 And I thought to myself, my gosh. 00:06:31.24 Someone who is that seasoned... 00:06:34.05 if she still needs that amount of preparation, 00:06:40.01 what about the rest of us? 00:06:43.11 Preparation -- that I would say is the second ingredient. 00:06:47.28 And then the third ingredient came from the world health statistician 00:06:52.29 Hans Rosling. 00:06:55.03 And Hans Rosling told me that, for him, 00:07:01.02 an important part of internal confidence was focus. 00:07:09.20 What Rosling said is before an important talk 00:07:12.07 he didn't want to... 00:07:15.12 in the minutes before, he didn't want to really speak to anybody. 00:07:17.27 He just wanted to be by himself. 00:07:21.11 And his best analogy for it 00:07:24.20 was to be like a downhill slalom skier 00:07:28.17 up at the top, just before you take off. 00:07:33.11 And a downhill slalom speak... 00:07:35.16 skier thinks about going here, going here, 00:07:39.03 going here, or going here. 00:07:42.01 And he said, for him, that was the same thing, 00:07:45.14 that he wanted to think about exactly 00:07:47.27 where he was going to go. 00:07:51.06 This idea of focus was also one 00:07:55.06 that a young presenter communicated to me -- 00:07:59.13 Sheila Patek, before one of her first presentations, 00:08:04.16 which was to more than 800 people. 00:08:08.16 And what she said is... 00:08:12.05 she said, if I had thought about the 800+ people 00:08:16.07 in the room that day, 00:08:18.19 I never could have done it. 00:08:21.05 Instead, I thought about the science 00:08:23.23 and my passion for the ideas, 00:08:25.28 and I let that carry me. 00:08:30.18 Strong content, preparation, focus. 00:08:36.23 If you will work on those three things, 00:08:39.22 you will bolster your internal confidence. 00:08:44.12 Now, I'm not saying that before a talk 00:08:47.23 you're not going to feel nervous. 00:08:50.29 There will be audiences and there will be situations 00:08:55.07 in which you will. 00:08:57.24 But... and I think something to realize 00:09:01.23 is that the internal confidence 00:09:04.12 is not an on/off switch. 00:09:06.29 A better analogy is that it's a bucket. 00:09:10.20 And the more that you work on those three, 00:09:13.18 the more fluid that is in that bucket. 00:09:18.00 And that... the nervousness that you will have 00:09:23.13 is not as much as if you had not done the strong content, 00:09:29.06 the preparation, 00:09:31.03 the focus. 00:09:33.21 So, that was a little bit about internal confidence. 00:09:37.01 How about projected confidence? 00:09:40.26 One thing that has amazed me 00:09:43.28 is that I have had students 00:09:46.19 who have given a presentation 00:09:49.09 -- former students -- 00:09:51.12 and I have watched them, 00:09:53.08 and I have thought, oh my gosh, 00:09:56.06 these students are doing great. 00:09:58.10 And then I think to myself, 00:10:00.24 I must be a really good teacher, 00:10:03.12 that my former students are presenting so well. 00:10:07.07 But then afterwards, I will talk to these students, 00:10:11.11 and they will tell me how nervous they were. 00:10:14.17 And I will think to myself, 00:10:16.21 I must not be a very good teacher, 00:10:19.07 because I couldn't see their nervousness. 00:10:22.22 But one thing that I've realized is that 00:10:27.07 even though inside you are hurting, 00:10:30.16 you can mask that discomfort 00:10:36.28 in certain ways and with certain steps that you can take. 00:10:40.04 And so, I'd like to come back to Twain 00:10:43.20 and talk about another quotation that he has. 00:10:45.23 And for the moment, disregard the religious implications 00:10:48.22 of this quotation. 00:10:50.24 But Twain talked about 00:10:54.04 the calm confidence of a Christian with four aces. 00:10:58.04 And I think as a presenter, 00:11:01.09 there are four aces that you can play in a presentation 00:11:05.02 to project more confidence. 00:11:07.27 The ace of spades: start strong. 00:11:13.06 On the day of the presentation, arrive early. 00:11:17.04 Make sure that all the equipment is working. 00:11:21.20 And not just your computer and projector, 00:11:25.08 but make sure that the clicker is working 00:11:27.27 so that you are ready to go. 00:11:31.03 One thing that is important to realize is that 00:11:36.10 different rooms have different ghosts. 00:11:38.05 And crazy things that will happen in one room 00:11:41.12 will not happen in another. 00:11:43.16 But in that other room, 00:11:46.05 something else crazy could happen. 00:11:48.17 Be prepared -- arrive early. 00:11:50.27 And then, when you've gotten everything ready to go, 00:11:54.28 don't hide out in the bathroom. 00:11:56.25 Rather, meet your audience. 00:12:00.21 Try to find out something from them 00:12:03.07 that you can incorporate into your talk. 00:12:06.29 I see speakers do that. 00:12:09.13 And even though I know exactly what it is that they're doing, 00:12:13.25 I'm always amazed at that particular move 00:12:17.21 that they make. 00:12:20.03 Just meet your audience, 00:12:22.20 try to find out something about them. 00:12:24.14 Even if you don't incorporate it, 00:12:27.01 you have strengthened your hand 00:12:29.18 by meeting that person, 00:12:31.23 becoming in a sense that person's friend 00:12:35.01 or that person becoming your ally. 00:12:38.18 And then, when it's time for you to go up, 00:12:43.18 move up to the stage, stand tall. 00:12:47.09 I like to feel the muscles in the backs of my calves and thighs. 00:12:50.28 I like to feel myself stand as tall as I can 00:12:55.11 in front of the audience. 00:12:57.23 Don't necessarily say anything for a moment. 00:13:00.29 Smile. 00:13:03.04 Smiling is a great thing to do 00:13:06.10 because what happens is the audience will smile back, 00:13:08.29 and that's a good feedback loop for you. 00:13:12.17 And then, when you're ready to speak, 00:13:15.11 make sure that the first syllable that you say 00:13:20.08 is not uh. 00:13:22.12 Oh my gosh, I can't tell you how many times 00:13:26.01 people have undercut such a wonderful moment 00:13:28.19 with an uh or um. 00:13:32.16 Know what you're going to say first. 00:13:35.24 I don't memorize much, 00:13:38.18 but I often have the first sentence committed to memory. 00:13:42.15 And when I say it, 00:13:45.00 I will pause somewhere in the middle at a natural place. 00:13:49.22 And when you pause 00:13:52.06 -- and this might sound odd -- 00:13:54.11 listen for the HVAC sound in the room 00:13:57.21 -- the heating, ventilation, and cooling sound of the room -- 00:14:00.29 and love that sound. 00:14:04.01 00:14:06.05 Embrace that sound. Feel comfortable with that sound 00:14:08.22 so that you don't have this urge 00:14:12.03 to fill that sound with uh or um. 00:14:16.25 And then, when you show that first slide, 00:14:19.28 know that first slide. 00:14:22.09 Don't turn to the slide 00:14:24.20 and read what the title is. 00:14:29.16 And for heaven's sake, 00:14:32.04 don't turn to the slide 00:14:35.24 when you say your name. 00:14:37.25 You ought to know your name. 00:14:39.18 Know that first slide. 00:14:41.22 Now, on that slide, you should have some kind of image 00:14:45.13 or a sequence of images 00:14:47.25 that orient the audience. 00:14:50.03 And turning to that image... 00:14:52.14 that's a natural turn, 00:14:54.19 because you want the audience to turn toward that image. 00:14:59.12 Ace of spades -- start strong. 00:15:03.07 Ace of hearts -- reduce the text on slides. 00:15:08.21 And so, don't follow PowerPoint's defaults 00:15:11.23 and have all these bullets. 00:15:14.09 Rather, use an approach 00:15:16.22 such as the assertion-evidence approach 00:15:19.03 that tries to minimize the text on slides 00:15:21.24 to just the essence of the argument 00:15:24.14 that you are giving. 00:15:26.22 Build your talk on messages, 00:15:28.25 not on topics. 00:15:31.01 Support your messages with visual evidence, 00:15:33.14 not bullet lists. 00:15:35.17 And that visual evidence could be photographs, drawings, 00:15:40.00 diagrams, graphs. 00:15:42.18 It could be a film, could be an equation, 00:15:44.25 could be a short table. 00:15:47.00 But reduce that text on slides 00:15:49.09 so that when you are speaking, 00:15:52.03 the words clearly come from you. 00:15:57.22 You show ownership of your research. 00:16:02.16 Ace of hearts -- reduce the text on slides. 00:16:06.16 Ace of diamonds -- know what comes next. 00:16:10.09 Now, for me, when I'm at a conference 00:16:15.17 that I'm the second speaker or third speaker 00:16:18.28 in a session, 00:16:20.26 I really don't have the confidence 00:16:23.18 to listen to the previous speakers 00:16:25.21 and to ask them questions. 00:16:28.03 I'm always amazed at those people who do. 00:16:30.16 But for me, I am sitting here 00:16:34.02 thinking about my own presentation. 00:16:36.11 And one thing that I like to do 00:16:38.22 -- and it's similar to what Rosling said -- 00:16:41.24 is that I like to think about each scene. 00:16:45.11 And so, I'll often take a blank sheet of paper 00:16:48.05 and I will sketch thumbnails of the slides, 00:16:52.14 and then the next slide, and then the next slide, 00:16:56.14 and the slide after that, 00:16:58.21 so that I know what comes next. 00:17:01.08 Because if I know what comes next 00:17:04.03 before I click to that slide, 00:17:07.01 I can make a transition to that scene. 00:17:11.26 And that making of a transition... 00:17:15.11 that projects confidence. 00:17:18.21 Ace of diamonds -- know what comes next. 00:17:22.28 Ace of clubs -- finish strong. 00:17:26.04 When you're at the end of your talk, 00:17:29.10 have a strong scene on which to end, 00:17:32.25 one that states your main conclusions 00:17:36.01 and then has images of your main arguments 00:17:41.15 to support that conclusion. 00:17:43.29 And then go through those arguments 00:17:48.00 in an efficient manner. 00:17:50.06 And when you get to the end of those arguments 00:17:53.18 and then you may your closure 00:17:55.27 to end the talk, pause. 00:18:01.08 And before you ask for questions, 00:18:05.29 say the word thank you. 00:18:10.23 Or if you're in Germany, danke schön. 00:18:13.23 Or if you're in China, xièxiè. 00:18:16.02 But give that audience 00:18:19.11 an opportunity to clap. 00:18:22.00 I find that in conference presentations 00:18:25.16 and research symposiums, 00:18:27.16 it's a coin flip. 00:18:29.10 The speaker might give the audience 00:18:31.17 a chance to clap. 00:18:33.25 But a lot of times, the speaker will just rush 00:18:36.08 and ask for questions. 00:18:38.12 And then the audience doesn't know what to do -- 00:18:41.01 do I clap, or do I ask a question? 00:18:43.02 And so, what you end up with is 00:18:46.25 you end up with some tepid applause as well as then 00:18:49.08 some people may be trying to voice a question. 00:18:52.22 Don't go there. 00:18:54.21 You want to receive the applause. 00:18:56.26 And in fact, I think it's important 00:18:59.10 that those people who are 00:19:03.14 perhaps not in your field 00:19:05.18 see the experts in your field... 00:19:07.22 see them applaud. 00:19:09.24 Say thank you. 00:19:11.19 And then, when that applause is dying down, 00:19:15.27 that's when you want to ask for questions. 00:19:19.14 And when you receive questions, 00:19:21.23 know that there are gonna be some questions 00:19:24.27 that you cannot fully answer. 00:19:27.21 And so, a lot of young scientists and engineers... 00:19:31.17 they will rush that answer 00:19:34.08 and say what it is that they know. 00:19:37.13 But then they'll have a big but. 00:19:41.10 And then they'll end that answer 00:19:44.15 with some negative news. 00:19:46.25 A better way to handle it is to pause 00:19:49.18 and start your answer with although. 00:19:53.04 Although I cannot answer everything 00:19:55.28 about the question that you asked, 00:19:57.22 I can say... 00:20:01.00 and then finish strong with what it is you do know. 00:20:05.07 Ace of clubs -- finish strong. 00:20:10.26 If you'll play those four aces in your presentation, 00:20:16.01 you will project confidence. 00:20:21.19 Thank you.

- Educators of H. School / Intro Undergrad
- Educators of Adv. Undergrad / Grad

Talk Overview
Michael Alley has been teaching scientists and engineers how to design presentation slides and deliver effective scientific talks for over three decades. In this three-part lecture, you will learn (a) how to design your PowerPoint or Keynote slides, (b) how to organize your talk, and (c) how to confidently deliver your research seminar. This series will help trainees and research scientists alike improve their presentation skills.
Speaker Bio
Michael alley.

Michael Alley holds a master of science in electrical engineering from Texas Tech University and a master of fine arts in writing from the University of Alabama. Alley is a teaching professor of Engineering Communication at Pennsylvania State University. He is the author of three popular textbooks: The Craft of Scientific Presentations (2013), The Craft… Continue Reading

Playlist: Tips for Science Trainees

Related Resources
Michael P Alley, 2013, The Craft of Scientific Presentations, Springer-Verlag, New York, NY, USA, pp. 286
J K Garner and Michael P Alley, 2016, Slide structure can influence the presenter’s understanding of the presentation’s content , International Journal of Engineering Education, 32, (1A), pp. 39-54
J K Garner and Michael P Alley, 2013, How the design of presentation slides affects audience comprehension: A case for the assertion-evidence approach , International Journal of Engineering Education 26 (6), pp. 1564-1579
PowerPoint templates for Assertion-evidence slides
Reader Interactions
Itizaz says
September 25, 2020 at 12:18 pm
Thank you sir i was really looking for this information…keep it up
Rick Salatino says
May 11, 2021 at 3:39 pm
In the early 2010s, I was a scientific training manager, a molecular biologist and learning professional, for a medium-sized biotechnology company. I tried to influence and train our R&D scientists to use Michael Alley’s Assertion-Evidence approach to improve slide design for better knowledge acquisition and retention. While scientists theoretically accepted the approach, they practically rejected it. Powerpoint defaults were the comfortable default for most scientific research presentations and, I suspect, they still are today.
Craig Hadden says
April 17, 2024 at 5:23 am
Thanks for sharing these videos, which offer a great way to improve the standard of fact-based presentations!
If you’re interested, I’ve summarised this approach in a short post – including a 5-minute video. (The post also discusses “hacking” the assertion-evidence approach, to make your slides more varied.)
Leave a Reply Cancel reply
Your email address will not be published. Required fields are marked *
Find a Video
- Search All Videos
- Search YouTube
- Short Films
- All Playlists
- All Speakers
- Videos in Other Languages
- Biochemistry
- Bioengineering
- Cell Biology
- CRISPR-Cas Technology
- Development and Stem Cells
- Genetics and Gene Regulation
- Human Disease
- Microbiology
- Neuroscience
- Plant Biology
- Professional Development
For Educators
- Educator Newsletter
- Educator Resources
- Flipped Courses
- Online Courses Resources
- All Educator Resources
Educators Recommend
- Cells of the Immune System
- Discovering Cell Cycle Regulators
- Genome Engineering with CRISPR-Cas9
- Hardy-Weinberg Equilibrium
- Next Generation Sequencing
- Semi-Conservative Replication of DNA
- Toll-Like Receptors in Adaptive Immunity
Free Courses
- Free Online Courses
- Take a Free Course
- Learn Microscopy
- Intro to Microscopy
Career Development
- Professional Development Courses
- Professional Development Talks
- Career Exploration
- Biomedical Workforce
- NRMN Resources
Best of iBiology
- Short Science Films
- Meselson and Stahl
- Doudna & Charpentier’s Nobel Prize
- Decoding Ancestral Knowledge
- Microscopy Series
- Short Microscopy Series
- Famous Discoveries
- Bench to Bedside
- Share Your Research
- Our Mission
- Science Communication Lab
- Financial Conflict of Interest Policy


- school Campus Bookshelves
- menu_book Bookshelves
- perm_media Learning Objects
- login Login
- how_to_reg Request Instructor Account
- hub Instructor Commons
Margin Size
- Download Page (PDF)
- Download Full Book (PDF)
- Periodic Table
- Physics Constants
- Scientific Calculator
- Reference & Cite
- Tools expand_more
- Readability
selected template will load here
This action is not available.

