- Research article
- Open access
- Published: 04 June 2021

Coronavirus disease (COVID-19) pandemic: an overview of systematic reviews
- Israel Júnior Borges do Nascimento 1 , 2 ,
- Dónal P. O’Mathúna 3 , 4 ,
- Thilo Caspar von Groote 5 ,
- Hebatullah Mohamed Abdulazeem 6 ,
- Ishanka Weerasekara 7 , 8 ,
- Ana Marusic 9 ,
- Livia Puljak ORCID: orcid.org/0000-0002-8467-6061 10 ,
- Vinicius Tassoni Civile 11 ,
- Irena Zakarija-Grkovic 9 ,
- Tina Poklepovic Pericic 9 ,
- Alvaro Nagib Atallah 11 ,
- Santino Filoso 12 ,
- Nicola Luigi Bragazzi 13 &
- Milena Soriano Marcolino 1
On behalf of the International Network of Coronavirus Disease 2019 (InterNetCOVID-19)
BMC Infectious Diseases volume 21 , Article number: 525 ( 2021 ) Cite this article
16k Accesses
29 Citations
13 Altmetric
Metrics details
Navigating the rapidly growing body of scientific literature on the SARS-CoV-2 pandemic is challenging, and ongoing critical appraisal of this output is essential. We aimed to summarize and critically appraise systematic reviews of coronavirus disease (COVID-19) in humans that were available at the beginning of the pandemic.
Nine databases (Medline, EMBASE, Cochrane Library, CINAHL, Web of Sciences, PDQ-Evidence, WHO’s Global Research, LILACS, and Epistemonikos) were searched from December 1, 2019, to March 24, 2020. Systematic reviews analyzing primary studies of COVID-19 were included. Two authors independently undertook screening, selection, extraction (data on clinical symptoms, prevalence, pharmacological and non-pharmacological interventions, diagnostic test assessment, laboratory, and radiological findings), and quality assessment (AMSTAR 2). A meta-analysis was performed of the prevalence of clinical outcomes.
Eighteen systematic reviews were included; one was empty (did not identify any relevant study). Using AMSTAR 2, confidence in the results of all 18 reviews was rated as “critically low”. Identified symptoms of COVID-19 were (range values of point estimates): fever (82–95%), cough with or without sputum (58–72%), dyspnea (26–59%), myalgia or muscle fatigue (29–51%), sore throat (10–13%), headache (8–12%) and gastrointestinal complaints (5–9%). Severe symptoms were more common in men. Elevated C-reactive protein and lactate dehydrogenase, and slightly elevated aspartate and alanine aminotransferase, were commonly described. Thrombocytopenia and elevated levels of procalcitonin and cardiac troponin I were associated with severe disease. A frequent finding on chest imaging was uni- or bilateral multilobar ground-glass opacity. A single review investigated the impact of medication (chloroquine) but found no verifiable clinical data. All-cause mortality ranged from 0.3 to 13.9%.
Conclusions
In this overview of systematic reviews, we analyzed evidence from the first 18 systematic reviews that were published after the emergence of COVID-19. However, confidence in the results of all reviews was “critically low”. Thus, systematic reviews that were published early on in the pandemic were of questionable usefulness. Even during public health emergencies, studies and systematic reviews should adhere to established methodological standards.
Peer Review reports
The spread of the “Severe Acute Respiratory Coronavirus 2” (SARS-CoV-2), the causal agent of COVID-19, was characterized as a pandemic by the World Health Organization (WHO) in March 2020 and has triggered an international public health emergency [ 1 ]. The numbers of confirmed cases and deaths due to COVID-19 are rapidly escalating, counting in millions [ 2 ], causing massive economic strain, and escalating healthcare and public health expenses [ 3 , 4 ].
The research community has responded by publishing an impressive number of scientific reports related to COVID-19. The world was alerted to the new disease at the beginning of 2020 [ 1 ], and by mid-March 2020, more than 2000 articles had been published on COVID-19 in scholarly journals, with 25% of them containing original data [ 5 ]. The living map of COVID-19 evidence, curated by the Evidence for Policy and Practice Information and Co-ordinating Centre (EPPI-Centre), contained more than 40,000 records by February 2021 [ 6 ]. More than 100,000 records on PubMed were labeled as “SARS-CoV-2 literature, sequence, and clinical content” by February 2021 [ 7 ].
Due to publication speed, the research community has voiced concerns regarding the quality and reproducibility of evidence produced during the COVID-19 pandemic, warning of the potential damaging approach of “publish first, retract later” [ 8 ]. It appears that these concerns are not unfounded, as it has been reported that COVID-19 articles were overrepresented in the pool of retracted articles in 2020 [ 9 ]. These concerns about inadequate evidence are of major importance because they can lead to poor clinical practice and inappropriate policies [ 10 ].
Systematic reviews are a cornerstone of today’s evidence-informed decision-making. By synthesizing all relevant evidence regarding a particular topic, systematic reviews reflect the current scientific knowledge. Systematic reviews are considered to be at the highest level in the hierarchy of evidence and should be used to make informed decisions. However, with high numbers of systematic reviews of different scope and methodological quality being published, overviews of multiple systematic reviews that assess their methodological quality are essential [ 11 , 12 , 13 ]. An overview of systematic reviews helps identify and organize the literature and highlights areas of priority in decision-making.
In this overview of systematic reviews, we aimed to summarize and critically appraise systematic reviews of coronavirus disease (COVID-19) in humans that were available at the beginning of the pandemic.
Methodology
Research question.
This overview’s primary objective was to summarize and critically appraise systematic reviews that assessed any type of primary clinical data from patients infected with SARS-CoV-2. Our research question was purposefully broad because we wanted to analyze as many systematic reviews as possible that were available early following the COVID-19 outbreak.
Study design
We conducted an overview of systematic reviews. The idea for this overview originated in a protocol for a systematic review submitted to PROSPERO (CRD42020170623), which indicated a plan to conduct an overview.
Overviews of systematic reviews use explicit and systematic methods for searching and identifying multiple systematic reviews addressing related research questions in the same field to extract and analyze evidence across important outcomes. Overviews of systematic reviews are in principle similar to systematic reviews of interventions, but the unit of analysis is a systematic review [ 14 , 15 , 16 ].
We used the overview methodology instead of other evidence synthesis methods to allow us to collate and appraise multiple systematic reviews on this topic, and to extract and analyze their results across relevant topics [ 17 ]. The overview and meta-analysis of systematic reviews allowed us to investigate the methodological quality of included studies, summarize results, and identify specific areas of available or limited evidence, thereby strengthening the current understanding of this novel disease and guiding future research [ 13 ].
A reporting guideline for overviews of reviews is currently under development, i.e., Preferred Reporting Items for Overviews of Reviews (PRIOR) [ 18 ]. As the PRIOR checklist is still not published, this study was reported following the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) 2009 statement [ 19 ]. The methodology used in this review was adapted from the Cochrane Handbook for Systematic Reviews of Interventions and also followed established methodological considerations for analyzing existing systematic reviews [ 14 ].
Approval of a research ethics committee was not necessary as the study analyzed only publicly available articles.
Eligibility criteria
Systematic reviews were included if they analyzed primary data from patients infected with SARS-CoV-2 as confirmed by RT-PCR or another pre-specified diagnostic technique. Eligible reviews covered all topics related to COVID-19 including, but not limited to, those that reported clinical symptoms, diagnostic methods, therapeutic interventions, laboratory findings, or radiological results. Both full manuscripts and abbreviated versions, such as letters, were eligible.
No restrictions were imposed on the design of the primary studies included within the systematic reviews, the last search date, whether the review included meta-analyses or language. Reviews related to SARS-CoV-2 and other coronaviruses were eligible, but from those reviews, we analyzed only data related to SARS-CoV-2.
No consensus definition exists for a systematic review [ 20 ], and debates continue about the defining characteristics of a systematic review [ 21 ]. Cochrane’s guidance for overviews of reviews recommends setting pre-established criteria for making decisions around inclusion [ 14 ]. That is supported by a recent scoping review about guidance for overviews of systematic reviews [ 22 ].
Thus, for this study, we defined a systematic review as a research report which searched for primary research studies on a specific topic using an explicit search strategy, had a detailed description of the methods with explicit inclusion criteria provided, and provided a summary of the included studies either in narrative or quantitative format (such as a meta-analysis). Cochrane and non-Cochrane systematic reviews were considered eligible for inclusion, with or without meta-analysis, and regardless of the study design, language restriction and methodology of the included primary studies. To be eligible for inclusion, reviews had to be clearly analyzing data related to SARS-CoV-2 (associated or not with other viruses). We excluded narrative reviews without those characteristics as these are less likely to be replicable and are more prone to bias.