8: Basic Techniques
- Last updated
- Save as PDF
- Page ID 7856

- Kevin Ahern, Indira Rajagopal, & Taralyn Tan
- Oregon State University
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
The environment of a cell is very complex, making it difficult to study individual reactions, enzymes, or pathways in situ. The traditional approach used by biochemists for the study of these things is to isolate molecules, enzymes, DNAs, RNAs, and other items of interest so they can be analyzed independently of the millions of other processes occurring simultaneously. Today, these approaches are used side by side with newer methods that allow us to understand events inside cells on a larger scale- for example, determining all the genes that are being expressed at a given time in specific cells. In this section we take a brief look at some commonly used methods used to study biological molecules and their interactions.
- 8.1: Cell Lysis To separate compounds from cellular environments, one must first break open (lyse) the cells. Cells are broken open, in buffered solutions, to obtain a lysate. There are several ways of accomplishing this.
- 8.2: Fractionation and Chromatography Techniques Fractionation of samples, as the name suggests, is a process of separating out the components or fractions of the lysate. Fractionation typically begins with centrifugation of the lysate. Using low-speed centrifugation, one can remove cell debris, leaving a supernatant containing the contents of the cell. By using successively higher centrifugation speeds (and resulting g forces) it is possible to separate out different cellular components, like nuclei, mitochondria, etc., from the cytoplasm.
- 8.3: Electrophoresis Electrophoresis uses an electric field applied across a gel matrix to separate large molecules such as DNA, RNA, and proteins by charge and size. Samples are loaded into the wells of a gel matrix that can separate molecules by size and an electrical field is applied across the gel. This field causes negatively charged molecules to move towards the positive electrode. The gel matrix, itself, acts as a sieve, through which the smallest molecules pass rapidly, while longer molecules are slower-movi
- 8.4: Detection, identification and quantitation of specific nucleic acids and proteins One way to detect the presence of a particular nucleic acid or protein is dependent on transferring the separated molecules from the gels onto a membrane made of nitrocellulose or nylon to create a “blot” and probing for the molecule(s) of interest using reagents that specifically bind to those molecules. The next section will discuss how this can be done for nucleic acids as well as for proteins.
- 8.5: Transcriptomics Consider a matrix containing all of the known gene sequences in a genome. To make such a matrix for analysis, one would need to make copies of every gene, either by chemical synthesis or by using PCR. The strands of the resulting DNAs would then be separated to obtain single-stranded sequences that could be attached to the chip. Each box of the grid would contain sequence from one gene. One could analyze the transcriptome - all of the mRNAs being made in selected cells at a given time.
- 8.6: Isolating Genes Methods to isolate genes were not available till the 1970s, when the discovery of restriction enzymes and the invention of molecular cloning provided, for the first time, ways to obtain large quantities of specific DNA fragments, for study. Although, for purposes of obtaining large amounts of a specific DNA fragment, molecular cloning has been largely replaced by direct amplification using the polymerase chain reaction described later, cloned DNAs are still very useful for a variety of reasons.
- 8.7: Polymerase Chain Reaction (PCR) The polymerase chain reaction (PCR) allows one to use the power of DNA replication to amplify DNA enormously in a short period of time. As you know, cells replicate their DNA before they divide, and in doing so, double the amount of the cell’s DNA. PCR essentially mimics cellular DNA replication in the test tube, repeatedly copying the target DNA over and over, to produce large quantities of the desired DNA.
- 8.8: Reverse Transcription In the central dogma, DNA codes for mRNA, which codes for protein. One known exception to the central dogma is exhibited by retroviruses. These RNA-encoded viruses have a phase in their life cycle in which their genomic RNA is converted back to DNA by a virally-encoded enzyme known as reverse transcriptase. The ability to convert RNA to DNA is a method that is desirable in the laboratory for numerous reasons.
- 8.9: FRET The fluorescence resonance energy transfer (FRET) technique is based on the observation that a molecule excited by the absorption of light can transfer energy to a nearby molecule if the emission spectrum of the first molecule overlaps with the excitation spectrum of the second. This transfer of energy can only take place if the two molecules are sufficiently close together (no more than a few nanometers apart.
- 8.10: Genome Editing (CRISPR) The development of tools that would allow scientists to make specific, targeted changes in the genome has been the Holy Grail of molecular biology. An ingenious new tool that is both simple and effective in making precise changes is poised to revolutionize the field, much as PCR did in the 1980s. Known as the CRISPR/Cas9 system, and often abbreviated simply as CRISPR, it is based on a sort of bacterial immune system that allows bacteria to recognize and inactivate viral invaders.
- 8.11: Protein Cleavage Because of their large size, intact proteins can be difficult to study using analytical techniques, such as mass spectrometry. Consequently, it is often desirable to break a large polypeptide down into smaller pieces. Proteases are enzymes that typically break peptide bonds by binding to specific amino acid sequences in a protein and catalyzing their hydrolysis.
- 8.12: Membrane Dynamics (FRAP) Understanding the dynamics of movement in the membranes of cells is the province of the Fluorescence Recovery After Photobleaching (FRAP) technique. This optical technique is used to measure the two dimensional lateral diffusion of molecules in thin films, like membranes, using fluorescently labeled probes. It also has applications in protein binding.
Thumbnail: A western blot. (CC BY-SA 3.0; Magnus Manske ).
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- My Account Login
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Open access
- Published: 03 June 2024
Correlations reveal the hierarchical organization of biological networks with latent variables
- Stefan Häusler ORCID: orcid.org/0000-0003-0772-8662 1
Communications Biology volume 7 , Article number: 678 ( 2024 ) Cite this article
Metrics details
- Computational biology and bioinformatics
- Neuroscience
Deciphering the functional organization of large biological networks is a major challenge for current mathematical methods. A common approach is to decompose networks into largely independent functional modules, but inferring these modules and their organization from network activity is difficult, given the uncertainties and incompleteness of measurements. Typically, some parts of the overall functional organization, such as intermediate processing steps, are latent. We show that the hidden structure can be determined from the statistical moments of observable network components alone, as long as the functional relevance of the network components lies in their mean values and the mean of each latent variable maps onto a scaled expectation of a binary variable. Whether the function of biological networks permits a hierarchical modularization can be falsified by a correlation-based statistical test that we derive. We apply the test to gene regulatory networks, dendrites of pyramidal neurons, and networks of spiking neurons.
Similar content being viewed by others
Network structure from a characterization of interactions in complex systems
Scarcity of scale-free topology is universal across biochemical networks
Third-order motifs are sufficient to fully and uniquely characterize spatiotemporal neural network activity
Introduction.
Modern recording techniques in neuroscience and cell biology are generating datasets of rapidly increasing dimensionality, posing a major challenge to current analytical methods for deciphering the function of the underlying biological systems 1 , 2 . A promising approach in graph theory 3 , 4 , 5 is to decompose and organize complex networks 6 into largely autonomous functional modules 7 , as found at all levels of biological organization 3 , 8 . Various heuristic algorithms have been proposed to detect functional modularity, many of them based on hierarchical clustering 9 , but a more rigorous analysis requires exact probabilistic inference 10 . Following this approach, a functional module can be conveniently formalized as a subnetwork that communicates or interacts with the rest of the network only through a particular variable, which we call interface variable. This interface variable may represent, for example, the firing rate of a population of sensory neurons that encodes all information about stimuli relevant to downstream areas. If the value of this interface variable is known, the internal and external components of a module are conditionally independent (Fig. 1 a).
a Undirected graphical model representing dependencies between network components (left). Right: Functional module consisting of the observable components s 1 and s 2 , which are independent of all other observable components given the interface variable y . b Examples of interface rate functions for the functional module shown in ( a ). c Left: Three of the interface rate functions shown in panel b are nonlinear, as illustrated for E [ s 1 ] = E [ s 2 ]. Right: Examples of nonlinear interface rate functions of three (black line) and five (gray line) arguments, illustrated for identical arguments E [ s i ]. d Three scenarios used for statistical testing.
However, probabilistic inference of functional modules in large biological networks is challenging. It is often not possible to record from the entire network, and interface variables may be inaccessible. Moreover, these variables can be abstract quantities such as sensory, associative, motor, or cognitive information encoded in the activity of cell populations 11 . And even if the interface variables are recorded, their identification for large networks is computationally intractable for combinatorial reasons. Here, we bypass these problems and investigate whether it is possible to infer functional modularizations from the distribution of observable network components alone, without information about the organization and values of interface variables.
Remarkably, an arbitrary scalar interface variable does not impose any experimentally testable conditions for continuous network states. This is easy to see, as each of the finitely many samples in a dataset can always be mapped to different values of a scalar variable, thus allowing any functional modularization. To avoid this trivial solution, we constrain the interface variables and focus on the simplest case of binary variables.
We show that the functional organization of networks with latent binary interface variables can be inferred from the statistical moments of observable network components alone, and derive a statistical test for hierarchical modularizations. Importantly, this test can also be applied to refute functional modularizations of networks consisting of continuous scalar interface variables if the following two conditions are met.
First, only the mean values of the continuous scalar interface variables and observable components are relevant for the function of the network and thus for its modularization. The actual distribution of the network states conditioned on these mean values is arbitrary as long as it is consistent with the modularization. For many stochastic biological systems, this condition is assumed to be satisfied, e.g., in molecular biology by the rate of gene transcription 12 and in neuroscience by the instantaneous firing rate of neurons 13 . Second, the mean of a variable downstream of an interface variable depends only linearly on the mean of that interface variable, where downstream refers to any sampling scheme (Fig. 1 b). The specific shape of this linear function may depend on other variables, allowing for distributed nonlinear computing (Fig. 1 c). In particular for modularizations where a subnetwork depends on the interface variables of several disjoint functional modules, this assumption is satisfied for arbitrary continuous interface variables as long as each of the interface variables contributes only linearly to the mean of each subnetwork component.
Although these assumptions limit the applicability of the method, it is relevant for a number of biological networks. Both assumptions are met by probabilistic Boolean networks, where uncertainties about binary network states are encoded by mean values. Moreover, these assumptions are reasonable when network components are well connected such that a single input has only a small, approximately linear effect on the overall nonlinear activity of a component. Here, we show that the statistical test for modularization is applicable to three biological networks at different spatial scales, and evaluate key hypotheses about their underlying functional organization (Fig. 1 d).
Functional modules
We describe observable network components by random vectors s = ( s 1 , …, s d ) in \({{\mathbb{R}}}^{d}\) and functional modules by sets S n for n = 1, 2,… that contain the indices of all observable components within a module. Associated with each functional module S n is a potentially hidden binary interface variable y n that separates its internal components from all other components such that all internal components indexed by S n are conditionally independent of all other components given y n (Fig. 1 a). A modularization consists of several functional modules and is described by a set \({{{{{{{\mathcal{M}}}}}}}}=\{{S}_{1},{S}_{2},\ldots \}\) .
The key question is how to infer functional modularizations from samples of s without information about the underlying interface variables y , which prevents direct testing of the corresponding conditional independencies. We assume that the observable states are bounded, so that all their moments are finite and uniquely determine the probability distribution of s . We first show that functional modules are reflected in pairwise correlations between network components.
Pairwise correlations indicate direct dependencies
Consider a large neural network where the spike counts of five neurons are observed within some time interval and described by the random components s 1 , …, s 5 . Furthermore, assume that the first two neurons are part of the same neuronal population such that their expected spike counts, i.e., E [ s 1 ] and E [ s 2 ], are proportional to an unknown population firing rate r , and only this rate drives the rest of the network. As the two components s 1 and s 2 are independent of all other observable components given r , they form a functional module. The dependencies between the five observable components are shown in the graphical model in Fig. 1 a.
We analyze pairwise correlations of an equivalent network where r is replaced by a binary interface variable y whose expectation, E [ y ], is proportional to r . By assumption, the expected components s 1 and s 2 are linear in the population rate r , and, thus, all pairwise correlations between the observable components remain unchanged. As the graphical model remains the same, the dependencies between components are also equivalent. Therefore, we can analyze the equivalent network to infer the dependencies in the original network. There are no restrictions on the dependence of the population rate r on the observable components outside of the functional module.
The effect of the functional module with binary interface variables on the properties of pairwise correlations can be visualized by a 2D vector representation (Fig. 2 ). According to the law of total expectation and the conditional independence statement of the module, the correlation of a component within the module, e.g., s 1 , and a component outside of the module, e.g., s 3 , can be written as a scalar product E [ s 1 s 3 ] = s 1 ⋅ s 3 of vectors
for i = 1, 2, …,5. Here, E [ s i ∣ y = 0] and P( y = 0) denote the conditional expectation of s i given y = 0 and the probability of y = 0, respectively.
a Components s 1 and s 3 of the undirected graphical model are conditionally independent given the interface variable y . b Their pairwise correlation, E [ s 1 s 3 ], corresponds to the scalar product of the vectors s 1 and s 3 . c All vector pairs associated with zero-mean network components are either parallel or antiparallel.
In general, the vector components are unknown because the interface variable y is unknown. Yet, if all components are normalized to zero mean E [ s i ] = 0, all pairs of vectors are either parallel or antiparallel (Fig. 2 right). Then, the ratio of s 3 ⋅ s i and s 4 ⋅ s i is independent of s i for i = 1, 2 and has an absolute value equal to the ratio ∣ s 3 ∣ / ∣ s 4 ∣ . Therefore, observable components with non-zero pairwise correlations, a functional module S = {1, 2} and a binary interface variable y exist only if
This condition can be tested to reject indirect dependencies between the components s 1 and s 2 and the components s 3 and s 4 via a rate function r . Based on estimators of pairwise correlations and estimators of their covariance, we derive an asymptotic test for direct dependencies that can be applied even when pairwise correlations are zero (see Methods).
Inferring direct interactions in gene regulatory networks
High-throughput technologies, such as RNA sequencing and microarrays, capture transcriptomes under a variety of experimental conditions to infer transcriptional gene regulation. Probabilistic Boolean networks have been successfully applied to infer the underlying gene regulatory networks 14 , 15 , where expression values of transcription factors (TFs) and target genes (TGs) form nodes (or components), and direct interactions between TFs and TGs form links. We therefore apply the proposed method to this inference task. In order to test in a competitive environment, we retroactively participate in the DREAM5 Challenge 16 , a comprehensive evaluation of 35 network inference methods on various datasets with established gold standards. Here, the interface variables of functional modules are not hidden, but correspond to recorded expression values of TFs, allowing a comparison with inference methods that rely on this information.
The reconstructed networks are compared to experimentally established gold standards for two datasets, Escherichia coli and an in-silico benchmark. The submission format of the DREAM5 Challenge is a ranked list of predicted regulatory interactions. Because TF–TF interactions are not organized hierarchically, we restrict the reconstruction to TF–TG interactions, which represent more than 94% of the gold standard. The performance is evaluated using the area under the precision-recall curve (AUPR), the receiver operating characteristic curve and an overall score that summarizes the performance across networks. For a fair comparison with previous results, we evaluate all performance measures against the full gold standard.
To apply the test, we start with a ranked list of direct TF–TG interactions, ordered by the absolute value of their Pearson correlation coefficient, and investigate whether some of these direct interactions can be explained by indirect dependencies through other TFs. More specifically, we investigate all subnetworks consisting of two TFs and two TGs and test for a functional module S containing both TFs. Based on the test, the rank of each TF–TG interaction is re-evaluated in such a way that evidence against a functional module, i.e., against an indirect interaction, shifts the rank towards more likely interactions, and reduced evidence shifts the rank in the opposite direction (see Methods). The test is only applied if a subnetwork is sufficiently connected such that at least three of the four putative TF–TG interactions are in the set of the most likely interactions. The size of this set is the only free parameter of the method.
For each subnetwork (Fig. 3 a), we denote the expression levels of the two TFs as s 1 and s 2 , the expression levels of the two TGs as s 3 and s 4 , and use their correlations E [ s k s l ] to test for the functional module S = {1, 2}, where k and l index the observable components inside and outside of the functional module, respectively. Only if the condition in Eq. ( 2 ) holds, TF–TG interactions in this subnetwork can be mediated by a single transcription factor with an expression level represented by the probability of a binary variable. We derive a statistical test for this condition, which can also be applied to zero-correlated expression levels (see Methods). Under certain assumptions, the method is asymptotically correct in the sense that the most likely inferred interactions are true TF–TG interactions. There are no constraints on the interface rate functions as long as the interdependence of the co-regulated TGs s 3 and s 4 is linear.
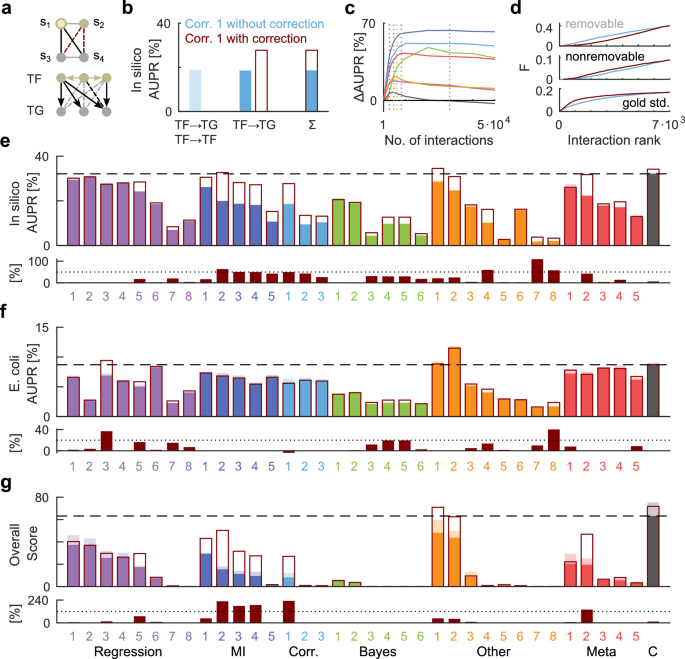
a Undirected graphical model of a subnetwork consisting of four genes (top). Black edges represent putative direct interactions, which are elements of the set of most likely interactions. Red edges represent putative indirect interactions to be tested. Bottom: Example of a directed graphical model representing causal interactions between genes (arrows). Dashed lines indicate removable (gray) and nonremovable (black) indirect interactions. b Performance of the uncorrected method based on Pearson correlation coefficients (Corr. 1) for all interactions (light blue) or only for TF-TG interactions (blue) of the in-silico benchmark. The correction based on the test improves the performance by 50% (dark red rectangle). c Performance as a function of the size of the set of most likely interactions for the network inference methods Regression 5, MI 2, Corr. 1, Bayes 4, Other 1, Meta 2 and the community network (color code as in e ). Dotted lines indicate the selected sizes of the sets, determined on a disjoint holdout set. d Cumulative distributions of removable, nonremovable and gold standard interactions as a function of the rank in the list of most likely interactions for the corrected (blue line) and uncorrected (dark red line) inference method Corr. 1. e Performance of all 36 inference methods of the DREAM5 challenge before (colored bars) and after their correction based on the test (dark red rectangles) for the in-silico benchmark (top). The dashed line indicates the performance of the community network (C). Bottom: Relative performance improvement in %. The dotted line indicates an improvement of 50%. f Same as ( e ), but for the Escherichia coli dataset. g Overall score summarizing the performance across networks and performance measures.
Figure 3 b shows the performance (AUPR) of the uncorrected reconstruction of the in-silico network based on all TF-TF and TF-TG interactions ordered by the absolute value of their Pearson correlation coefficients. Omitting all TF-TF interactions from the reconstruction results in about the same performance, while correcting according to the test improves the AUPR by 50%. The test doesn’t require fine-tuning of its only free parameter, the number of most likely interactions that determine whether a subnetwork is sufficiently connected, which is optimized on a holdout set (Fig. 3 c).
The effect of the correction can be analyzed in terms of the rates of true negatives and false positives (type I errors). If a TG is regulated by several interdependent TFs, the test might fail to refute a false direct TF–TG interaction because the corresponding functional module has not one but several interface variables. If this is the case according to the gold standard, we call indirect TF-TG interactions nonremovable (Fig. 3 a), which account for less than 30% of all TF-TG dependencies. Figure 3 d shows that the improvement in performance is due to a majority of removable indirect interactions, whose rank distribution is correctly shifted towards less likely interactions. In contrast, the rank distribution of nonremovable indirect interactions is shifted towards more likely interactions, introducing more likely false positives (type I errors). Overall, the rank distribution of the gold standard is shifted toward more likely interactions.
As the correction only requires a ranked list of predicted regulatory interactions, we apply it to each of the inference methods of the DREAM5 Challenge for the in-silico (Fig. 3 e) and E. coli microarray data (Fig. 3 f). In general, the correction improves most of the 36 inference methods, suggesting that it takes advantage of otherwise unexploited information. In particular, the correction improves the overall score of the community network, which is about the same as that of the single corrected inference method Genie3 17 , denoted as Other 1. However, our aim is not to develop a single best inference method for gene regulatory networks, which will probably be a combination of different inference methods. Rather, we show that this inference method is generally suitable for reconstructing gene regulatory networks and propose it for datasets with missing or unknown regulatory TFs.
Moment ratios indicate functional modules
To allow the construction of an efficient statistical test, we consider only hierarchically organized modularizations that are either flat or nested. We call a modularization \({{{{{{{\mathcal{M}}}}}}}}\) flat if all functional modules contained in \({{{{{{{\mathcal{M}}}}}}}}\) are disjoint (Fig. 1 d). And we call a modularization \({{{{{{{\mathcal{M}}}}}}}}\) nested if all functional modules contained in \({{{{{{{\mathcal{M}}}}}}}}\) are either disjoint or a subset of another functional module in \({{{{{{{\mathcal{M}}}}}}}}\) (Fig. 1 d). To clearly distinguish between flat and non-flat nested modularizations, we single out one component, denoted as s ref , that is not part of any functional module. In the following, s ref refers to s d .
We show that moments of s uniquely determine whether the observable states form a particular modularization or not (see Methods). Let P n for n = 1, 2, … denote an infinite sequence of all monomials in the observable components within a given functional module, e.g., for the module in Fig. 1 a, the sequence P 1 = 1, P 2 = s 1 , P 3 = s 1 s 2 , …. Moreover, let Q m for m = 1, 2, … denote the corresponding sequence of all monomials in the observable components outside of the functional module, e.g., for the module in Fig. 1 a, the sequence Q 1 = 1, Q 2 = s 3 , Q 3 = s 3 s 4 , Q 4 = s 3 s 5 , ….
As in the case of pairwise correlations, the moment E [ P n Q m ] is equal to the scalar product p n ⋅ q m of the two vectors p n and q m defined analogous to Eq. ( 1 ). If all observable states have zero mean, all pairs of vectors s k are either parallel or antiparallel (Fig. 4 b), where k indexes all observable components inside of the module. Hence, ratios of scalar products of different q m and the same s k have equal values for all k . For a single functional module S , we use the monomials s k for P k , where k indexes all observable components inside of the module. In addition, we use s ref for Q ref and the monomials s l s ref for Q l , where l indexes all observable components outside of the module. To enable efficient testing, we introduce a matrix B with elements
for 1 ≤ k < d and 1 ≤ l < d , where we divide each element by E [ s l s ref ] to obtain a symmetric matrix. Observable components s with (finite) moments as above, a functional module S and a binary interface variable exist if and only if for each l indexing an observable component outside of the functional module S , the moment ratios B k l have the same value for all k in S (Fig. 4 c, d).
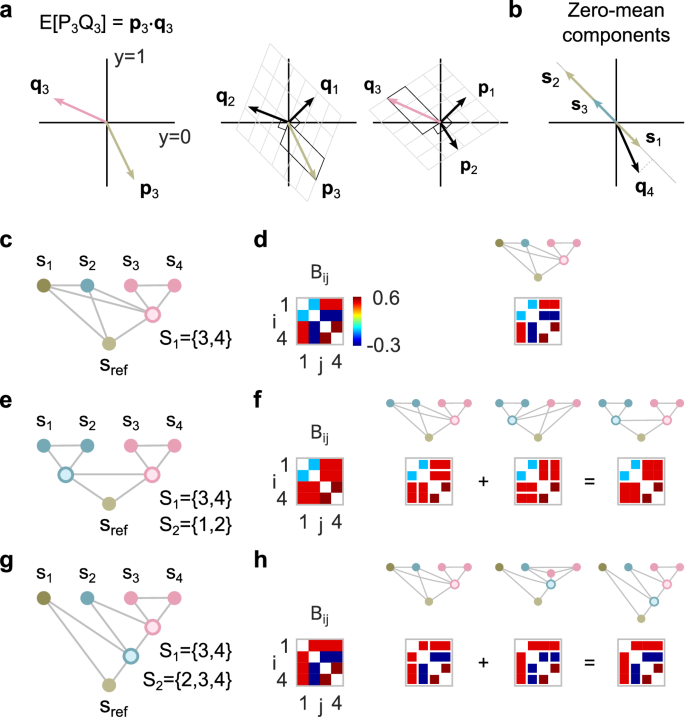
a Left: The moment E [ P 3 Q 3 ] corresponds to the scalar product of the associated vectors p 3 and q 3 . Middle: Vector p 3 expressed in the skewed coordinate system with axes perpendicular to q 1 and q 2 . Right: Vector q 3 expressed in the skewed coordinate system with axes perpendicular to p 1 and p 2 . b All vector pairs associated with zero-mean network components are either parallel or antiparallel. The vector q 4 is associated with a mixed moment of order greater than one and can point in any direction. c Single functional module. d Example of a moment ratio matrix for the modularization shown in ( c ) (left). Right: Matrix elements with the same value due to the single functional module are shown as connected. e Flat modularization. f Example of a moment ratio matrix for the modularization shown in panel e (left). Right: Each of the two modules implies that different elements of the moment ratio matrix have the same value. The conditions for the moment ratio matrix resulting from the overall modularization are obtained by combining the conditions for the individual modules. g Nested modularization. h The conditions for the moment ratio matrix resulting from the nested modularization shown in ( g ) are obtained by combining the conditions for the individual modules.
Moreover, for flat and nested modularizations consisting of several functional modules, the combined conditions can be derived directly from the conditions of the individual functional modules (Fig. 4 e–h). Observable components s with moments analogous to those above, a nested (flat) modularization \({{{{{{{\mathcal{M}}}}}}}}\) and a binary interface variable exist only if (if and only if) the corresponding combined conditions are met.
Based on estimators \(\hat{{{{{{{{\bf{B}}}}}}}}}\) of the moment ratio matrix B and estimators of the covariance matrix of the elements of \(\hat{{{{{{{{\bf{B}}}}}}}}}\) , we derive an asymptotic test for the conditions in Eq. ( 3 ) that can be applied even if some moment ratios do not exist (see Methods). Furthermore, we consolidate numerically that this test is not only asymptotically correct but holds for sufficiently many samples such that the moment estimates are approximately joint normal. For the investigated datasets, a few hundred samples turn out to be sufficient to fulfill these requirements. A more general test using arbitrary moments is given in the Methods.
As a typical use case, the test is applied to multiple candidate modularizations and needs to be corrected for multiple testing (see Methods). Note that the method does not require parameter optimization, only the calculation of the test statistic for each candidate modularization. However, the number of potential single functional modules already grows exponentially with the number of observable components, which can lead to combinatorial problems. We expect that current conventional computing resources can handle up to 24 observable components, resulting in approximately 10 7 candidate modules. To overcome the combinatorial explosion for flat modularizations, we propose to first identify all single functional modules and then test for all of their combinations. For nested modularizations, we suggest starting with an educated guess of a detailed nested modularization consisting of n functional modules and testing for all 2 n potential nested modularizations obtained by combining these functional modules.
Inferring modularizations in neural networks: proof of principle
As a case study illustrating the power of the approach, we infer the hierarchical organization of a neural network from simultaneously recorded spiking activity. It is a classical hypothesis in neuroscience that neurons communicate information only through their instantaneous firing rates, typically characterized by the spike count within a certain time window or population 13 , 18 , 19 . Accordingly, we simulate the spiking activity of five populations of 10 neurons each with dependencies as shown in the undirected graphical model in Fig. 5 a. The corresponding interface rate functions are shown in Fig. 1 b (see Methods) and can be implemented by a probabilistic Boolean network.
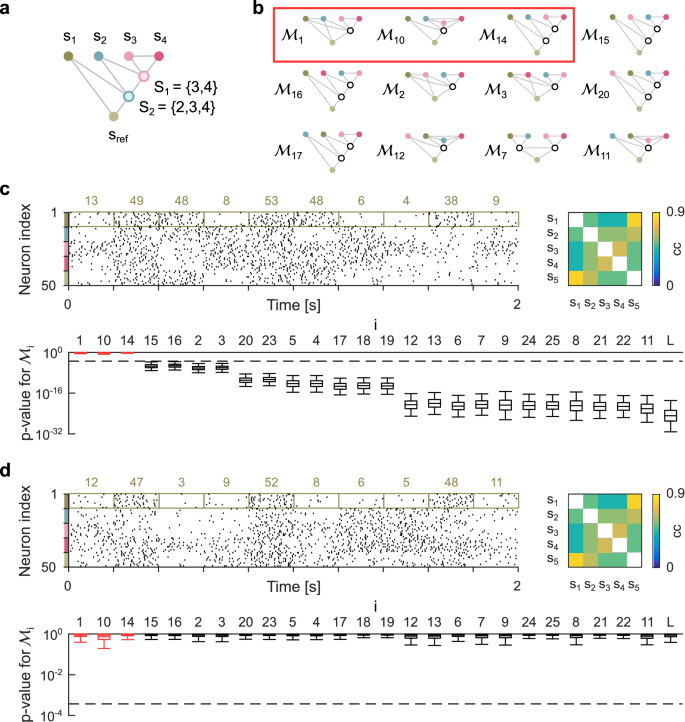
a Nested modularization consisting of five populations of neurons. b 12 out of 26 potential modularizations. Modularizations consistent with the modularization shown in panel a are marked with a red rectangle. c Spiking activity of all neurons and correlation matrix of observable components (top). Observable states are spike counts within populations and time intervals. Colored numbers represent samples of the observable component s 1 . Bottom: Boxplot showing the distribution of p values for testing each of the 26 modularizations. Boxes represent the first through third quartiles, and whiskers indicate the 2.5 and 97.5 percentiles. d Same as ( c ), but for a linear modularization with the same correlation matrix. Dashed lines represent overall significance levels of 0.01.
The task is to infer all modularizations that are consistent with the data (Fig. 5 b), given recorded spiking activity over 15 min. For simplicity, we follow a classical population coding approach and define the observable network components as the total number of spikes of all neurons recorded within a population during consecutive 200 ms time intervals (Fig. 5 c). Likewise, the interface variables are spike counts for latent populations. The corresponding moment ratio matrix \(\hat{{{{{{{{\bf{B}}}}}}}}}\) (Eq. ( 3 )) is shown in Fig. 4 h, which is consistent with the three modularizations \({{{{{{{{\mathcal{M}}}}}}}}}_{1}\) , \({{{{{{{{\mathcal{M}}}}}}}}}_{10}\) and \({{{{{{{{\mathcal{M}}}}}}}}}_{14}\) . In contrast, all other modularizations, including a purely linear model \({{{{{{{{\mathcal{M}}}}}}}}}_{{{{{{{{\rm{L}}}}}}}}}\) , are rejected by the test at an overall significance level of 0.01 (Fig. 5 c).
The test relies on a small number of correlations between observable components to efficiently investigate potential nonlinear dependencies with s ref . For this approach, pairwise correlations alone are not sufficient to refute any nonlinear modularization. There always exists a random vector with the same pairwise correlations as the observable states that allows any modularization (Fig. 5 d).
Inferring functional modules in dendrites of pyramidal neurons
Pyramidal neurons exhibit complex morphologies and spatially modulated distributions of ion channels 20 , 21 that generate regenerative events, such as Na + or NMDA spikes, localized to specific branches or subtrees 22 . Previous work has suggested that these branches act as independent functional modules 23 , 24 , 25 , 26 , 27 , whose responses to local synaptic inputs are linearly summed at the soma. Simulation studies have confirmed that the resulting flat modularization is indeed an accurate description for computations on firing rates 28 , 29 , where the input and the response are encoded by the rate of synaptic inputs and somatic action potentials, respectively.
However, these studies required complete information about synaptic inputs and were limited to paired branch stimulation. It is still unclear whether the functional modularization breaks down in scenarios in which a large number of branches are stimulated at the same time. For pulse stimulation applied to pairs of branches in the dendritic tuft, it has been shown that cross-talk between the branches prevents precise functional modules 29 . Here, we investigate the formation of functional modules in proximal and oblique apical dendrites when the pyramidal neuron is excited by strong input to all its terminal branches. In particular, we apply the proposed method not only to infer single functional modules, but the total overall modularization of the proximal apical dendrites.
More specifically, we simulate a detailed multi-compartment model of a CA1 pyramidal neuron 24 and stimulate excitatory synapses at terminal branches at a constant rate. The network components correspond to subthreshold membrane potentials at 26 locations in the proximal and oblique apical dendrites, recorded at 50 ms intervals over 20 or 60 min (Fig. 6 a, b) to investigate the statistical power of the test for different sample sizes. The interface variables are non-binary, latent and correspond to membrane potentials within functionally independent dendritic compartments downstream of the recording sites that are linearly summed at the soma. As the somatic module is linear and we are testing for flat modularizations, there are no constraints on the interface rate functions.
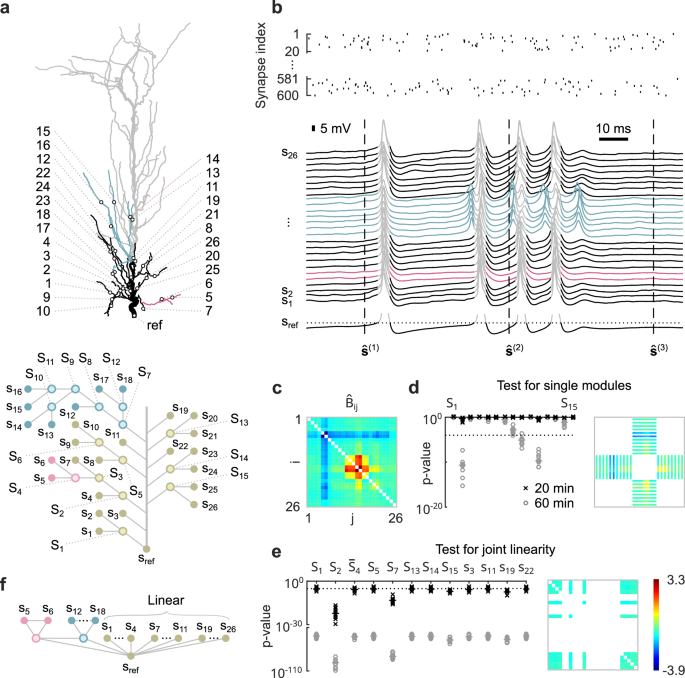
a Model of the apical dendrites of a CA1 pyramidal neuron (top). The investigated subtree is shown in color and black. Black circles indicate recording locations. Bottom: Undirected graphical model of the largest possible modularization consistent with the morphology of the investigated subtree. b Synaptic input to terminal branches (top). Bottom: Observable states are membrane potentials recorded at the corresponding locations shown in panel a , sampled every 50 ms. Spiking activity at the soma (gray traces) is excluded from the analysis. c Estimated moment ratio matrix. d p values for testing each of the 15 functional modules shown in panel a on a log scale (left). Right: Elements of the moment ratio matrix expected to have the same value according to the single module S 7 , represented as contiguous blocks. e p values for testing whether individual functional modules or dendritic branches originating from the trunk participate in a large linear somatic module (on log scales). All tests are repeated ten times on independent datasets obtained from 20 and 60 min recordings. Dotted lines represent overall significance levels of 0.01. Horizontal bars indicate medians. Right: Elements of the moment ratio matrix expected to have the same value according to a linear module consisting of S 1 , S 14 , S 15 , s 3 , s 11 , s 22 and \({\bar{S}}_{4}=\{{s}_{4},{s}_{7}\}\) . f Approximate flat modularization of the proximal apical and oblique dendrites.
The estimated moment ratio matrix \(\hat{{{{{{{{\bf{B}}}}}}}}}\) (Eq. ( 3 )), obtained from 60 min recordings, shows three nonlinear functional modules, i.e., S 4 , S 7 and S 11 (Fig. 6 c). The test rejects only three individual functional modules (Fig. 6 d) at an overall significance level of 0.01. The first, S 2 , is rejected because its two most proximal observable components, s 4 and s 7 , are part of a large linear somatic module, while its two distal observable components, s 5 and s 6 , form the nonlinear module S 4 . The other two rejected modules, S 9 and S 11 , are part of the large nonlinear module S 7 .
In addition, we investigate which of the functional modules are purely linear, as these can be integrated into a large somatic module. For a purely linear module S , the square submatrix of B indexed by S has identical off-diagonal elements. Then, any functional modularization within S is possible, reflecting the commutative property of addition, as multiplication can be excluded for somatic integration. For each functional module, we test whether it is part of a larger linear module consisting of 11 observable components. The constraint ensures the same degrees of freedom for all tests. For 20 min recordings, only modules S 2 and S 7 are not part of the large linear somatic module, in contrast to the proximal part of module S 2 labeled \({\bar{S}}_{4}\) (Fig. 6 e).
The resulting flat functional modularization of the proximal apical dendrite consists of a linear somatic module and the two nonlinear modules S 4 and S 7 (Fig. 6 f). However, the large linear somatic module is only approximate and rejected for large enough sample sizes.
Unraveling the functional organization of large biological networks is challenged by incomplete information and combinatorial problems. We present an asymptotic test for hierarchical functional organization of network components based on observable correlations alone, which requires no information about latent network components.
The method presented here differs significantly from previous approaches to inferring functional structure in large networks because it is not based on optimization. In neuroscience, network connectivity and hidden variables are traditionally inferred from neuronal activity based on principles such as maximum a posteriori estimation, Bayesian inference, or information theory 2 , 30 , 31 . However, such optimization paradigms require regularization in the form of prior information or otherwise prefer more complex structures due to overfitting. Moreover, there is no indication of whether unexplained network activity is due to noise or an inadequate (hierarchical) structure of the latent variables.
In contrast, constraint-based approaches, which first test for conditional independence in the data and then find appropriate network structures, provide intuitive results such as p values, but cannot be applied to incomplete data 10 . The method presented here combines the advantages of constraint-based approaches and probabilistic models by providing a statistical test for partially latent network structures. In particular, the method tests not only necessary conditions, potentially refuting any false nested modularization, but also sufficient conditions for flat modularizations of observable network components with the same correlations as used for the test.
In molecular biology, probabilistic Boolean network models have been successfully applied to infer gene regulatory networks 32 , 33 , 34 , but these methods aim at a complete network reconstruction including all logical relationships between genes (logic gates). In contrast, the statistical test presented here does not estimate model parameters. For flat modularizations, only a minimal set of sufficient and necessary conditions is considered, in the sense that omitting a single correlation estimate renders the test inconclusive. We therefore believe it is particularly well suited for small sample sizes, or equivalently, for inferring large functional organizations from datasets of a given size. In particular, the method is useful as a first step in the analysis process to gain an initial understanding of the functional organization of a network.
We test the method against a benchmark for reconstructing gene regulatory networks and show that it outperforms previously superior community-based methods when used as a correction to the single inference method Genie3 17 . In addition, we apply the method to a detailed model of a pyramidal neuron and show that its proximal apical dendrites form multiple functional modules in response to distributed and strong driving synaptic input.
Previous work 29 analyzed the response of the same pyramidal neuron model to synaptic stimulation of pairs of dendritic branches, allowing a comparison of the functional modularization for the two input types. In this study, only three of the proximal branches are analyzed, but not the branch with the observable components s 12 to s 18 (see Fig. 5 a). These branches show a similar, but not identical, flat modularization with a single nonlinear functional module containing the components s 5 to s 7 . The larger functional module for paired-branch stimulation may be due to the strong local synaptic input required to activate the neuron, whereas weaker distributed stimulation results in a smaller functional module.
The functional modularization of the pyramidal neuron model can be identified without any information about the synaptic inputs or the electrophysiological properties of the neuron. This makes the method suitable for conventional two-photon microscopy with voltage indicators, which measures subthreshold membrane potentials at unspecific locations defined by the intersection of the neuron and the imaging plane.
The test is based on correlations between network components, which are usually reflected in correlated moment ratios. The stronger these correlations are, the lower is the statistical power of the test. For moderately correlated network components, as in the neural network inference example, a recording duration of about 15 min is sufficient to achieve adequate statistical power. However, the highly correlated network components of the pyramidal neuron model require recording durations of 60 min. In particular, for synaptic integration in the tuft dendrites, the statistical power is too low to refute any functional module for the chosen recording durations.
Probabilistic Boolean networks with univariate interface variables can not capture the complex dependencies between components of many biological neural networks. However, they may be suitable for modeling sensory systems if neuronal populations implement optimal coding schemes for information processing on short time scales 35 , 36 . Then, the optimal neuronal response functions are binary and intermediate rates reflect states of uncertainty. In particular, binary response functions have been shown to be reasonable approximations for various sensory domains 37 .
Univariate binary interface variables allow information to flow in only one direction. Interface variables with a larger number of values can capture more complex network dependencies that exhibit bidirectional information flow, multidimensional interface variables or noise correlations 38 . In particular, the lower performance in reconstructing the E. coli gene regulatory network compared to the in-silico benchmark may be due to noise correlations of expression levels caused by sample preparation, array fabrication, and array processing 39 , 40 . An extension to interface variables with four or more values seems feasible and promising, since the necessary conditions for corresponding functional modularizations are already derived in the Supp. Methods .
In general, exact probabilistic inference is intractable in large biological networks. We provide a hypothesis-driven statistical method that efficiently tests for selected functional modularizations and does not require complete information about the entire network. With recent advances in high-throughput single-cell technology 41 , multi-electrode array technology 42 , 43 , two-photon microscopy 44 , 45 and genetically encoded voltage indicators 46 , 47 , our mathematical framework can be applied to a wide range of datasets to facilitate the analysis of complex biological systems.
Moments indicate functional modules
We show that moments of s uniquely determine whether the observable states form a particular modularization or not (see Supp. Methods) . We consider raw mixed moments defined as an expectation of the corresponding monomials in s , e.g., E [ s 1 s 2 ].
The method is based on the following property of single functional modules. Let P n for n = 1, 2, … and Q m for m = 1, 2, … denote infinite sequences of all monomials in the observable components inside and outside of a given functional module, respectively. If the 2 × 2 matrix M i j = E [ P i Q j ] for i , j = 1, 2 is invertible, then a necessary and sufficient condition for the existence of a binary interface variable y is
for all n , m (see Lemma 1 , Lemma 2 and Theorem 6 in the Supp. Methods) .
Intuitively, the expectation E [ P n Q m ] is equal to the scalar product p n ⋅ q m of the two vectors p n and q m defined analogous to Eq. ( 1 ). The vector components are unknown because the interface variable y is unknown. However, p n can be expressed in the skewed coordinate system with axes perpendicular to q 1 and q 2 and, likewise, q m can be expressed in the skewed coordinate system with axes perpendicular to p 1 and p 2 (Fig. 4 a). Evaluation of these coordinates by means of scalar products, transformation of the resulting vectors into the original orthogonal coordinate system by means of the matrix M and calculation of the scalar product results in Eq. ( 4 ).
A necessary and sufficient condition for the existence of a nested modularization is that this condition holds for each functional module in the modularization. Although a precise identification of modularizations requires an infinite number of moments, these conditions can be tested for finitely many moments to potentially falsify modularizations. Moreover, if the conditions for flat modularizations with moments as used in Eq. ( 3 ) hold, then there is always a flat modularization with these moments.
A statistical test for modularizations
Let N (t) denote the total number of i.i.d. samples of s and let \({\hat{{{{{{{{\bf{s}}}}}}}}}}^{(n)}\) for n ∈ {1, …, N (t) } denote the n -th sample. We assume N (t) is even and the samples are normalized to zero mean. In this case, the test can be further simplified to the condition that certain ratios of moments of s have identical values if they are finite.