Scoping reviews and rapid reviews were eligible for inclusion in this overview if they met our pre-defined inclusion criteria noted above. We included reviews that addressed SARS-CoV-2 and other coronaviruses if they reported separate data regarding SARS-CoV-2.
Information sources
Nine databases were searched for eligible records published between December 1, 2019, and March 24, 2020: Cochrane Database of Systematic Reviews via Cochrane Library, PubMed, EMBASE, CINAHL (Cumulative Index to Nursing and Allied Health Literature), Web of Sciences, LILACS (Latin American and Caribbean Health Sciences Literature), PDQ-Evidence, WHO’s Global Research on Coronavirus Disease (COVID-19), and Epistemonikos.
The comprehensive search strategy for each database is provided in Additional file 1 and was designed and conducted in collaboration with an information specialist. All retrieved records were primarily processed in EndNote, where duplicates were removed, and records were then imported into the Covidence platform [ 23 ]. In addition to database searches, we screened reference lists of reviews included after screening records retrieved via databases.
Study selection
All searches, screening of titles and abstracts, and record selection, were performed independently by two investigators using the Covidence platform [ 23 ]. Articles deemed potentially eligible were retrieved for full-text screening carried out independently by two investigators. Discrepancies at all stages were resolved by consensus. During the screening, records published in languages other than English were translated by a native/fluent speaker.
Data collection process
We custom designed a data extraction table for this study, which was piloted by two authors independently. Data extraction was performed independently by two authors. Conflicts were resolved by consensus or by consulting a third researcher.
We extracted the following data: article identification data (authors’ name and journal of publication), search period, number of databases searched, population or settings considered, main results and outcomes observed, and number of participants. From Web of Science (Clarivate Analytics, Philadelphia, PA, USA), we extracted journal rank (quartile) and Journal Impact Factor (JIF).
We categorized the following as primary outcomes: all-cause mortality, need for and length of mechanical ventilation, length of hospitalization (in days), admission to intensive care unit (yes/no), and length of stay in the intensive care unit.
The following outcomes were categorized as exploratory: diagnostic methods used for detection of the virus, male to female ratio, clinical symptoms, pharmacological and non-pharmacological interventions, laboratory findings (full blood count, liver enzymes, C-reactive protein, d-dimer, albumin, lipid profile, serum electrolytes, blood vitamin levels, glucose levels, and any other important biomarkers), and radiological findings (using radiography, computed tomography, magnetic resonance imaging or ultrasound).
We also collected data on reporting guidelines and requirements for the publication of systematic reviews and meta-analyses from journal websites where included reviews were published.
Quality assessment in individual reviews
Two researchers independently assessed the reviews’ quality using the “A MeaSurement Tool to Assess Systematic Reviews 2 (AMSTAR 2)”. We acknowledge that the AMSTAR 2 was created as “a critical appraisal tool for systematic reviews that include randomized or non-randomized studies of healthcare interventions, or both” [ 24 ]. However, since AMSTAR 2 was designed for systematic reviews of intervention trials, and we included additional types of systematic reviews, we adjusted some AMSTAR 2 ratings and reported these in Additional file 2 .
Adherence to each item was rated as follows: yes, partial yes, no, or not applicable (such as when a meta-analysis was not conducted). The overall confidence in the results of the review is rated as “critically low”, “low”, “moderate” or “high”, according to the AMSTAR 2 guidance based on seven critical domains, which are items 2, 4, 7, 9, 11, 13, 15 as defined by AMSTAR 2 authors [ 24 ]. We reported our adherence ratings for transparency of our decision with accompanying explanations, for each item, in each included review.
One of the included systematic reviews was conducted by some members of this author team [ 25 ]. This review was initially assessed independently by two authors who were not co-authors of that review to prevent the risk of bias in assessing this study.
Synthesis of results
For data synthesis, we prepared a table summarizing each systematic review. Graphs illustrating the mortality rate and clinical symptoms were created. We then prepared a narrative summary of the methods, findings, study strengths, and limitations.
For analysis of the prevalence of clinical outcomes, we extracted data on the number of events and the total number of patients to perform proportional meta-analysis using RStudio© software, with the “meta” package (version 4.9–6), using the “metaprop” function for reviews that did not perform a meta-analysis, excluding case studies because of the absence of variance. For reviews that did not perform a meta-analysis, we presented pooled results of proportions with their respective confidence intervals (95%) by the inverse variance method with a random-effects model, using the DerSimonian-Laird estimator for τ 2 . We adjusted data using Freeman-Tukey double arcosen transformation. Confidence intervals were calculated using the Clopper-Pearson method for individual studies. We created forest plots using the RStudio© software, with the “metafor” package (version 2.1–0) and “forest” function.
Managing overlapping systematic reviews
Some of the included systematic reviews that address the same or similar research questions may include the same primary studies in overviews. Including such overlapping reviews may introduce bias when outcome data from the same primary study are included in the analyses of an overview multiple times. Thus, in summaries of evidence, multiple-counting of the same outcome data will give data from some primary studies too much influence [ 14 ]. In this overview, we did not exclude overlapping systematic reviews because, according to Cochrane’s guidance, it may be appropriate to include all relevant reviews’ results if the purpose of the overview is to present and describe the current body of evidence on a topic [ 14 ]. To avoid any bias in summary estimates associated with overlapping reviews, we generated forest plots showing data from individual systematic reviews, but the results were not pooled because some primary studies were included in multiple reviews.
Our search retrieved 1063 publications, of which 175 were duplicates. Most publications were excluded after the title and abstract analysis ( n = 860). Among the 28 studies selected for full-text screening, 10 were excluded for the reasons described in Additional file 3 , and 18 were included in the final analysis (Fig. 1 ) [ 25 , 26 , 27 , 28 , 29 , 30 , 31 , 32 , 33 , 34 , 35 , 36 , 37 , 38 , 39 , 40 , 41 , 42 ]. Reference list screening did not retrieve any additional systematic reviews.
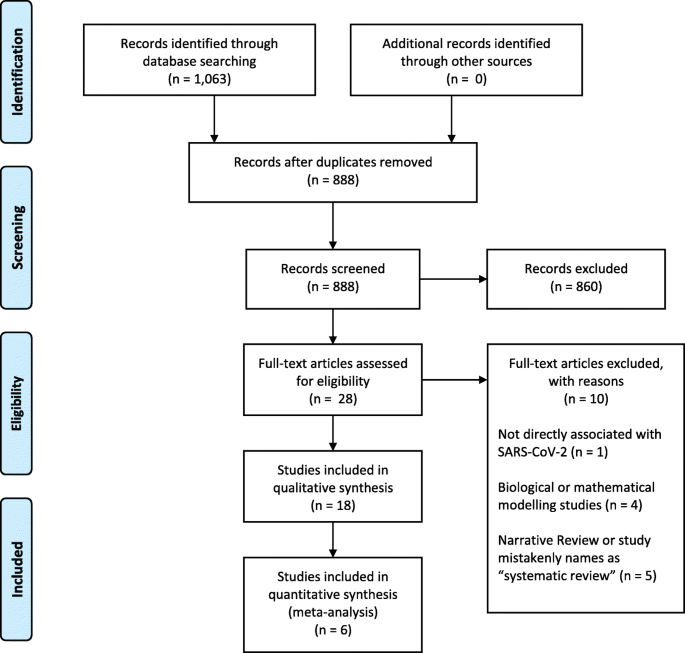
PRISMA flow diagram
Characteristics of included reviews
Summary features of 18 systematic reviews are presented in Table 1 . They were published in 14 different journals. Only four of these journals had specific requirements for systematic reviews (with or without meta-analysis): European Journal of Internal Medicine, Journal of Clinical Medicine, Ultrasound in Obstetrics and Gynecology, and Clinical Research in Cardiology . Two journals reported that they published only invited reviews ( Journal of Medical Virology and Clinica Chimica Acta ). Three systematic reviews in our study were published as letters; one was labeled as a scoping review and another as a rapid review (Table 2 ).
All reviews were published in English, in first quartile (Q1) journals, with JIF ranging from 1.692 to 6.062. One review was empty, meaning that its search did not identify any relevant studies; i.e., no primary studies were included [ 36 ]. The remaining 17 reviews included 269 unique studies; the majority ( N = 211; 78%) were included in only a single review included in our study (range: 1 to 12). Primary studies included in the reviews were published between December 2019 and March 18, 2020, and comprised case reports, case series, cohorts, and other observational studies. We found only one review that included randomized clinical trials [ 38 ]. In the included reviews, systematic literature searches were performed from 2019 (entire year) up to March 9, 2020. Ten systematic reviews included meta-analyses. The list of primary studies found in the included systematic reviews is shown in Additional file 4 , as well as the number of reviews in which each primary study was included.