Let \({d}^{({{{{{{{{\mathcal{X}}}}}}}}}_{0})}\) denote the total number of moment ratios used for the test and let the components of \({{{{{{{\bf{b}}}}}}}}=({b}_{1},\ldots ,{b}_{{d}^{({{{{{{{{\mathcal{X}}}}}}}}}_{0})}})\) denote these ratios (see Eqs. ( 2 ) and ( 3 )). For a given modularization \({{{{{{{\mathcal{M}}}}}}}}\) , we introduce \({d}^{({{{{{{{{\mathcal{X}}}}}}}}})}\) index sets \({X}_{c}^{({{{{{{{\mathcal{M}}}}}}}})}\) for \(c\in \{1,\ldots ,{d}^{({{{{{{{\mathcal{X}}}}}}}})}\}\) such that all components of b indexed by a set have the same value. The specific choice of moments, their ratios used for the test and the definition of the index sets \({X}_{c}^{({{{{{{{\mathcal{M}}}}}}}})}\) depend on the type of modularization, i.e., single functional module, flat or nested modularization, and is described in the sections below (see Supp. Methods for details).
For each sample moment ratio \({\hat{b}}_{v}\) for \(v\in \{1,\ldots ,{d}^{({{{{{{{{\mathcal{X}}}}}}}}}_{0})}\}\) we define a vector \({\hat{{{{{{{{\bf{b}}}}}}}}}}^{(v)}\in {{\mathbb{R}}}^{2}\) containing the numerator and the denominator of \({\hat{b}}_{v}\) . We estimate the moment ratio b v by
where a cutoff \({\theta }_{\hat{\mu }}=5\) ensures finite expectations of \({\hat{b}}_{v}\) for joint normal \({\hat{{{{{{{{\bf{b}}}}}}}}}}^{\left(v\right)}\) .
The simplest possible moments for testing nested modularizations are \({b}_{1}^{\left(v\right)}=E[{s}_{k} \, {s}_{l}\, {s}_{d}]\) and \({b}_{2}^{\left(v\right)}=E[{s}_{k}\, {s}_{d}]E[{s}_{l} \, {s}_{d}]\) for k ≠ l , which are estimated by the sample moments
for i ∈ {1, 2}, k ∈ {1, …, l − 1}, l ∈ {2, …, d − 1} and vectorization
The corresponding sample covariance matrices are
for i , j ∈ {1, 2} and \(v,\omega \in \{1,\ldots ,{d}^{({{{{{{{{\mathcal{X}}}}}}}}}_{0})}\}\) , where ω ( k , l ) = v ( k , l ). The covariance matrix of the sample moment ratios \(\hat{{{{{{{{\bf{b}}}}}}}}}\) can be approximated by
for \(v,\omega \in \{1,\ldots ,{d}^{({{{{{{{{\mathcal{X}}}}}}}}}_{0})}\}\) .
The simplest possible moments for testing single functional modules (in gene regulatory networks) are \({b}_{1}^{\left(v\right)}=E[{s}_{k}\,{s}_{l}]\) and \({b}_{2}^{\left(v\right)}=E[{s}_{k}\, {s}_{d}]\) for k ≠ l estimated by
We show that the statistic
can be used to test for modularization \({{{{{{{\mathcal{M}}}}}}}}\) even if some moment ratios don’t exist because of zero denominators. The test is asymptotically correct for λ v = 1. However, we consolidate numerically that the test also applies to finitely many samples if the moment estimates are approximately joint normal, \({N}^{({{{{{{{\rm{t}}}}}}}})}\hat{{{{{{{{\bf{\Sigma }}}}}}}}}\, \approx \, {N}^{({{{{{{{\rm{t}}}}}}}})}E[\hat{{{{{{{{\bf{\Sigma }}}}}}}}}]\) and
where ( ξ 0 , …, ξ 5 ) = (1.367, 2.047, 4.735, −1.923, −1.231, 2.790).
For the most general test, we introduce a scaling factor \({\lambda }^{(\max )}\) , which corrects for correlated components of \(\hat{{{{{{{{\bf{b}}}}}}}}}\) , and constrain the nominal significance level α (*) to be larger than a minimal nominal significance level \({\alpha }^{(\min )}\) , which corrects for potential zero correlations between s ref and other observable components.
Let \({{{{{{{{\mathcal{M}}}}}}}}}_{i}\) denote the i -th of \({d}^{({{{{{{{\mathcal{H}}}}}}}})}\) modularizations tested on the same samples \(\hat{{{{{{{{\bf{s}}}}}}}}}\) . If the observable states form the modularization \({{{{{{{{\mathcal{M}}}}}}}}}_{i}\) , the probability of sampling \({{{{{{{{\mathcal{T}}}}}}}}}_{{{{{{{{{\mathcal{M}}}}}}}}}_{i}}(\hat{{{{{{{{\bf{b}}}}}}}}},{\lambda }^{(\max )}\hat{{{{{{{{\bf{\Sigma }}}}}}}}})\) at least as extreme as observed is less than α (*) if
where \({p}_{i}^{(\Gamma )}\,{{\mbox{-value}}}\,\) denotes the probability of sampling \({{{{{{{{\mathcal{T}}}}}}}}}_{{{{{{{{{\mathcal{M}}}}}}}}}_{i}}(\hat{{{{{{{{\bf{b}}}}}}}}},{\lambda }^{(\max )}\hat{{{{{{{{\bf{\Sigma }}}}}}}}})\) at least as extreme as observed when distributed according to the one-sided right-tail gamma distribution \(\Gamma ({\zeta }_{{{{{{{{{\mathcal{M}}}}}}}}}_{i}},1)\) for shape parameter \({\zeta }_{{{{{{{{\mathcal{M}}}}}}}}_{i}}=\frac{1}{2}\left({d}^{({{{{{{{{\mathcal{X}}}}}}}}}_{0})}-{d}^{({{{{{{{\mathcal{X}}}}}}}})}\right)\) and scale parameter 1. For \({\zeta }_{{{{{{{{\mathcal{M}}}}}}}}_{i}}=0\) , \({{{{{{{{\mathcal{T}}}}}}}}}_{{{{{{{{\mathcal{M}}}}}}}}_{i}}=0\) . \({\zeta }_{{{{{{{{\mathcal{M}}}}}}}}_{i}}\) corresponds to the degrees of freedom.
For potentially correlated components of the sample moment ratios \(\hat{{{{{{{{\bf{b}}}}}}}}}\) , we set \({\lambda }^{(\max )}\) such that the statistical test remains conservative. The smallest possible \({\lambda }^{(\max )}\) with this property is the largest eigenvalue of a submatrix of \(\hat{{{{{{{{\bf{\Sigma }}}}}}}}}\) normalized to unit diagonal, i.e.,
where the matrix \(\grave{{{{{\mathbf{\Sigma}}}}}}\) has the same size as \(\hat{{{{{{{{\bf{\Sigma }}}}}}}}}\) , is diagonal and \({\grave{\Sigma }}_{vv}={\hat{\Sigma }}_{vv}^{-1/2}\) if v is element of an index set of size larger than one and \({\grave{\Sigma }}_{vv}=0\) , otherwise. A more power full test is derived in the Supp. Methods (see Section 2.4) . The minimal nominal significance level
where erf denotes the error function.
If all moment rations are finite, the test is asymptotically consistent for flat modularizations, i.e., its power for any incorrect modularization converges asymptotically to one. If in addition all moment ratios are uncorrelated, the test statistic is asymptotically minimal sufficient, i.e., it most efficiently captures all information about a modularization contained in the sample moment ratios. More details and the derivative of the statistical test can be found in the Supp. Methods . The performance of the test for detecting deviations from ideal functional modules, such as non-binary interface variables and additional dependencies, and for the case where moment ratios do not exist is shown in the Supp. Methods (see chapter 2.6) .
Finally, if both conditions in the introduction are met, the inference for modularizations with binary and continuous interface variables is equivalent. Given that s ref is considered as a single module, a modularization with continuous interface variables exists if and only if a corresponding modularization with binary interface variables and the same correlations as used for the test exist.
Inference in gene regulatory networks
We participated in the transcriptional network inference challenge from DREAM5 that compares 35 methods for inference of gene regulatory networks: 29 submitted by participants and additional 6 off-the-shelf methods classified into six categories: Regression, Mutual information, Correlation, Bayesian networks, Meta (combinations of several different approaches), and Other (methods not belonging to any of the previous categories). The design of the challenge, detailed methods and results are reported elsewhere 16 .
We evaluate the network reconstruction from gene expression microarray datasets for E. coli and an in-silico benchmark using the area under the precision-recall (PR) curve, the area under the receiver operating characteristic (ROC) curve, and an overall score defined as the mean of the (log-transformed) network-specific p values (obtained by simulating a null distribution for 25000 random networks),
We omit the third dataset of the challenge for S. cerevisiae due to technical reasons, i.e., the size of the network is too large for the algorithm and computer hardware in use.
The Escherichia coli dataset consists of 4511 genes (334 TF) and a gold standard of 3766 TF-TG interactions (94% of 4012 total), according to which 89% of all indirect TF-TG interactions are removable. The in-silico datasets consist of 1643 genes (195 TF) and a gold standard of 1923 TF-TG interactions (94% of 2066 total), according to which 72% of all indirect TF-TG interactions are removable.
Given a sorted list of regulatory interaction p values in ascending order, we apply the test to any four-node subnetwork consisting of two TFs and two TGs. We call a certain number of regulatory interactions with the lowest ranks in the list the set of most likely interactions. We call the TF with the most likely interaction in a four-node subnetwork the putative interface variable. We call a subnetwork sufficiently connected if at least three of the four TF–TG interactions, including both with the putative interface variable, are in the set of the most likely interactions. If a subnetwork is sufficiently connected, then each p value of interactions with the other TF (not the putative interface variable) is changed by
where p (test) -value denotes the p value of the test. For a scaling factor c = 1, the test is conservative for the null hypothesis (no direct interaction), which corresponds to the hypothesis ((no dependency) or (functional module)). We heuristically set c to the smallest p value of any interaction in the set of most likely interactions, ensuring that missing evidence against a functional module is not weighted more heavily than the evidence for interactions that determine whether the test is performed at all. The size of the set of most likely interactions is determined by a holdout set consisting of every 8th sample.
To combine tests for the same interaction in different subnetworks, we again take the maximum of the individual p values, which corresponds to a combined conservative test for the logic or operation of the individual hypotheses. If the gene regulatory network is i) nested such that each TG can only by reached from any TF via a single interface variable, ii) all TF–TG interactions are essential, i.e., the removal of a single TF-TG interaction results in additional independencies in s , and iii) indirect TF–TG interactions are less correlated than direct TF–TG interactions, then the network reconstruction is asymptotically correct in the sense that the most likely inferred interactions are all true TF–TG interactions.
More precisely, we test for the modularization \({{{{{{{\mathcal{M}}}}}}}}=\{S\}\) consisting of the single functional module S = {1, 2} (Fig. 3 a) resulting in a single set \({X}^{({{{{{{{\mathcal{M}}}}}}}})}=\{1,2\}\) that indexes the only two components of the moment ratio vector b k = E [ s k s 3 ]/ E [ s k s ref ] for k ∈ {1, 2}. To avoid corrections for correlated components of the moment ratio vector, we estimate both b k using disjoint sets of samples. Both datasets consist of N (t) = 804 samples (microarrays). Furthermore, we use the uncorrected asymptotic version of the test, where λ v = 1, and a small cutoff \({\theta }_{\hat{\mu }} \, < \, 1\) .
To apply the method to a ranked list of N regulatory interactions, we artificially assign p values r a n k / N . For the sorted list of Pearson correlation coefficients, this procedure results in roughly the same AUPR, with even a slight performance improvement of 0.29%.
Inference in pyramidal neurons
We simulated the detailed compartmental model of a CA1 pyramidal neuron developed by ref. 24 in the simulation environment NEURON. The model includes various active and passive membrane mechanisms, such as sodium and potassium currents, A-type potassium currents, m-type potassium currents, hyperpolarization-activated h-current, voltage-dependent calcium currents, and Ca 2+ -dependent potassium currents. The densities and distributions of these currents are based on published data. We are interested in subthreshold synaptic integration and block all spike-generating currents at the soma.
Synaptic inputs consist of an NMDA and an AMPA-type conductance with a ratio of their peak values of 2.5. Each of the 60 terminal branches contains ten synapses, with equal distances between adjacent synapses or branch ends. Each synapse is stimulated by a Poisson process at a constant rate of 32 Hz. The dendritic spike rate is ~28 Hz, which is in the range of values observed experimentally in neocortical pyramidal neurons from freely behaving rats 22 .
The datasets consist of the membrane potentials at the soma and the centers of the 26 most proximal terminal branches of the apical dendrites. Samples are recorded for 20 or 60 min at 50 ms time intervals, ensuring that their normalized autocovariance is less than 0.05. N (t) is either 24,000 or 72,000.
The moment ratio vector \(\hat{{{{{{{{\bf{b}}}}}}}}}\) and the index sets \({X}^{({{{{{{{\mathcal{M}}}}}}}})}\) are derived from the corresponding elements above the diagonal of \(\hat{{{{{{{{\bf{B}}}}}}}}}\) (see Fig. 4 f and Supp. Methods) . To test for a purely linear functional module S , we define an additional set \({X}^{({{{{{{{\mathcal{M}}}}}}}})}\) that indexes all elements above the diagonal of the square submatrix of \(\hat{{{{{{{{\bf{B}}}}}}}}}\) that is indexed by S . For each module or branch originating from the trunk, the largest p value is calculated over all combinations of modules and branches containing that module and consisting of 11 observable components to ensure identical degrees of freedoms.
The resulting flat modularization, which consists of the nonlinear functional modules S 4 and S 7 and a complementary linear somatic module (Fig. 6 f), cannot be rejected at an overall significance level of 0.01. In contrast, the flat modularization consisting of the functional modules S 2 and S 7 and a complementary linear somatic module can be rejected at an overall significance level of 0.01.
Inference in neural networks
The neural network consists of d = 5 populations, each with 10 neurons. For every 200 ms time interval, the firing rates of all neurons in the i -th population are chosen according to a binary random variable x i ∈ {1, 2} for i ∈ {1, …, d } such that they are set to 5 Hz if x i = 1 or 25 Hz, otherwise. The total number of spikes in the i -th population and the n -th time interval defines the observable component \({\hat{s}}_{i}^{(n)}\) (normalized to zero mean). N (t) = 4500.
We implement the modularization shown in Fig. 5 a, which consists of \({d}^{({{{{{{{\mathcal{M}}}}}}}})}=2\) functional modules, where s ref refers to s 5 . The binary random vector x and the binary interface variables y c ∈ {1, 2} for c ∈ {1, 2} are distributed according to
The rate functions for y 1 and x 5 are shown in the bottom left panel and the rate function for y 2 is shown in the bottom right panel of Fig. 1 b. An optimal linear model predicting x 5 from the other components of x has a coefficient of determination of R 2 = 0.79.
The moment ratio vector \(\hat{{{{{{{{\bf{b}}}}}}}}}\) and the index sets \({X}^{({{{{{{{\mathcal{M}}}}}}}})}\) are derived from the corresponding elements above the diagonal of \(\hat{{{{{{{{\bf{B}}}}}}}}}\) (see Fig. 4 h and Supp. Methods) . To test for an arbitrary linear modularization \({{{{{{{{\mathcal{M}}}}}}}}}_{{{{{{{{\rm{L}}}}}}}}}\) , we define an additional set \({X}^{({{{{{{{{\mathcal{M}}}}}}}}}_{{{{{{{{\rm{L}}}}}}}}})}\) that indexes all elements above the diagonal of \(\hat{{{{{{{{\bf{B}}}}}}}}}\) .
All components of \(\hat{{{{{{{{\bf{s}}}}}}}}}\) are correlated with Pearson correlation coefficients greater than 0.41 (Fig. 5 c). To generate a linear modularization with the covariance matrix \({\hat{{{{{{{{\bf{\Sigma }}}}}}}}}}^{{{{{{{{\rm{(s)}}}}}}}}}\) of \(\hat{{{{{{{{\bf{s}}}}}}}}}\) , we apply the linear transformation L obtained from the Cholesky decomposition \({\hat{{{{{{{{\bf{\Sigma }}}}}}}}}}^{{{{{{{{\rm{(s)}}}}}}}}}={{{{{{{\bf{L}}}}}}}}{{{{{{{{\bf{L}}}}}}}}}^{T}\) to the time-shuffled observable states (shuffled independently for each population). All tests are repeated 10 4 times on independent datasets. All 25 modularizations are shown in Fig. 7 .
All 25 modularizations \({{{{{{{{\mathcal{M}}}}}}}}}_{1}\) to \({{{{{{{{\mathcal{M}}}}}}}}}_{25}\) of an undirected graphical model consisting of five components s 1 to s 5 , such that each functional model has at least two components.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The datasets analyzed during the current study are available at Zenodo 48 .
Code availability
An implementation of the statistical test in MATLAB (R2015a) is available at Zenodo 48 .
Urai, A. E., Doiron, B., Leifer, A. M. & Churchland, A. K. Large-scale neural recordings call for new insights to link brain and behavior. Nat. Neurosci. 25 , 11–19 (2022).
Article CAS PubMed Google Scholar
Paninski, L. & Cunningham, J. P. Neural data science: accelerating the experiment-analysis-theory cycle in large-scale neuroscience. Curr. Opin. Neurobiol. 50 , 232–241 (2018).
Bullmore, E. & Sporns, O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat. Rev. Neurosci. 10 , 186–198 (2009).
Bullmore, E. T. & Bassett, D. S. Brain graphs: graphical models of the human brain connectome. Annu. Rev. Clin. Psychol. 7 , 113–140 (2011).
Article PubMed Google Scholar
Bassett, D. S. & Sporns, O. Network neuroscience. Nat. Neurosci. 20 , 353–364 (2017).
Article CAS PubMed PubMed Central Google Scholar
Newman, M. E. The structure and function of complex networks. SIAM Rev. 45 , 167–256 (2003).
Article Google Scholar
Newman, M. E. Communities, modules and large-scale structure in networks. Nat. Phys. 8 , 25–31 (2012).
Article CAS Google Scholar
Wagner, G. P., Pavlicev, M. & Cheverud, J. M. The road to modularity. Nat. Rev. Genet. 8 , 921–931 (2007).
Girvan, M. & Newman, M. E. Community structure in social and biological networks. Proc. Natl Acad. Sci. 99 , 7821–7826 (2002).
Koller, D. & Friedman, N. Probabilistic Graphical Models: Principles and Techniques (MIT Press, 2009).
Panzeri, S., Moroni, M., Safaai, H. & Harvey, C. D. The structures and functions of correlations in neural population codes. Nat. Rev. Neurosci. 23 , 551–567 (2022).
Latchman, D. S. Transcription factors: an overview. Int. J. Biochem. Cell Biol. 29 , 1305–1312 (1997).
Gerstner, W. & Kistler, W. M. Spiking Neuron Models: Single Neurons, Populations, Plasticity (Cambridge University Press, 2002).
Karlebach, G. & Shamir, R. Modelling and analysis of gene regulatory networks. Nat. Rev. Mol. Cell Biol. 9 , 770–780 (2008).
Le Novere, N. Quantitative and logic modelling of molecular and gene networks. Nat. Rev. Genet. 16 , 146–158 (2015).
Article PubMed PubMed Central Google Scholar
Marbach, D. et al. Wisdom of crowds for robust gene network inference. Nat. Methods 9 , 796–804 (2012).
Huynh-Thu, V. A., Irrthum, A., Wehenkel, L. & Geurts, P. Inferring regulatory networks from expression data using tree-based methods. PloS one 5 , e12776 (2010).
Decharms, R. C. & Zador, A. Neural representation and the cortical code. Annu. Rev. Neurosci. 23 , 613–647 (2000).
Theunissen, F. & Miller, J. P. Temporal encoding in nervous systems: a rigorous definition. J. Comput. Neurosci. 2 , 149–162 (1995).
Stuart, G. J. & Spruston, N. Dendritic integration: 60 years of progress. Nat. Neurosci. 18 , 1713–1721 (2015).
Stuart, G., Spruston, N. & Häusser, M. Dendrites (Oxford University Press, 2016).
Moore, J. J. et al. Dynamics of cortical dendritic membrane potential and spikes in freely behaving rats. Science 355 , eaaj1497 (2017).
Archie, K. A. & Mel, B. W. A model for intradendritic computation of binocular disparity. Nat. Neurosci. 3 , 54–63 (2000).
Poirazi, P., Brannon, T. & Mel, B. W. Pyramidal neuron as two-layer neural network. Neuron 37 , 989–999 (2003).
Polsky, A., Mel, B. W. & Schiller, J. Computational subunits in thin dendrites of pyramidal cells. Nat. Neurosci. 7 , 621–627 (2004).
Katz, Y. et al. Synapse distribution suggests a two-stage model of dendritic integration in ca1 pyramidal neurons. Neuron 63 , 171–177 (2009).
Branco, T., Clark, B. A. & Häusser, M. Dendritic discrimination of temporal input sequences in cortical neurons. Science 329 , 1671–1675 (2010).
Behabadi, B. F. & Mel, B. W. Mechanisms underlying subunit independence in pyramidal neuron dendrites. Proc. Natl Acad. Sci. 111 , 498–503 (2014).
Eberhardt, F., Herz, A. V. & Häusler, S. Tuft dendrites of pyramidal neurons operate as feedback-modulated functional subunits. PLoS Comput. Biol. 15 , e1006757 (2019).
Ver Steeg, G. & Galstyan, A. Discovering structure in high-dimensional data through correlation explanation. Adv. Neural Inf. Process. Syst. 27 , 577–585 (2014).
Bielza, C. & Larrañaga, P. Bayesian networks in neuroscience: a survey. Front. Comput. Neurosci. 8 , 131 (2014).
Grieb, M. et al. Predicting variabilities in cardiac gene expression with a boolean network incorporating uncertainty. PloS One 10 , e0131832 (2015).
Malekpour, S. A., Alizad-Rahvar, A. R. & Sadeghi, M. Logicnet: probabilistic continuous logics in reconstructing gene regulatory networks. BMC Bioinforma. 21 , 1–21 (2020).
Malekpour, S. A., Shahdoust, M., Aghdam, R. & Sadeghi, M. wplogicnet: logic gate and structure inference in gene regulatory networks. Bioinformatics 39 , btad072 (2023).
Bethge, M., Rotermund, D. & Pawelzik, K. Optimal neural rate coding leads to bimodal firing rate distributions. Netw. Comput. Neural Syst. 14 , 303 (2003).
Nikitin, A. P., Stocks, N. G., Morse, R. P. & McDonnell, M. D. Neural population coding is optimized by discrete tuning curves. Phys. Rev. Lett. 103 , 138101 (2009).
Gjorgjieva, J., Meister, M. & Sompolinsky, H. Functional diversity among sensory neurons from efficient coding principles. PLoS Comput. Biol. 15 , e1007476 (2019).
Averbeck, B. B., Latham, P. E. & Pouget, A. Neural correlations, population coding and computation. Nat. Rev. Neurosci. 7 , 358–366 (2006).
Hartemink, A. J., Gifford, D. K., Jaakkola, T. S. & Young, R. A. Maximum-likelihood estimation of optimal scaling factors for expression array normalization. In Microarrays: Optical Technologies and Informatics , 4266 , 132–140 (SPIE, 2001).
Bolstad, B. M., Irizarry, R. A., Åstrand, M. & Speed, T. P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19 , 185–193 (2003).
Badia-i Mompel, P. et al. Gene regulatory network inference in the era of single-cell multi-omics. Nat. Rev. Genet. 24 , 739–754 (2023).
Abbott, J. et al. A nanoelectrode array for obtaining intracellular recordings from thousands of connected neurons. Nat. Biomed. Eng. 4 , 232–241 (2020).
Paulk, A. C. et al. Large-scale neural recordings with single neuron resolution using neuropixels probes in human cortex. Nat. Neurosci. 25 , 252–263 (2022).
Adesnik, H. & Abdeladim, L. Probing neural codes with two-photon holographic optogenetics. Nat. Neurosci. 24 , 1356–1366 (2021).
Grienberger, C., Giovannucci, A., Zeiger, W. & Portera-Cailliau, C. Two-photon calcium imaging of neuronal activity. Nat. Rev. Methods Prim. 2 , 67 (2022).
Bando, Y., Wenzel, M. & Yuste, R. Simultaneous two-photon imaging of action potentials and subthreshold inputs in vivo. Nat. Commun. 12 , 7229 (2021).
Cornejo, V. H., Ofer, N. & Yuste, R. Voltage compartmentalization in dendritic spines in vivo. Science 375 , 82–86 (2022).
Häusler, S. Zenodo, https://doi.org/10.5281/zenodo.8190172 (2023).
Download references
Acknowledgements
I am grateful for the support of the Bernstein Center Munich and Andreas V. M. Herz. I also thank Martin Stemmler and Kay Thurley for helpful feedback on the manuscript.
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and affiliations.
Faculty of Biology and Bernstein Center for Computational Neuroscience, Ludwig-Maximilians-Universität München, Munich, Germany
Stefan Häusler
You can also search for this author in PubMed Google Scholar
Corresponding author
Correspondence to Stefan Häusler .
Ethics declarations
Competing interests.
The author declares no competing interests.
Peer review
Peer review information.
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Enzo Tagliazucchi and Luke R. Grinham.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information, reporting summary, rights and permissions.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ .
Reprints and permissions
About this article
Cite this article.
Häusler, S. Correlations reveal the hierarchical organization of biological networks with latent variables. Commun Biol 7 , 678 (2024). https://doi.org/10.1038/s42003-024-06342-y
Download citation
Received : 19 September 2023
Accepted : 16 May 2024
Published : 03 June 2024
DOI : https://doi.org/10.1038/s42003-024-06342-y
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
By submitting a comment you agree to abide by our Terms and Community Guidelines . If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing newsletter — what matters in science, free to your inbox daily.
- Search Menu
- Sign in through your institution
- Advance articles
- Author Guidelines
- Submission Site
- Open Access
- Why publish with this journal?
- About Bioinformatics
- Journals Career Network
- Editorial Board
- Advertising and Corporate Services
- Self-Archiving Policy
- Dispatch Dates
- Journals on Oxford Academic
- Books on Oxford Academic