Population and study designs
Most of the reviews analyzed data from patients with COVID-19 who developed pneumonia, acute respiratory distress syndrome (ARDS), or any other correlated complication. One review aimed to evaluate the effectiveness of using surgical masks on preventing transmission of the virus [ 36 ], one review was focused on pediatric patients [ 34 ], and one review investigated COVID-19 in pregnant women [ 37 ]. Most reviews assessed clinical symptoms, laboratory findings, or radiological results.
Systematic review findings
The summary of findings from individual reviews is shown in Table 2 . Overall, all-cause mortality ranged from 0.3 to 13.9% (Fig. 2 ).
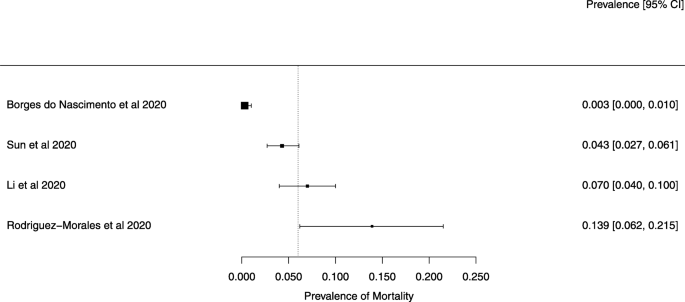
A meta-analysis of the prevalence of mortality
Clinical symptoms
Seven reviews described the main clinical manifestations of COVID-19 [ 26 , 28 , 29 , 34 , 35 , 39 , 41 ]. Three of them provided only a narrative discussion of symptoms [ 26 , 34 , 35 ]. In the reviews that performed a statistical analysis of the incidence of different clinical symptoms, symptoms in patients with COVID-19 were (range values of point estimates): fever (82–95%), cough with or without sputum (58–72%), dyspnea (26–59%), myalgia or muscle fatigue (29–51%), sore throat (10–13%), headache (8–12%), gastrointestinal disorders, such as diarrhea, nausea or vomiting (5.0–9.0%), and others (including, in one study only: dizziness 12.1%) (Figs. 3 , 4 , 5 , 6 , 7 , 8 and 9 ). Three reviews assessed cough with and without sputum together; only one review assessed sputum production itself (28.5%).
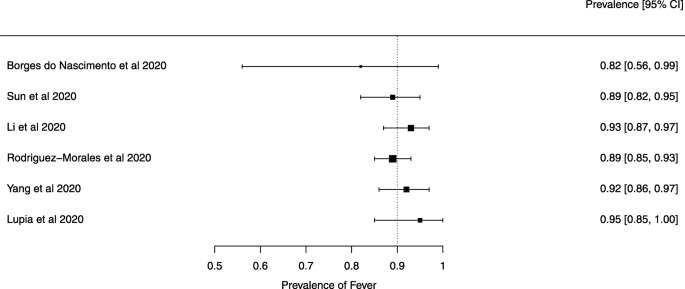
A meta-analysis of the prevalence of fever
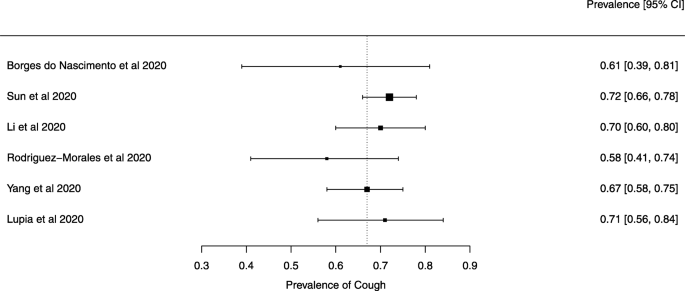
A meta-analysis of the prevalence of cough
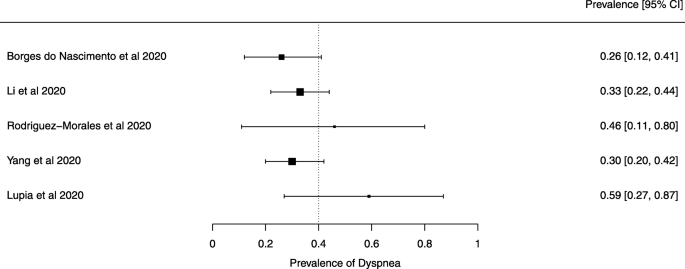
A meta-analysis of the prevalence of dyspnea
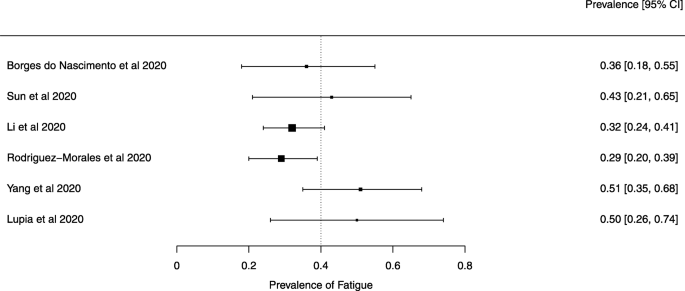
A meta-analysis of the prevalence of fatigue or myalgia
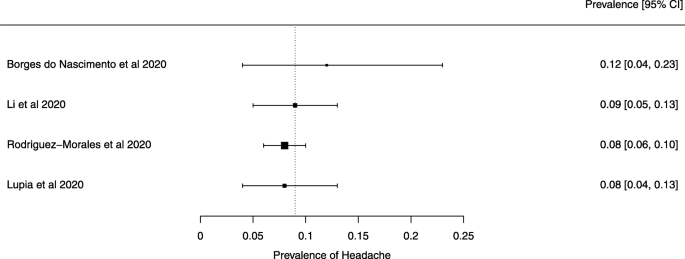
A meta-analysis of the prevalence of headache
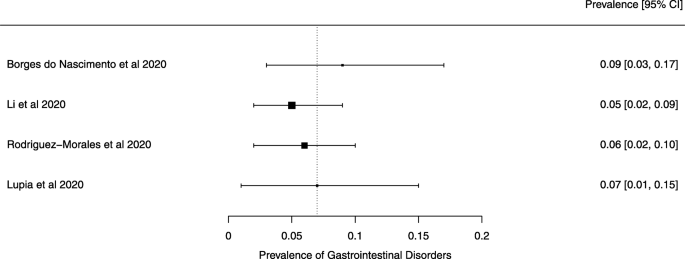
A meta-analysis of the prevalence of gastrointestinal disorders
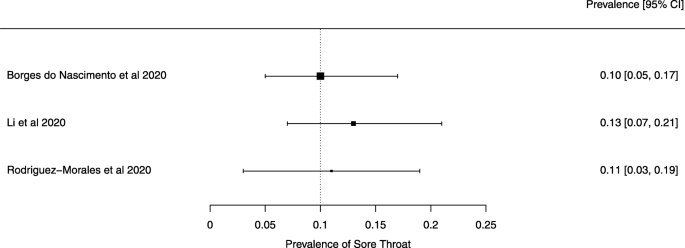
A meta-analysis of the prevalence of sore throat
Diagnostic aspects
Three reviews described methodologies, protocols, and tools used for establishing the diagnosis of COVID-19 [ 26 , 34 , 38 ]. The use of respiratory swabs (nasal or pharyngeal) or blood specimens to assess the presence of SARS-CoV-2 nucleic acid using RT-PCR assays was the most commonly used diagnostic method mentioned in the included studies. These diagnostic tests have been widely used, but their precise sensitivity and specificity remain unknown. One review included a Chinese study with clinical diagnosis with no confirmation of SARS-CoV-2 infection (patients were diagnosed with COVID-19 if they presented with at least two symptoms suggestive of COVID-19, together with laboratory and chest radiography abnormalities) [ 34 ].
Therapeutic possibilities
Pharmacological and non-pharmacological interventions (supportive therapies) used in treating patients with COVID-19 were reported in five reviews [ 25 , 27 , 34 , 35 , 38 ]. Antivirals used empirically for COVID-19 treatment were reported in seven reviews [ 25 , 27 , 34 , 35 , 37 , 38 , 41 ]; most commonly used were protease inhibitors (lopinavir, ritonavir, darunavir), nucleoside reverse transcriptase inhibitor (tenofovir), nucleotide analogs (remdesivir, galidesivir, ganciclovir), and neuraminidase inhibitors (oseltamivir). Umifenovir, a membrane fusion inhibitor, was investigated in two studies [ 25 , 35 ]. Possible supportive interventions analyzed were different types of oxygen supplementation and breathing support (invasive or non-invasive ventilation) [ 25 ]. The use of antibiotics, both empirically and to treat secondary pneumonia, was reported in six studies [ 25 , 26 , 27 , 34 , 35 , 38 ]. One review specifically assessed evidence on the efficacy and safety of the anti-malaria drug chloroquine [ 27 ]. It identified 23 ongoing trials investigating the potential of chloroquine as a therapeutic option for COVID-19, but no verifiable clinical outcomes data. The use of mesenchymal stem cells, antifungals, and glucocorticoids were described in four reviews [ 25 , 34 , 35 , 38 ].
Laboratory and radiological findings
Of the 18 reviews included in this overview, eight analyzed laboratory parameters in patients with COVID-19 [ 25 , 29 , 30 , 32 , 33 , 34 , 35 , 39 ]; elevated C-reactive protein levels, associated with lymphocytopenia, elevated lactate dehydrogenase, as well as slightly elevated aspartate and alanine aminotransferase (AST, ALT) were commonly described in those eight reviews. Lippi et al. assessed cardiac troponin I (cTnI) [ 25 ], procalcitonin [ 32 ], and platelet count [ 33 ] in COVID-19 patients. Elevated levels of procalcitonin [ 32 ] and cTnI [ 30 ] were more likely to be associated with a severe disease course (requiring intensive care unit admission and intubation). Furthermore, thrombocytopenia was frequently observed in patients with complicated COVID-19 infections [ 33 ].
Chest imaging (chest radiography and/or computed tomography) features were assessed in six reviews, all of which described a frequent pattern of local or bilateral multilobar ground-glass opacity [ 25 , 34 , 35 , 39 , 40 , 41 ]. Those six reviews showed that septal thickening, bronchiectasis, pleural and cardiac effusions, halo signs, and pneumothorax were observed in patients suffering from COVID-19.
Quality of evidence in individual systematic reviews
Table 3 shows the detailed results of the quality assessment of 18 systematic reviews, including the assessment of individual items and summary assessment. A detailed explanation for each decision in each review is available in Additional file 5 .
Using AMSTAR 2 criteria, confidence in the results of all 18 reviews was rated as “critically low” (Table 3 ). Common methodological drawbacks were: omission of prospective protocol submission or publication; use of inappropriate search strategy: lack of independent and dual literature screening and data-extraction (or methodology unclear); absence of an explanation for heterogeneity among the studies included; lack of reasons for study exclusion (or rationale unclear).
Risk of bias assessment, based on a reported methodological tool, and quality of evidence appraisal, in line with the Grading of Recommendations Assessment, Development, and Evaluation (GRADE) method, were reported only in one review [ 25 ]. Five reviews presented a table summarizing bias, using various risk of bias tools [ 25 , 29 , 39 , 40 , 41 ]. One review analyzed “study quality” [ 37 ]. One review mentioned the risk of bias assessment in the methodology but did not provide any related analysis [ 28 ].
This overview of systematic reviews analyzed the first 18 systematic reviews published after the onset of the COVID-19 pandemic, up to March 24, 2020, with primary studies involving more than 60,000 patients. Using AMSTAR-2, we judged that our confidence in all those reviews was “critically low”. Ten reviews included meta-analyses. The reviews presented data on clinical manifestations, laboratory and radiological findings, and interventions. We found no systematic reviews on the utility of diagnostic tests.
Symptoms were reported in seven reviews; most of the patients had a fever, cough, dyspnea, myalgia or muscle fatigue, and gastrointestinal disorders such as diarrhea, nausea, or vomiting. Olfactory dysfunction (anosmia or dysosmia) has been described in patients infected with COVID-19 [ 43 ]; however, this was not reported in any of the reviews included in this overview. During the SARS outbreak in 2002, there were reports of impairment of the sense of smell associated with the disease [ 44 , 45 ].
The reported mortality rates ranged from 0.3 to 14% in the included reviews. Mortality estimates are influenced by the transmissibility rate (basic reproduction number), availability of diagnostic tools, notification policies, asymptomatic presentations of the disease, resources for disease prevention and control, and treatment facilities; variability in the mortality rate fits the pattern of emerging infectious diseases [ 46 ]. Furthermore, the reported cases did not consider asymptomatic cases, mild cases where individuals have not sought medical treatment, and the fact that many countries had limited access to diagnostic tests or have implemented testing policies later than the others. Considering the lack of reviews assessing diagnostic testing (sensitivity, specificity, and predictive values of RT-PCT or immunoglobulin tests), and the preponderance of studies that assessed only symptomatic individuals, considerable imprecision around the calculated mortality rates existed in the early stage of the COVID-19 pandemic.
Few reviews included treatment data. Those reviews described studies considered to be at a very low level of evidence: usually small, retrospective studies with very heterogeneous populations. Seven reviews analyzed laboratory parameters; those reviews could have been useful for clinicians who attend patients suspected of COVID-19 in emergency services worldwide, such as assessing which patients need to be reassessed more frequently.
All systematic reviews scored poorly on the AMSTAR 2 critical appraisal tool for systematic reviews. Most of the original studies included in the reviews were case series and case reports, impacting the quality of evidence. Such evidence has major implications for clinical practice and the use of these reviews in evidence-based practice and policy. Clinicians, patients, and policymakers can only have the highest confidence in systematic review findings if high-quality systematic review methodologies are employed. The urgent need for information during a pandemic does not justify poor quality reporting.
We acknowledge that there are numerous challenges associated with analyzing COVID-19 data during a pandemic [ 47 ]. High-quality evidence syntheses are needed for decision-making, but each type of evidence syntheses is associated with its inherent challenges.
The creation of classic systematic reviews requires considerable time and effort; with massive research output, they quickly become outdated, and preparing updated versions also requires considerable time. A recent study showed that updates of non-Cochrane systematic reviews are published a median of 5 years after the publication of the previous version [ 48 ].
Authors may register a review and then abandon it [ 49 ], but the existence of a public record that is not updated may lead other authors to believe that the review is still ongoing. A quarter of Cochrane review protocols remains unpublished as completed systematic reviews 8 years after protocol publication [ 50 ].
Rapid reviews can be used to summarize the evidence, but they involve methodological sacrifices and simplifications to produce information promptly, with inconsistent methodological approaches [ 51 ]. However, rapid reviews are justified in times of public health emergencies, and even Cochrane has resorted to publishing rapid reviews in response to the COVID-19 crisis [ 52 ]. Rapid reviews were eligible for inclusion in this overview, but only one of the 18 reviews included in this study was labeled as a rapid review.
Ideally, COVID-19 evidence would be continually summarized in a series of high-quality living systematic reviews, types of evidence synthesis defined as “ a systematic review which is continually updated, incorporating relevant new evidence as it becomes available ” [ 53 ]. However, conducting living systematic reviews requires considerable resources, calling into question the sustainability of such evidence synthesis over long periods [ 54 ].
Research reports about COVID-19 will contribute to research waste if they are poorly designed, poorly reported, or simply not necessary. In principle, systematic reviews should help reduce research waste as they usually provide recommendations for further research that is needed or may advise that sufficient evidence exists on a particular topic [ 55 ]. However, systematic reviews can also contribute to growing research waste when they are not needed, or poorly conducted and reported. Our present study clearly shows that most of the systematic reviews that were published early on in the COVID-19 pandemic could be categorized as research waste, as our confidence in their results is critically low.
Our study has some limitations. One is that for AMSTAR 2 assessment we relied on information available in publications; we did not attempt to contact study authors for clarifications or additional data. In three reviews, the methodological quality appraisal was challenging because they were published as letters, or labeled as rapid communications. As a result, various details about their review process were not included, leading to AMSTAR 2 questions being answered as “not reported”, resulting in low confidence scores. Full manuscripts might have provided additional information that could have led to higher confidence in the results. In other words, low scores could reflect incomplete reporting, not necessarily low-quality review methods. To make their review available more rapidly and more concisely, the authors may have omitted methodological details. A general issue during a crisis is that speed and completeness must be balanced. However, maintaining high standards requires proper resourcing and commitment to ensure that the users of systematic reviews can have high confidence in the results.
Furthermore, we used adjusted AMSTAR 2 scoring, as the tool was designed for critical appraisal of reviews of interventions. Some reviews may have received lower scores than actually warranted in spite of these adjustments.
Another limitation of our study may be the inclusion of multiple overlapping reviews, as some included reviews included the same primary studies. According to the Cochrane Handbook, including overlapping reviews may be appropriate when the review’s aim is “ to present and describe the current body of systematic review evidence on a topic ” [ 12 ], which was our aim. To avoid bias with summarizing evidence from overlapping reviews, we presented the forest plots without summary estimates. The forest plots serve to inform readers about the effect sizes for outcomes that were reported in each review.
Several authors from this study have contributed to one of the reviews identified [ 25 ]. To reduce the risk of any bias, two authors who did not co-author the review in question initially assessed its quality and limitations.