Article Contents
- 1 Introduction
- 2 Materials and methods
- 4 Conclusion and discussion
- Acknowledgements
- Supplementary data
- Conflict of interest
- Data availability
- < Previous
Pathformer: a biological pathway informed transformer for disease diagnosis and prognosis using multi-omics data
= Xiaofan Liu and Yuhuan Tao equal contribution.
- Article contents
- Figures & tables
- Supplementary Data
Xiaofan Liu, Yuhuan Tao, Zilin Cai, Pengfei Bao, Hongli Ma, Kexing Li, Mengtao Li, Yunping Zhu, Zhi John Lu, Pathformer: a biological pathway informed transformer for disease diagnosis and prognosis using multi-omics data, Bioinformatics , Volume 40, Issue 5, May 2024, btae316, https://doi.org/10.1093/bioinformatics/btae316
- Permissions Icon Permissions
Multi-omics data provide a comprehensive view of gene regulation at multiple levels, which is helpful in achieving accurate diagnosis of complex diseases like cancer. However, conventional integration methods rarely utilize prior biological knowledge and lack interpretability.
To integrate various multi-omics data of tissue and liquid biopsies for disease diagnosis and prognosis, we developed a biological pathway informed Transformer, Pathformer. It embeds multi-omics input with a compacted multi-modal vector and a pathway-based sparse neural network. Pathformer also leverages criss-cross attention mechanism to capture the crosstalk between different pathways and modalities. We first benchmarked Pathformer with 18 comparable methods on multiple cancer datasets, where Pathformer outperformed all the other methods, with an average improvement of 6.3%–14.7% in F1 score for cancer survival prediction, 5.1%–12% for cancer stage prediction, and 8.1%–13.6% for cancer drug response prediction. Subsequently, for cancer prognosis prediction based on tissue multi-omics data, we used a case study to demonstrate the biological interpretability of Pathformer by identifying key pathways and their biological crosstalk. Then, for cancer early diagnosis based on liquid biopsy data, we used plasma and platelet datasets to demonstrate Pathformer’s potential of clinical applications in cancer screening. Moreover, we revealed deregulation of interesting pathways (e.g. scavenger receptor pathway) and their crosstalk in cancer patients’ blood, providing potential candidate targets for cancer microenvironment study.
Pathformer is implemented and freely available at https://github.com/lulab/Pathformer .
Email alerts
Citing articles via, looking for your next opportunity.
- Recommend to your Library
Affiliations
- Online ISSN 1367-4811
- Copyright © 2024 Oxford University Press
- About Oxford Academic
- Publish journals with us
- University press partners
- What we publish
- New features
- Open access
- Institutional account management
- Rights and permissions
- Get help with access
- Accessibility
- Advertising
- Media enquiries
- Oxford University Press
- Oxford Languages
- University of Oxford
Oxford University Press is a department of the University of Oxford. It furthers the University's objective of excellence in research, scholarship, and education by publishing worldwide
- Copyright © 2024 Oxford University Press
- Cookie settings
- Cookie policy
- Privacy policy
- Legal notice
This Feature Is Available To Subscribers Only
Sign In or Create an Account
This PDF is available to Subscribers Only
For full access to this pdf, sign in to an existing account, or purchase an annual subscription.