Finally, we note that the systematic reviews included in our overview may have had issues that our analysis did not identify because we did not analyze their primary studies to verify the accuracy of the data and information they presented. We give two examples to substantiate this possibility. Lovato et al. wrote a commentary on the review of Sun et al. [ 41 ], in which they criticized the authors’ conclusion that sore throat is rare in COVID-19 patients [ 56 ]. Lovato et al. highlighted that multiple studies included in Sun et al. did not accurately describe participants’ clinical presentations, warning that only three studies clearly reported data on sore throat [ 56 ].
In another example, Leung [ 57 ] warned about the review of Li, L.Q. et al. [ 29 ]: “ it is possible that this statistic was computed using overlapped samples, therefore some patients were double counted ”. Li et al. responded to Leung that it is uncertain whether the data overlapped, as they used data from published articles and did not have access to the original data; they also reported that they requested original data and that they plan to re-do their analyses once they receive them; they also urged readers to treat the data with caution [ 58 ]. This points to the evolving nature of evidence during a crisis.
Our study’s strength is that this overview adds to the current knowledge by providing a comprehensive summary of all the evidence synthesis about COVID-19 available early after the onset of the pandemic. This overview followed strict methodological criteria, including a comprehensive and sensitive search strategy and a standard tool for methodological appraisal of systematic reviews.
In conclusion, in this overview of systematic reviews, we analyzed evidence from the first 18 systematic reviews that were published after the emergence of COVID-19. However, confidence in the results of all the reviews was “critically low”. Thus, systematic reviews that were published early on in the pandemic could be categorized as research waste. Even during public health emergencies, studies and systematic reviews should adhere to established methodological standards to provide patients, clinicians, and decision-makers trustworthy evidence.
Availability of data and materials
All data collected and analyzed within this study are available from the corresponding author on reasonable request.
World Health Organization. Timeline - COVID-19: Available at: https://www.who.int/news/item/29-06-2020-covidtimeline . Accessed 1 June 2021.
COVID-19 Dashboard by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (JHU). Available at: https://coronavirus.jhu.edu/map.html . Accessed 1 June 2021.
Anzai A, Kobayashi T, Linton NM, Kinoshita R, Hayashi K, Suzuki A, et al. Assessing the Impact of Reduced Travel on Exportation Dynamics of Novel Coronavirus Infection (COVID-19). J Clin Med. 2020;9(2):601.
Chinazzi M, Davis JT, Ajelli M, Gioannini C, Litvinova M, Merler S, et al. The effect of travel restrictions on the spread of the 2019 novel coronavirus (COVID-19) outbreak. Science. 2020;368(6489):395–400. https://doi.org/10.1126/science.aba9757 .
Article CAS PubMed PubMed Central Google Scholar
Fidahic M, Nujic D, Runjic R, Civljak M, Markotic F, Lovric Makaric Z, et al. Research methodology and characteristics of journal articles with original data, preprint articles and registered clinical trial protocols about COVID-19. BMC Med Res Methodol. 2020;20(1):161. https://doi.org/10.1186/s12874-020-01047-2 .
EPPI Centre . COVID-19: a living systematic map of the evidence. Available at: http://eppi.ioe.ac.uk/cms/Projects/DepartmentofHealthandSocialCare/Publishedreviews/COVID-19Livingsystematicmapoftheevidence/tabid/3765/Default.aspx . Accessed 1 June 2021.
NCBI SARS-CoV-2 Resources. Available at: https://www.ncbi.nlm.nih.gov/sars-cov-2/ . Accessed 1 June 2021.
Gustot T. Quality and reproducibility during the COVID-19 pandemic. JHEP Rep. 2020;2(4):100141. https://doi.org/10.1016/j.jhepr.2020.100141 .
Article PubMed PubMed Central Google Scholar
Kodvanj, I., et al., Publishing of COVID-19 Preprints in Peer-reviewed Journals, Preprinting Trends, Public Discussion and Quality Issues. Preprint article. bioRxiv 2020.11.23.394577; doi: https://doi.org/10.1101/2020.11.23.394577 .
Dobler CC. Poor quality research and clinical practice during COVID-19. Breathe (Sheff). 2020;16(2):200112. https://doi.org/10.1183/20734735.0112-2020 .
Article Google Scholar
Bastian H, Glasziou P, Chalmers I. Seventy-five trials and eleven systematic reviews a day: how will we ever keep up? PLoS Med. 2010;7(9):e1000326. https://doi.org/10.1371/journal.pmed.1000326 .
Lunny C, Brennan SE, McDonald S, McKenzie JE. Toward a comprehensive evidence map of overview of systematic review methods: paper 1-purpose, eligibility, search and data extraction. Syst Rev. 2017;6(1):231. https://doi.org/10.1186/s13643-017-0617-1 .
Pollock M, Fernandes RM, Becker LA, Pieper D, Hartling L. Chapter V: Overviews of Reviews. In: Higgins JPT, Thomas J, Chandler J, Cumpston M, Li T, Page MJ, Welch VA (editors). Cochrane Handbook for Systematic Reviews of Interventions version 6.1 (updated September 2020). Cochrane. 2020. Available from www.training.cochrane.org/handbook .
Higgins JPT, Thomas J, Chandler J, Cumpston M, Li T, Page MJ, et al. Cochrane handbook for systematic reviews of interventions version 6.1 (updated September 2020). Cochrane. 2020; Available from www.training.cochrane.org/handbook .
Pollock M, Fernandes RM, Newton AS, Scott SD, Hartling L. The impact of different inclusion decisions on the comprehensiveness and complexity of overviews of reviews of healthcare interventions. Syst Rev. 2019;8(1):18. https://doi.org/10.1186/s13643-018-0914-3 .
Pollock M, Fernandes RM, Newton AS, Scott SD, Hartling L. A decision tool to help researchers make decisions about including systematic reviews in overviews of reviews of healthcare interventions. Syst Rev. 2019;8(1):29. https://doi.org/10.1186/s13643-018-0768-8 .
Hunt H, Pollock A, Campbell P, Estcourt L, Brunton G. An introduction to overviews of reviews: planning a relevant research question and objective for an overview. Syst Rev. 2018;7(1):39. https://doi.org/10.1186/s13643-018-0695-8 .
Pollock M, Fernandes RM, Pieper D, Tricco AC, Gates M, Gates A, et al. Preferred reporting items for overviews of reviews (PRIOR): a protocol for development of a reporting guideline for overviews of reviews of healthcare interventions. Syst Rev. 2019;8(1):335. https://doi.org/10.1186/s13643-019-1252-9 .
Moher D, Liberati A, Tetzlaff J, Altman DG, PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Open Med. 2009;3(3):e123–30.
Krnic Martinic M, Pieper D, Glatt A, Puljak L. Definition of a systematic review used in overviews of systematic reviews, meta-epidemiological studies and textbooks. BMC Med Res Methodol. 2019;19(1):203. https://doi.org/10.1186/s12874-019-0855-0 .
Puljak L. If there is only one author or only one database was searched, a study should not be called a systematic review. J Clin Epidemiol. 2017;91:4–5. https://doi.org/10.1016/j.jclinepi.2017.08.002 .
Article PubMed Google Scholar
Gates M, Gates A, Guitard S, Pollock M, Hartling L. Guidance for overviews of reviews continues to accumulate, but important challenges remain: a scoping review. Syst Rev. 2020;9(1):254. https://doi.org/10.1186/s13643-020-01509-0 .
Covidence - systematic review software. Available at: https://www.covidence.org/ . Accessed 1 June 2021.
Shea BJ, Reeves BC, Wells G, Thuku M, Hamel C, Moran J, et al. AMSTAR 2: a critical appraisal tool for systematic reviews that include randomised or non-randomised studies of healthcare interventions, or both. BMJ. 2017;358:j4008.
Borges do Nascimento IJ, et al. Novel Coronavirus Infection (COVID-19) in Humans: A Scoping Review and Meta-Analysis. J Clin Med. 2020;9(4):941.
Article PubMed Central Google Scholar
Adhikari SP, Meng S, Wu YJ, Mao YP, Ye RX, Wang QZ, et al. Epidemiology, causes, clinical manifestation and diagnosis, prevention and control of coronavirus disease (COVID-19) during the early outbreak period: a scoping review. Infect Dis Poverty. 2020;9(1):29. https://doi.org/10.1186/s40249-020-00646-x .
Cortegiani A, Ingoglia G, Ippolito M, Giarratano A, Einav S. A systematic review on the efficacy and safety of chloroquine for the treatment of COVID-19. J Crit Care. 2020;57:279–83. https://doi.org/10.1016/j.jcrc.2020.03.005 .
Li B, Yang J, Zhao F, Zhi L, Wang X, Liu L, et al. Prevalence and impact of cardiovascular metabolic diseases on COVID-19 in China. Clin Res Cardiol. 2020;109(5):531–8. https://doi.org/10.1007/s00392-020-01626-9 .
Article CAS PubMed Google Scholar
Li LQ, Huang T, Wang YQ, Wang ZP, Liang Y, Huang TB, et al. COVID-19 patients’ clinical characteristics, discharge rate, and fatality rate of meta-analysis. J Med Virol. 2020;92(6):577–83. https://doi.org/10.1002/jmv.25757 .
Lippi G, Lavie CJ, Sanchis-Gomar F. Cardiac troponin I in patients with coronavirus disease 2019 (COVID-19): evidence from a meta-analysis. Prog Cardiovasc Dis. 2020;63(3):390–1. https://doi.org/10.1016/j.pcad.2020.03.001 .
Lippi G, Henry BM. Active smoking is not associated with severity of coronavirus disease 2019 (COVID-19). Eur J Intern Med. 2020;75:107–8. https://doi.org/10.1016/j.ejim.2020.03.014 .
Lippi G, Plebani M. Procalcitonin in patients with severe coronavirus disease 2019 (COVID-19): a meta-analysis. Clin Chim Acta. 2020;505:190–1. https://doi.org/10.1016/j.cca.2020.03.004 .
Lippi G, Plebani M, Henry BM. Thrombocytopenia is associated with severe coronavirus disease 2019 (COVID-19) infections: a meta-analysis. Clin Chim Acta. 2020;506:145–8. https://doi.org/10.1016/j.cca.2020.03.022 .
Ludvigsson JF. Systematic review of COVID-19 in children shows milder cases and a better prognosis than adults. Acta Paediatr. 2020;109(6):1088–95. https://doi.org/10.1111/apa.15270 .
Lupia T, Scabini S, Mornese Pinna S, di Perri G, de Rosa FG, Corcione S. 2019 novel coronavirus (2019-nCoV) outbreak: a new challenge. J Glob Antimicrob Resist. 2020;21:22–7. https://doi.org/10.1016/j.jgar.2020.02.021 .
Marasinghe, K.M., A systematic review investigating the effectiveness of face mask use in limiting the spread of COVID-19 among medically not diagnosed individuals: shedding light on current recommendations provided to individuals not medically diagnosed with COVID-19. Research Square. Preprint article. doi : https://doi.org/10.21203/rs.3.rs-16701/v1 . 2020 .
Mullins E, Evans D, Viner RM, O’Brien P, Morris E. Coronavirus in pregnancy and delivery: rapid review. Ultrasound Obstet Gynecol. 2020;55(5):586–92. https://doi.org/10.1002/uog.22014 .
Pang J, Wang MX, Ang IYH, Tan SHX, Lewis RF, Chen JIP, et al. Potential Rapid Diagnostics, Vaccine and Therapeutics for 2019 Novel coronavirus (2019-nCoV): a systematic review. J Clin Med. 2020;9(3):623.
Rodriguez-Morales AJ, Cardona-Ospina JA, Gutiérrez-Ocampo E, Villamizar-Peña R, Holguin-Rivera Y, Escalera-Antezana JP, et al. Clinical, laboratory and imaging features of COVID-19: a systematic review and meta-analysis. Travel Med Infect Dis. 2020;34:101623. https://doi.org/10.1016/j.tmaid.2020.101623 .
Salehi S, Abedi A, Balakrishnan S, Gholamrezanezhad A. Coronavirus disease 2019 (COVID-19): a systematic review of imaging findings in 919 patients. AJR Am J Roentgenol. 2020;215(1):87–93. https://doi.org/10.2214/AJR.20.23034 .
Sun P, Qie S, Liu Z, Ren J, Li K, Xi J. Clinical characteristics of hospitalized patients with SARS-CoV-2 infection: a single arm meta-analysis. J Med Virol. 2020;92(6):612–7. https://doi.org/10.1002/jmv.25735 .
Yang J, Zheng Y, Gou X, Pu K, Chen Z, Guo Q, et al. Prevalence of comorbidities and its effects in patients infected with SARS-CoV-2: a systematic review and meta-analysis. Int J Infect Dis. 2020;94:91–5. https://doi.org/10.1016/j.ijid.2020.03.017 .
Bassetti M, Vena A, Giacobbe DR. The novel Chinese coronavirus (2019-nCoV) infections: challenges for fighting the storm. Eur J Clin Investig. 2020;50(3):e13209. https://doi.org/10.1111/eci.13209 .
Article CAS Google Scholar
Hwang CS. Olfactory neuropathy in severe acute respiratory syndrome: report of a case. Acta Neurol Taiwanica. 2006;15(1):26–8.
Google Scholar
Suzuki M, Saito K, Min WP, Vladau C, Toida K, Itoh H, et al. Identification of viruses in patients with postviral olfactory dysfunction. Laryngoscope. 2007;117(2):272–7. https://doi.org/10.1097/01.mlg.0000249922.37381.1e .
Rajgor DD, Lee MH, Archuleta S, Bagdasarian N, Quek SC. The many estimates of the COVID-19 case fatality rate. Lancet Infect Dis. 2020;20(7):776–7. https://doi.org/10.1016/S1473-3099(20)30244-9 .
Wolkewitz M, Puljak L. Methodological challenges of analysing COVID-19 data during the pandemic. BMC Med Res Methodol. 2020;20(1):81. https://doi.org/10.1186/s12874-020-00972-6 .
Rombey T, Lochner V, Puljak L, Könsgen N, Mathes T, Pieper D. Epidemiology and reporting characteristics of non-Cochrane updates of systematic reviews: a cross-sectional study. Res Synth Methods. 2020;11(3):471–83. https://doi.org/10.1002/jrsm.1409 .
Runjic E, Rombey T, Pieper D, Puljak L. Half of systematic reviews about pain registered in PROSPERO were not published and the majority had inaccurate status. J Clin Epidemiol. 2019;116:114–21. https://doi.org/10.1016/j.jclinepi.2019.08.010 .
Runjic E, Behmen D, Pieper D, Mathes T, Tricco AC, Moher D, et al. Following Cochrane review protocols to completion 10 years later: a retrospective cohort study and author survey. J Clin Epidemiol. 2019;111:41–8. https://doi.org/10.1016/j.jclinepi.2019.03.006 .
Tricco AC, Antony J, Zarin W, Strifler L, Ghassemi M, Ivory J, et al. A scoping review of rapid review methods. BMC Med. 2015;13(1):224. https://doi.org/10.1186/s12916-015-0465-6 .
COVID-19 Rapid Reviews: Cochrane’s response so far. Available at: https://training.cochrane.org/resource/covid-19-rapid-reviews-cochrane-response-so-far . Accessed 1 June 2021.
Cochrane. Living systematic reviews. Available at: https://community.cochrane.org/review-production/production-resources/living-systematic-reviews . Accessed 1 June 2021.
Millard T, Synnot A, Elliott J, Green S, McDonald S, Turner T. Feasibility and acceptability of living systematic reviews: results from a mixed-methods evaluation. Syst Rev. 2019;8(1):325. https://doi.org/10.1186/s13643-019-1248-5 .
Babic A, Poklepovic Pericic T, Pieper D, Puljak L. How to decide whether a systematic review is stable and not in need of updating: analysis of Cochrane reviews. Res Synth Methods. 2020;11(6):884–90. https://doi.org/10.1002/jrsm.1451 .
Lovato A, Rossettini G, de Filippis C. Sore throat in COVID-19: comment on “clinical characteristics of hospitalized patients with SARS-CoV-2 infection: a single arm meta-analysis”. J Med Virol. 2020;92(7):714–5. https://doi.org/10.1002/jmv.25815 .
Leung C. Comment on Li et al: COVID-19 patients’ clinical characteristics, discharge rate, and fatality rate of meta-analysis. J Med Virol. 2020;92(9):1431–2. https://doi.org/10.1002/jmv.25912 .
Li LQ, Huang T, Wang YQ, Wang ZP, Liang Y, Huang TB, et al. Response to Char’s comment: comment on Li et al: COVID-19 patients’ clinical characteristics, discharge rate, and fatality rate of meta-analysis. J Med Virol. 2020;92(9):1433. https://doi.org/10.1002/jmv.25924 .
Download references
Acknowledgments
We thank Catherine Henderson DPhil from Swanscoe Communications for pro bono medical writing and editing support. We acknowledge support from the Covidence Team, specifically Anneliese Arno. We thank the whole International Network of Coronavirus Disease 2019 (InterNetCOVID-19) for their commitment and involvement. Members of the InterNetCOVID-19 are listed in Additional file 6 . We thank Pavel Cerny and Roger Crosthwaite for guiding the team supervisor (IJBN) on human resources management.
This research received no external funding.
Author information
Authors and affiliations.