How To Get Free Access To Microsoft PowerPoint
E very time you need to present an overview of a plan or a report to a whole room of people, chances are you turn to Microsoft PowerPoint. And who doesn't? It's popular for its wide array of features that make creating effective presentations a walk in the park. PowerPoint comes with a host of keyboard shortcuts for easy navigation, subtitles and video recordings for your audience's benefit, and a variety of transitions, animations, and designs for better engagement.
But with these nifty features comes a hefty price tag. At the moment, the personal plan — which includes other Office apps — is at $69.99 a year. This might be the most budget-friendly option, especially if you plan to use the other Microsoft Office apps, too. Unfortunately, you can't buy PowerPoint alone, but there are a few workarounds you can use to get access to PowerPoint at no cost to you at all.
Read more: The 20 Best Mac Apps That Will Improve Your Apple Experience
Method #1: Sign Up For A Free Microsoft Account On The Office Website
Microsoft offers a web-based version of PowerPoint completely free of charge to all users. Here's how you can access it:
- Visit the Microsoft 365 page .
- If you already have a free account with Microsoft, click Sign in. Otherwise, press "Sign up for the free version of Microsoft 365" to create a new account at no cost.
- On the Office home page, select PowerPoint from the side panel on the left.
- Click on "Blank presentation" to create your presentation from scratch, or pick your preferred free PowerPoint template from the options at the top (there's also a host of editable templates you can find on the Microsoft 365 Create site ).
- Create your presentation as normal. Your edits will be saved automatically to your Microsoft OneDrive as long as you're connected to the internet.
It's important to keep in mind, though, that while you're free to use this web version of PowerPoint to create your slides and edit templates, there are certain features it doesn't have that you can find on the paid version. For instance, you can access only a handful of font styles and stock elements like images, videos, icons, and stickers. Designer is also available for use on up to three presentations per month only (it's unlimited for premium subscribers). When presenting, you won't find the Present Live and Always Use Subtitles options present in the paid plans. The biggest caveat of the free version is that it won't get any newly released features, unlike its premium counterparts.
Method #2: Install Microsoft 365 (Office) To Your Windows
Don't fancy working on your presentation in a browser? If you have a Windows computer with the Office 365 apps pre-installed or downloaded from a previous Office 365 trial, you can use the Microsoft 365 (Office) app instead. Unlike the individual Microsoft apps that you need to buy from the Microsoft Store, this one is free to download and use. Here's how to get free PowerPoint on the Microsoft 365 (Office) app:
- Search for Microsoft 365 (Office) on the Microsoft Store app.
- Install and open it.
- Sign in with your Microsoft account. Alternatively, press "Create free account" if you don't have one yet.
- Click on Create on the left side panel.
- Select Presentation.
- In the PowerPoint window that opens, log in using your account.
- Press Accept on the "Free 5-day pass" section. This lets you use PowerPoint (and Word and Excel) for five days — free of charge and without having to input any payment information.
- Create your presentation as usual. As you're using the desktop version, you can access the full features of PowerPoint, including the ability to present in Teams, export the presentation as a video file, translate the slides' content to a different language, and even work offline.
The only downside of this method is the time limit. Once the five days are up, you can no longer open the PowerPoint desktop app. However, all your files will still be accessible to you. If you saved them to OneDrive, you can continue editing them on the web app. If you saved them to your computer, you can upload them to OneDrive and edit them from there.
Method #3: Download The Microsoft PowerPoint App On Your Android Or iOS Device
If you're always on the move and need the flexibility of creating and editing presentations on your Android or iOS device, you'll be glad to know that PowerPoint is free and available for offline use on your mobile phones. But — of course, there's a but — you can only access the free version if your device is under 10.1 inches. Anything bigger than that requires a premium subscription. If your phone fits the bill, then follow these steps to get free PowerPoint on your device:
- Install Microsoft PowerPoint from the App Store or Google Play Store .
- Log in using your existing Microsoft email or enter a new email address to create one if you don't already have an account.
- On the "Get Microsoft 365 Personal Plan" screen, press Skip For Now.
- If you're offered a free trial, select Try later (or enjoy the free 30-day trial if you're interested).
- To make a new presentation, tap the plus sign in the upper right corner.
- Change the "Create in" option from OneDrive - Personal to a folder on your device. This allows you to save the presentation to your local storage and make offline edits.
- Press "Set as default" to set your local folder as the default file storage location.
- Choose your template from the selection or use a blank presentation.
- Edit your presentation as needed.
Do note that PowerPoint mobile comes with some restrictions. There's no option to insert stock elements, change the slide size to a custom size, use the Designer feature, or display the presentation in Immersive Reader mode. However, you can use font styles considered premium on the web app.
Method #4: Use Your School Email Address
Office 365 Education is free for students and teachers, provided they have an email address from an eligible school. To check for your eligibility, here's what you need to do:
- Go to the Office 365 Education page .
- Type in your school email address in the empty text field.
- Press "Get Started."
- On the next screen, verify your eligibility. If you're eligible, you'll be asked to select whether you're a student or a teacher. If your school isn't recognized, however, you'll get a message telling you so.
- For those who are eligible, proceed with creating your Office 365 Education account. Make sure your school email can receive external mail, as Microsoft will send you a verification code for your account.
- Once you're done filling out the form, press "Start." This will open your Office 365 account page.
You can then start making your PowerPoint presentation using the web app. If your school's plan supports it, you can also install the Office 365 apps to your computer by clicking the "Install Office" button on your Office 365 account page and running the downloaded installation file. What sets the Office 365 Education account apart from the regular free account is that you have unlimited personal cloud storage and access to other Office apps like Word, Excel, and Outlook.
Read the original article on SlashGear .