University Hospital and School of Medicine, Universidade Federal de Minas Gerais, Belo Horizonte, Minas Gerais, Brazil
Israel Júnior Borges do Nascimento & Milena Soriano Marcolino
Medical College of Wisconsin, Milwaukee, WI, USA
Israel Júnior Borges do Nascimento
Helene Fuld Health Trust National Institute for Evidence-based Practice in Nursing and Healthcare, College of Nursing, The Ohio State University, Columbus, OH, USA
Dónal P. O’Mathúna
School of Nursing, Psychotherapy and Community Health, Dublin City University, Dublin, Ireland
Department of Anesthesiology, Intensive Care and Pain Medicine, University of Münster, Münster, Germany
Thilo Caspar von Groote
Department of Sport and Health Science, Technische Universität München, Munich, Germany
Hebatullah Mohamed Abdulazeem
School of Health Sciences, Faculty of Health and Medicine, The University of Newcastle, Callaghan, Australia
Ishanka Weerasekara
Department of Physiotherapy, Faculty of Allied Health Sciences, University of Peradeniya, Peradeniya, Sri Lanka
Cochrane Croatia, University of Split, School of Medicine, Split, Croatia
Ana Marusic, Irena Zakarija-Grkovic & Tina Poklepovic Pericic
Center for Evidence-Based Medicine and Health Care, Catholic University of Croatia, Ilica 242, 10000, Zagreb, Croatia
Livia Puljak
Cochrane Brazil, Evidence-Based Health Program, Universidade Federal de São Paulo, São Paulo, Brazil
Vinicius Tassoni Civile & Alvaro Nagib Atallah
Yorkville University, Fredericton, New Brunswick, Canada
Santino Filoso
Laboratory for Industrial and Applied Mathematics (LIAM), Department of Mathematics and Statistics, York University, Toronto, Ontario, Canada
Nicola Luigi Bragazzi
You can also search for this author in PubMed Google Scholar
Contributions
IJBN conceived the research idea and worked as a project coordinator. DPOM, TCVG, HMA, IW, AM, LP, VTC, IZG, TPP, ANA, SF, NLB and MSM were involved in data curation, formal analysis, investigation, methodology, and initial draft writing. All authors revised the manuscript critically for the content. The author(s) read and approved the final manuscript.
Corresponding author
Correspondence to Livia Puljak .
Ethics declarations
Ethics approval and consent to participate.
Not required as data was based on published studies.
Consent for publication
Not applicable.
Competing interests
The authors declare no conflict of interest.
Additional information
Publisher’s note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: appendix 1..
Search strategies used in the study.
Additional file 2: Appendix 2.
Adjusted scoring of AMSTAR 2 used in this study for systematic reviews of studies that did not analyze interventions.
Additional file 3: Appendix 3.
List of excluded studies, with reasons.
Additional file 4: Appendix 4.
Table of overlapping studies, containing the list of primary studies included, their visual overlap in individual systematic reviews, and the number in how many reviews each primary study was included.
Additional file 5: Appendix 5.
A detailed explanation of AMSTAR scoring for each item in each review.
Additional file 6: Appendix 6.
List of members and affiliates of International Network of Coronavirus Disease 2019 (InterNetCOVID-19).
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ . The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
Reprints and permissions
About this article
Cite this article.
Borges do Nascimento, I.J., O’Mathúna, D.P., von Groote, T.C. et al. Coronavirus disease (COVID-19) pandemic: an overview of systematic reviews. BMC Infect Dis 21 , 525 (2021). https://doi.org/10.1186/s12879-021-06214-4
Download citation
Received : 12 April 2020
Accepted : 19 May 2021
Published : 04 June 2021
DOI : https://doi.org/10.1186/s12879-021-06214-4
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Coronavirus
- Evidence-based medicine
- Infectious diseases
BMC Infectious Diseases
ISSN: 1471-2334
- Submission enquiries: [email protected]
- General enquiries: [email protected]
- - Google Chrome
Intended for healthcare professionals
- My email alerts
- BMA member login
- Username * Password * Forgot your log in details? Need to activate BMA Member Log In Log in via OpenAthens Log in via your institution

Search form
- Advanced search
- Search responses
- Search blogs
- News & Views
- What are the latest...
What are the latest covid drugs and treatments?
- Related content
- Peer review
- Mun-Keat Looi , international features editor
- mlooi{at}bmj.com
Vaccines have taken up much of the spotlight, but where have we got to with covid-19 treatments, asks Mun-Keat Looi —and is there a global standard of care?
What are the best treatments for covid-19?
Written in cooperation with the World Health Organization, The BMJ ’s living systematic review is a meta-analysis comparing the effects of treatments for covid-19, 1 using data from more than 400 randomised clinical trials worldwide.
At the time of writing, it states that systemic corticosteroids (particularly dexamethasone), interleukin-6 receptor antagonists (such as tocilizumab), and Janus kinase inhibitors (such as baricitinib) reduce mortality and have other benefits in patients with severe covid-19, such as reducing the length of hospital stay and the time needed on a ventilator. It also notes that the antivirals molnupiravir (Lagevrio), nirmatrelvir/ritonavir (Paxlovid), and remdesivir (Veklury) have also been shown to be effective against non-severe covid-19.
How has treatment advice changed during the pandemic?
What is considered the “best” treatment continues to change as the pandemic progresses. Where previously the primary aim was to prevent death, the world’s exposure to covid-19 now means that outcomes are increasingly viewed in terms of reducing hospital admissions, disease severity, and perhaps even transmission.
Molnupiravir is a case in point. A study published in December 2022 involving 25 000 people confirmed that oral molnupiravir was associated with reduced viral detection and load, and patients recovered around four days more quickly than those who received usual care. However, it didn’t reduce hospital admissions or deaths among vaccinated high risk patients, which was the primary outcome the trial was set up to test. 2
Chris Butler, clinical director of the University of Oxford’s Primary Care Clinical Trials Unit and co-chief investigator of the study, tells The BMJ that although the trial found no benefit from molnupiravir for its primary outcome (to reduce the likelihood of hospital admission or death), it could have other benefits such as a faster recovery time and reduced follow-up with health services. “This could help to ease the burden on UK health services through the treatment of selected patients at home, during times of high disease burden and pressure on key services,” he says.
Janet Scott, clinical lecturer in infectious diseases at the University of Glasgow, says, “The vaccines are now doing their job and reducing the severity of infection in the high risk groups, so the benefit of molnupiravir is now more about time to recovery than reducing hospitalisation.” Whether the benefits are worth the £577 it costs for the five day course will depend on whether it reduces the number of people who go on to develop long covid, and those results are still being analysed.
“In my view there are currently two major challenges in covid-19 treatment,” adds Scott. “The prevention and treatment of long covid, and the prevention and treatment of acute covid-19 in the highest risk groups including immunosuppressed people. This immunosuppressed group is likely going to require a bespoke study focusing on this issue.”
Does the standard of care differ around the world?
Although there are recommended standard treatments for acute covid-19 in line with WHO’s advice, huge differences in access mean that countries and regions are not consistent.
“The consistency around the globe is probably not what we would want at this point,” says Janet Diaz, who leads clinical management at the WHO Health Emergencies Programme. “Of all the drugs that we have available, the one that’s most consistently available and used globally is corticosteroids—what we use for patients who have severe or critical covid-19. But I think for the remainder of the drugs that WHO has recommended—such as interleukin-6 receptor blockers, tocilizumab or baricitinib, and oral antivirals—the availability and access is limited in many low and middle income countries, and that has unfortunately probably impacted their use.”
There are many reasons behind this, but the upshot is that with limited access and supplies the cost becomes a major factor, as governments apply more scrutiny over evidence of efficacy. With antivirals, for instance, it comes down to how much a government has invested in buying up the various licensed therapies (mainly Paxlovid and molnupiravir), says Stephen Griffin, reader at the University of Leeds. He points out that the European Union still hasn’t approved molnupiravir, which shows mixed efficacy data.
Some places are still widely using drugs that have been shown to be ineffective, such as antibiotics and ivermectin—the latter still commonly used in Brazil, for instance. 3 Butler says that this variation in care can be justified to some extent by different vaccination rates, deprivation and nutrition, coinfection with other organisms, and problems in accessing modern antivirals. “But overall, I think there’s a lot of practice that is still not evidence based going on around the world,” he says.
Butler adds, “It’s also really important not to assume that the evidence from small trials done by the pharma company translates into evidence at scale in every other context in every other country, particularly since the phenotype of the illness varies so much: covid is a very different illness when the population is vaccinated and when there’s a different strain around.
“We’ve got to do the research to make sure that we are generating evidence from within the context. We need evidence from the intended use population before we start giving out drugs at scale.”
He cites inhaled budesonide, a steroid, which does have a benefit in terms of recovery and shows a high probability of reducing the need for hospital admission. 4 “That drug is being used in some places, though it wasn’t approved in the UK,” he says. “But it is an option in other places.”