- Share full article
Advertisement
Supported by
Why Did North Korea Bombard the South With Trash Balloons?
The unusual offensive, across the world’s most heavily fortified border, is a revival of a Cold War-era tactic. The South has threatened to respond by blasting K-pop.

By Choe Sang-Hun
Reporting from Seoul
North Korea launched 720 balloons across the world’s most heavily armed border overnight Saturday, hitting South Korea with their payloads: plastic bags full of cigarette butts and other trash.
Since last Tuesday, North Korea has sent roughly 1,000 of these trash balloons across the Demilitarized Zone separating the two Koreas. Once the balloons reached South Korean airspace, their timers released the plastic bags containing assorted rubbish, including scraps of used paper and cloth.
The South Korean military dismissed initial reports that the balloons were carrying human waste, but it did note that some of the trash appeared to be compost.
So far, the authorities in the South have found “nothing hazardous” in the payloads. On Sunday, the office of President Yoon Suk Yeol accused North Korea of “dirty provocations no normal country would think of.” It said South Korea would start taking “steps that North Korea would find unbearable.”
Two days later, South Korea suspended an agreement signed with North Korea in 2018 that called for the two Koreas to cease all hostile activities, such as military drills and reconnaissance flights, along their border. North Korea had already suspended the accord last year, calling it a “mere scrap of paper.”
South Korea said Tuesday that it would “revive all military activities” restricted under the 2018 agreement, until “inter-Korean mutual trust is restored.” It did not elaborate on what it planned to do. But one of the options its officials were considering was to switch on their loudspeakers along the inter-Korean border to blare K-pop music, which the North’s leader, Kim Jong-un, has found so threatening that he once called it a “ vicious cancer .”
The North has cast the floating offensive as “ tit-for-tat action .” It has accused North Korean defectors living in South Korea of “scattering leaflets and various dirty things” over its border counties in recent days.
Here’s what to know about the unusual offensive.
It has been unsettling but not disruptive.
When South Korea reports objects launched from North Korea, they are usually rockets carrying satellites or ballistic missiles of a kind the North says is capable of delivering nuclear warheads. But the North’s actions in the past week have been a revival of a Cold War era tactic: propaganda balloons as psychological warfare.
Last week’s balloon offensive triggered some confusion and public complaints when the government mistakenly warned people near the border of an “air raid.”
Mostly South Koreans remained calm, treating the episode as little more than irritating antics from the North. On social media, people posted pictures of the North Korean balloons in trees, on farmland or on urban side streets bursting with trash. One plastic bag dropped from a balloon was heavy enough to destroy the windshield of a parked car, according to photos carried by local news media.
But there was an ominous undertone when South Korea urged people not to touch the balloons and to report them to the authorities immediately. North Korea is known to hold large stockpiles of biological and chemical weapons, which its agents once used to assassinate Mr. Kim’s estranged half brother, Kim Jong-nam.
Photos and video footage released by the South Korean military on Sunday showed officers clad in biohazard and bomb-disposal gear inspecting the trash piles.
The balloon rivalry goes back decades.
During the Cold War, North and South Korea waged psychological warfare. They tried to influence each other’s citizens with shortwave radio broadcasts laden with propaganda. Along the DMZ, loudspeakers bombarded rival soldiers day and night with propaganda songs. Billboards urged the soldiers to defect to a “people’s paradise” in the North or to the “free and democratic” South.
And the two Koreas launched leaflet-laden balloons into each other’s airspace. Millions of such leaflets vilifying the other side’s government were scattered over the Korean Peninsula, material that both Koreas banned their people from reading or keeping. In the South, the police rewarded children with pencils and other school supplies when they found the leaflets in the hills and reported them.
But until fairly recently, balloons from North Korea seldom carried common trash.
A court decision allowed the balloons to fly again.
By the 1990s, it was clear that the North’s propaganda was losing its relevance as the South’s economy pulled ahead. The South had become a vibrant democracy and a global export powerhouse, while the North suffered chronic food shortages and relied on a personality cult and a total information blackout to control its people.
When their leaders held the first inter-Korean summit meeting in 2000, the two Koreas agreed to end government-sponsored efforts to influence each other’s citizens. But North Korean defectors and conservative and Christian activists in the South carried on the information war , sending balloons laden with mini-Bibles, transistor radios, household medicine, computer thumb drives containing K-pop music and drama, and leaflets that called Mr. Kim a “pig.”
To them, their payloads contained “truth” and “freedom of expression” that would help awaken North Koreans from their government’s brainwashing. To Pyongyang, they were nothing more than political “filth,” and North Korean leaders vowed to retaliate in kind.
Then the government in Seoul enacted a law that banned the sending of leaflets to the North, saying they did little more than provoke Pyongyang. But a few years later, in 2023, a court ruled the law unconstitutional, and last month the activists resumed launching balloons.
“We have tried something they have always been doing, but I cannot understand why they are making a fuss as if they were hit by a shower of bullets,” Kim Yo-jong, Mr. Kim’s sister and spokeswoman, said last week. “If they experience how unpleasant the feeling of picking up filth is and how tired it is, they will know that it is not easy to dare talk about freedom of expression.”
Choe Sang-Hun is the lead reporter for The Times in Seoul, covering South and North Korea. More about Choe Sang-Hun
RSC Advances
Exploration of novel cationic amino acid-enriched short peptides: design, spps, biological evaluation and in silico study †.

* Corresponding authors
a Department of Chemistry, Marwadi University, Rajkot-360003, Gujarat, India E-mail: [email protected]
b Red de Estudios Moleculares Avanzados, Instituto de Ecología, A.C. Carretera Antigua a Coatepec 351, Xalapa, Veracruz, Mexico
c Departamento de Química, Universidad de Guanajuato, Noria Alta S/N, Guanajuato-36050, Guanajuato, Mexico
d Department of Pharmaceutical Chemistry, R. C. Patel Institute of Pharmaceutical Education and Research, Shirpur District Dhule-425405, Maharashtra, India
e Micro and Nanotechnology Research Center, Universidad Veracruzana, Blvd. Av. Ruiz Cortines No. 455 Fracc. Costa Verde, Boca del Río 94294, Mexico
f Department of Chemistry, S. S. C. College, Junnar, Pune-410502, Maharashtra, India E-mail: [email protected]
Antimicrobial resistance (AMR) represents a critical challenge worldwide, necessitating the pursuit of novel approaches to counteract bacterial and fungal pathogens. In this context, we explored the potential of cationic amino acid-enriched short peptides, synthesized via solid-phase methods, as innovative antimicrobial candidates. Our comprehensive evaluation assessed the antibacterial and antifungal efficacy of these peptides against a panel of significant pathogens, including Escherichia coli , Pseudomonas aeruginosa , Staphylococcus aureus , Streptococcus pyogenes , Candida albicans , and Aspergillus niger . Utilizing molecular docking techniques, we delved into the molecular interactions underpinning the peptides' action against these microorganisms. The results revealed a spectrum of inhibitory activities, with certain peptide sequences displaying pronounced effectiveness across various pathogens. These findings underscore the peptides' potential as promising antimicrobial agents, with molecular docking offering valuable insights into their mechanisms of action. This study enriches antimicrobial peptide (AMP) research by identifying promising candidates for further refinement and development toward therapeutic application, highlighting their significance in addressing the urgent issue of AMR.

Supplementary files
- Supplementary information PDF (889K)
- Supplementary movie MPEG (103617K)
Article information
Download Citation
Permissions.
Exploration of novel cationic amino acid-enriched short peptides: design, SPPS, biological evaluation and in silico study
P. K. Chandole, T. J. Pawar, J. L. Olivares-Romero, S. R. Tivari, B. Garcia Lara, H. Patel, I. Ahmad, E. Delgado-Alvarado, S. V. Kokate and Y. Jadeja, RSC Adv. , 2024, 14 , 17710 DOI: 10.1039/D3RA08313F
This article is licensed under a Creative Commons Attribution-NonCommercial 3.0 Unported Licence . You can use material from this article in other publications, without requesting further permission from the RSC, provided that the correct acknowledgement is given and it is not used for commercial purposes.
To request permission to reproduce material from this article in a commercial publication , please go to the Copyright Clearance Center request page .
If you are an author contributing to an RSC publication, you do not need to request permission provided correct acknowledgement is given.
If you are the author of this article, you do not need to request permission to reproduce figures and diagrams provided correct acknowledgement is given. If you want to reproduce the whole article in a third-party commercial publication (excluding your thesis/dissertation for which permission is not required) please go to the Copyright Clearance Center request page .
Read more about how to correctly acknowledge RSC content .
Social activity
Search articles by author, advertisements.

An official website of the United States government, Department of Justice.
Here's how you know
Official websites use .gov A .gov website belongs to an official government organization in the United States.
Secure .gov websites use HTTPS A lock ( Lock A locked padlock ) or https:// means you’ve safely connected to the .gov website. Share sensitive information only on official, secure websites.
Quantitative Analysis Method for Tetrahydrocannabinol Isomers in Biological Matrices
Of recent, forensic toxicology laboratories have been grappling with the emergence of tetrahydrocannabinol isomers within biological specimens. Traditional methods for the identification and quantitation of cannabinoids only include the evaluation of Δ9-tetrahydrocannabinol (Δ9-THC) and its metabolites in biological matrices. Upon analysis of additional tetrahydrocannabinol isomers, laboratories often find co-elution or minimal separation between Δ9-THC, exo-THC, Δ8-THC, Δ10-THC, and Δ6a,10a-THC. These emerging isomers are commonly observed in the seized drug community, particularly in manufactured cannabis products (e.g., edibles, electronic cigarette cartridges). Trends within the seized drug community and legislative changes to include tetrahydrocannabinol isomers dictate the need for change within forensic toxicology. Traditional methods require adaptation to the ever-changing climate surrounding tetrahydrocannabinol. To accomplish this, additional method development with subsequent validation to meet ANSI/ASB Standard 036, Standard Practices for Method Validation in Forensic Toxicology, is often required.
A dual chromatographic column method was developed and optimized for the separation of tetrahydrocannabinol isomers. An Agilent Technologies 1290 Infinity liquid chromatograph was coupled independently to both a 6460 and 6470 quadrupole mass spectrometer for development and validation. Two independent chromatographic methods were developed using different analytical columns, mobile phase conditions, chromatographic gradients, and flow rates. The qualitative analytical method utilized an Agilent Technologies Poroshell 120 PFP 3.0 x 100 mm, 2.7 µm column held at 50°C. The quantitative analytical method used an Agilent Technologies Poroshell 120 EC-C18 3.0 x 50 mm, 2.7 µm column held at 50°C. The dual column methodology was used to enhance the separation of tetrahydrocannabinol isomers.
The sample preparation procedure consisted of a supported liquid extraction (SLE) using 0.5 mL of biological specimen. The biological specimen was acidified with 200 µL of formic acid in water prior to placement onto the SLE cartridge. Each specimen was allowed to incubate for 5 minutes prior to the addition of ethyl acetate (3.0 mL). After elution and collection, n-hexane (3.0 mL) was added to each cartridge. Samples were evaporated to dryness at approximately 50°C prior to reconstitution in 50 µL of methanol.
The optimized method was validated for quantitation of Δ9-THC, (±)-11-hydroxy-Δ9-THC (Δ9-OH-THC), (±)-11-nor-9-carboxy-Δ9-THC (Δ9-carboxy-THC), (-)-Δ8-tetrahydrocannabinol (Δ8-THC), and cannabidiol to meet ANSI/ASB Standard 036, Standard Practices for Method Validation in Forensic Toxicology. All other isomers were validated to meet qualitative identification criteria. During validation, samples were evaluated on both analytical methods to ensure congruence in results. The calibration range was 1/2/5 ng/mL to 100/200/500 ng/mL (Δ9-THC, Δ8-THC/Δ9-OH-THC, cannabidiol/Δ9-carboxy-THC). All compounds were within ±20% for bias and precision when evaluating pooled fortified samples of blank blood, antemortem blood, and postmortem blood. The estimated limit of detection and lower limit of quantitation were assessed for each matrix type within the validation. Significant ionization suppression (>25%) was noted for antemortem blood, postmortem blood, and urine. Further, interferences were evaluated including other isomers and derivatives of Δ9-THC.
An extensive validation was performed on the optimized SLE extraction with subsequent analytical analysis using liquid chromatography tandem mass spectrometry. The validated method optimizes chromatographic separation between tetrahydrocannabinols providing enhanced identification of these isobaric compounds.
Detailed Learning Objectives
- Attendees will be introduced to some of the analytical challenges associated with emerging tetrahydrocannabinol isomers in biological specimens.
- Attendees will be introduced to different analytical strategies for the chromatographic separation of tetrahydrocannabinol isomers.
- Attendees will be provided with a validated extraction procedure and analytical parameters that meet ANSI/ASB Standard 036, Standard Practices for Method Validation in Forensic Toxicology.
Certificate of completion
Read more about

IMAGES
VIDEO
COMMENTS
Qurratulain Kamil. This document discusses the scientific method used in biology. It begins by outlining the steps of the scientific method: observation, hypothesis, experiment, data collection, conclusion, and retesting. It then goes into more detail about observations, distinguishing between qualitative and quantitative observations.
Biological method. Jun 14, 2020 • Download as PPTX, PDF •. 3 likes • 2,841 views. U. Ubaid MaLik. Here is slideshare on the topic biological method .Here students will be able to solve biological problem.So slides are important for students searching for topics biological method or how to solve biological problems.
Biology Scientific Method. Apr 1, 2009 •. 9 likes • 7,756 views. M, Michelle Jeannite. Education Technology Self Improvement. 1 of 16. Biology Scientific Method - Download as a PDF or view online for free.
The scientific method. At the core of biology and other sciences lies a problem-solving approach called the scientific method. The scientific method has five basic steps, plus one feedback step: Make an observation. Ask a question. Form a hypothesis, or testable explanation. Make a prediction based on the hypothesis.
Biology is the branch of science concerned with the study of living things, or organisms. Biologists have identified traits common to all the living organisms that we know. Although nonliving things may show some of these properties, in order for something to be considered living, it must meet all of them.
Biology is a natural science concerned with the study of life and living organisms. Modern biology is a vast and eclectic field composed of many specialized disciplines that study the structure, function, growth, distribution, evolution, or other features of living organisms. However, despite the broad scope of biology, there are certain ...
The scientific method is a method of research with defined steps that include experiments and careful observation. The steps of the scientific method will be examined in detail later, but one of the most important aspects of this method is the testing of hypotheses by means of repeatable experiments. A hypothesis is a suggested explanation for ...
develop an understanding of the logical and philosophical underpinnings of the scientific method, learn how biological research is commonly conducted and disseminated, and develop a skillset of data ... 10% final presentation; 35% total). This project is graded using the group scores as a baseline, adjusted by individual contributions if needed
The scientific method was first documented by Sir Francis Bacon (1561-1626) of England, and can be applied to almost all fields of study. The scientific method is founded upon observation, which then leads to a question and the development of a hypothesis which answers that question. The scientist can then design an experiment to test the ...
2.1 Biological Method Questions about living things have provided problems that man has investigated to aid his own survival and to satisfy his desire to know. The scientific method in which biological problems are solved, is termed as biological method. It comprises the steps a biologist adopts in order to solve a biological problem.
Biological method is commonly viewed as the best, eco-accommodating, and less vital concentrated to eliminate the greater part of poisons from wastewater. The total disintegration of colors by fungal microorganism is done in two phases (Imran et al., 2015).In the main stage, minimal enzymatic severance of diazene bond brings about creation of lackluster metabolites.
The biological synthesis method uses biological catalysts, resulting in an environmentally safe way to synthesize (Hebbalalu et al. 2013; Kruis et al. 2000; Sastry et al. 2003; ... elemental composition in ppt units, and the elements' internal chemical and electrical states. XPS is carried out in a high-vacuum environment.
The textbook on biological techniques brings into focus, global best practices, methods and techniques for the collection, preparation and preservation of biological samples and specimens. The ...
Consequently, biological synthesis methods are favored over physical and chemical routes for synthesizing nanoparticles. The nanoparticles' fabrication through biological method involves the utilization of various microorganisms, including bacteria [ 20 ], fungi [ 21 ], algae, and plants [ 22, 23, 24 ]. However, the use of microorganisms for ...
In this method, biological samples are mixed with protein precipitating agent followed by dilution with solvent (3-5 times the sample amount). ... or whole blood during BA. There are two types of PPT methods, one is salting out, in which ammonium sulfate is used as protein precipitating agent, and the other one is solvent precipitation, where ...
•Description of the validation of bioanalytical methods and study sample analysis. •Applicable to bioanalytical methods used to measure concentrations of chemical and biological drugs in biological samples from: - nonclinical toxicokinetic studies conducted according to the principles of Good Laboratory Practice.
Download the "Biology Infographics" template for PowerPoint or Google Slides and discover this set of editable infographics for education presentations. These resources, from graphs to tables, can be combined with other presentations or used independently. The most important thing is what you will achieve: transmit information in an orderly and ...
Module 1 Scientific Method Ppt. ... • Analytes in biological liquids such as serum or urine are frequently assayed (i.e., measured) using immunoassay methods. • In essence, the method depends upon the fact that the analyte in question is known to undergo a unique immune reaction with a second substance, which is used to determine the ...
Abstract. Mosquitoes represent the major arthropod vectors of human disease worldwide transmitting malaria, lymphatic filariasis, and arboviruses such as dengue virus and Zika virus. Unfortunately, no treatment (in the form of vaccines or drugs) is available for most of these diseases and vector control is still the main form of prevention.
In this three-part lecture, you will learn (a) how to design your PowerPoint or Keynote slides, (b) how to organize your talk, and (c) how to confidently deliver your research seminar. This series will help trainees and research scientists alike improve their presentation skills.
In this section we take a brief look at some commonly used methods used to study biological molecules and their interactions. 8.1: Cell Lysis To separate compounds from cellular environments, one must first break open (lyse) the cells. Cells are broken open, in buffered solutions, to obtain a lysate. There are several ways of accomplishing this.
Chemical And Biological Methods Of Food Preservation. 25. Preservation Using Chemicals. Salt and sugar. Salt and sugar have long been used as effective means of extending shelf life of various products as these solutes bind water, leaving less water available for the growth of microorganisms. Essentially the water activity (aw) of the product ...
Deciphering the functional organization of large biological networks is a major challenge for current mathematical methods. A common approach is to decompose networks into largely independent ...
It introduces a new embedding method to incorporate biological multi-modal data at both gene level and pathway level: it initiates the process by uniformly transforming different modalities to the gene level through a series of statistical indicators, then concatenates these modalities into compacted multi-modal vectors to define gene embedding ...
Click on "Blank presentation" to create your presentation from scratch, or pick your preferred free PowerPoint template from the options at the top (there's also a host of editable templates you ...
North Korea is known to hold large stockpiles of biological and chemical weapons, which its agents once used to assassinate Mr. Kim's estranged half brother, Kim Jong-nam.
Accurate prediction of complex traits is an important task in quantitative genetics that has become increasingly relevant for personalized medicine. Genotypes have traditionally been used for trait prediction using a variety of methods such as mixed models, Bayesian methods, penalized regressions, dimension reductions, and machine learning methods. Recent studies have shown that gene ...
Antimicrobial resistance (AMR) represents a critical challenge worldwide, necessitating the pursuit of novel approaches to counteract bacterial and fungal pathogens. In this context, we explored the potential of cationic amino acid-enriched short peptides, synthesized via solid-phase methods, as innovative antimicr
The document discusses various methods for controlling plant diseases, including regulatory, cultural, biological, physical, and chemical methods. Regulatory methods aim to prevent the spread of pathogens through quarantines and inspections. Cultural methods manipulate the environment and genetics of plants, such as host eradication, crop ...
Of recent, forensic toxicology laboratories have been grappling with the emergence of tetrahydrocannabinol isomers within biological specimens. Traditional methods for the identification and quantitation of cannabinoids only include the evaluation of Δ9-tetrahydrocannabinol (Δ9-THC) and its metabolites in biological matrices. Upon analysis of additional tetrahydrocannabinol isomers ...