Why don’t we have better data on covid treatments?
“We have few head-to-head trials of medications, or comparisons of different combinations of medications,” says Tari Turner, director of the National Covid-19 Clinical Evidence Taskforce at Monash University in Australia. “As a result, we have a small shopping list of effective drug treatment options, and little reliable information to guide decisions about which drugs should be used first or in which sequence or combination they should be used.”
Griffin says that the development of direct acting drugs was hampered by the initial response to covid-19, which focused on repurposing existing drugs since that was a faster route. “Back in 2020, we had to try and find any antiviral that worked against this virus—that’s why remdesivir and molnupiravir was used, as they had been tried before on different sorts of viruses,” he says. “There was data on things like interferon beta combined with lopinavir and ritonavir [having efficacy] in vitro, and there was a paper that showed favipiravir worked, but not very well.
“Basically, everything that was in a fairly bare antivirals cupboard was thrown at it in cell culture. That was fine at the time, as it identified lots of decent hits. But what they didn’t do was really carry through the validation process particularly well. And we ended up with things like hydroxychloroquine and ivermectin that, rather than repurposed, were mis-purposed.”
Antivirals have become caught up in this confusion because, says Griffin, their pricing in comparison with drugs such as ivermectin means that “some quarters believe that the pharmacy companies are trying to thrust expensive drugs down our throats, rather than use cheap, effective alternatives.” The monopoly of western drug companies—Pfizer with Paxlovid, for instance—hasn’t helped.
However, Diaz says that big pharma is playing its part. She says that the US Food and Drug Administration’s partnership for covid-19 drugs “has a therapeutic pillar, and many partners have been trying to advance on negotiations with manufacturers to have fair, transparent pricing and to secure doses and treatment courses for people in poorer, low middle income countries, and also to increase generic production of products.
“I think next year there will be more generic products on the horizon, which will be associated with better pricing of these drugs—and, I think at that point, more access.”
How might treatment advice change further in the coming months?
The BMJ ’s living systematic review is updated regularly as evidence continues to be published. 1 For instance, in December 2022 the Remap-Cap study of long term (180 day) outcomes in critically ill patients with covid-19 found that the benefit of interleukin-6 receptor antagonists persisted at six months. 5 Martin Landray, professor of medicine and epidemiology at Oxford Population Health, University of Oxford, says that while the results raised the possibility that antiplatelet treatment in patients with severe covid-19 would reduce long term mortality, this was not “conclusive.”
“It would be wise to wait for the results of the [10 times larger] study of aspirin in the Recovery trial,” he said. “These results, including around 18 months of follow-up, should be available early in 2023, along with the results for four treatments that have previously been shown to reduce 28 day mortality: dexamethasone, tocilizumab [an interleukin-6 receptor antagonist], baricitinib, and monoclonal antibody treatment.”
Do you have a “Covid Unanswered Question”? Email mlooi{at}bmj.com , and we’ll try to cover it in a future instalment.
Competing interests: None.
Provenance and peer review: Commissioned; externally peer reviewed.
This article is made freely available for personal use in accordance with BMJ's website terms and conditions for the duration of the covid-19 pandemic or until otherwise determined by BMJ. You may download and print the article for any lawful, non-commercial purpose (including text and data mining) provided that all copyright notices and trade marks are retained.
- Siemieniuk RAC ,
- Bartoszko JJ ,
- Zeraatkar D ,
- Gbinigie O ,
- Hentschke-Lopes M ,
- Botton MR ,
- Freitas M ,
- Mancuso ACB ,
- Higgins AM ,
- Lorenzi E ,
- Writing Committee for the REMAP-CAP Investigators
COVID-19 Research
Stanford Medicine scientists have launched dozens of research projects as part of the global response to COVID-19. Some aim to prevent, diagnose and treat the disease; others aim to understand how it spreads and how people’s immune systems respond to it.
Below is a curated selection, including summaries, of the projects.
To participate in research , browse COVID-19 studies . Our research registry also connects people like you with teams conducting research to make advances in health care. If you are eligible for a study, researchers may contact you to provide additional details on how to participate.
By participating in clinical research, you help accelerate medical science by providing valuable insights into potential treatments and methons of prevention.
Stanford COVID-19 Study Directory Stanford Medicine Research Registry
To improve our ability to determine who has COVID-19 and treat those infected.
Transmission
To better prevent and understand the transmission of the coronavirus.
Vaccination and Treatment
To improve our ability to prevent COVID-19 and treat those infected.
Epidemiology
To better understand how the coronavirus is spreading.
Data Science and Modeling
To better predict medical, fiscal and resource-related outcomes of the COVID-19 pandemic.
To better understand immune responses to the coronavirus.
Cardiovascular
To better understand the way the virus affects the cardiovascular system.
To better enable the workforce to achieve its goals during the COVID-19 pandemic.
Miscellaneous
A variety of other research projects related to the COVID-19 pandemic.
The list isn’t comprehensive and instead represents a portion of Stanford Medicine research on COVID-19. If you are a Stanford Medicine scientist and would like to see your research included here, please send a note to: [email protected].
The Stanford Institute for Human-Centered Artificial Intelligence has also created a webpage for COVID-19 research collaborations and other opportunities, such as research positions, internships and funding. If you would like to submit an opening please use the following form and they will post it on their website.
Support Stanford Medicine’s response to COVID-19 by making a gift .
COVID-19 Research Projects
The COVID-19 research landscape: Measuring topics and collaborations using scientific literature
Affiliations.
- 1 Institute of Medical Information, Chinese Academy of Medical Sciences.
- 2 Digital China Health Technologies Co. Ltd., Beijing, China.
- PMID: 33120818
- PMCID: PMC7581087
- DOI: 10.1097/MD.0000000000022849
Objectives: The Coronavirus Disease 2019 (COVID-19) caused heavy burdens and brought tremendous challenges to global public health. This study aimed to investigate collaboration relationships, research topics, and research trends on COVID-19 using scientific literature.
Method: COVID-19-related articles published from January 1 to July 1, 2020 were retrieved from PubMed database. A total of 27,370 articles were included. Excel 2010, Medical Text Indexer (MTI), VOSviewer, and D3.js were used to summarize bibliometric features.
Results: The number of the COVID-19 research publications has been continuously increasing after its break. United States was the most productive and active country for COVID-19 research, with the largest number of publications and collaboration relationships. Huazhong University of Science and Technology from China was the most productive institute on the number of publications, and University of Toronto from Canada ranked as Top 1 institute for global research collaboration. Four key research topics were identified, of which the topic of epidemiology and public health interventions has gathered highest attentions. Topic of virus infection and immunity has been more focused during the early stage of COVID-19 outbreak compared with later stage. The topic popularity of clinical symptoms and diagnosis has been steady.
Conclusions: Our topic analysis results revealed that the study of drug treatment was insufficient. To achieve critical breakthroughs of this research area, more interdisciplinary, multi-institutional, and global research collaborations are needed.
Publication types
- Systematic Review
- Betacoronavirus*
- Bibliometrics*
- Biomedical Research / trends*
- Coronavirus Infections* / diagnosis
- Coronavirus Infections* / epidemiology
- Coronavirus Infections* / therapy
- Global Health
- Intersectoral Collaboration
- Pneumonia, Viral* / diagnosis
- Pneumonia, Viral* / epidemiology
- Pneumonia, Viral* / therapy
- Publishing / trends*
- Research Design / trends
Marie Skłodowska-Curie Actions
Projects researching covid–19, sars-cov-2 and related topics.
The current COVID-19 outbreak has not caught EU-funded research off guard. The Marie Skłodowska-Curie Actions (MSCA) of the European Commission are supporting outstanding researchers in finding solutions to challenges posed by the novel coronavirus disease COVID-19 and other infectious diseases.
This page will be regularly updated with MSCA projects, results and testimonials relevant to COVID-19, SARS-CoV-2 and related topics.
DIAGNOSTICS AND TREATMENTS (including vaccines)
Diabetic nephropathy modelling in hesc-derived 3d kidney organoids.
EPIORGABOLISM is studying how SARS-Co-V2, the coronavirus responsible for the 2019 novel coronavirus disease (COVID-19), interacts with and infects kidney cells. Together with the lung, the kidney is one of the main organs affected by the COVID-19 disease. Dr Carmen Hurtado, researcher of EPIORGABOLISM, is currently generating human kidney organoids from human pluripotent stem cells.
The use of human organoids allows to test treatments against coronavirus in an agile way, dramatically reducing the time human drug trials take. Hurtado is part of international research team has identified a drug capable of blocking the effects of the SARS-CoV-2 virus. The findings have been partially supported by EPIORGABOLISM and published in the journal ‘Cell’.
Find out more
- Trial drug shows promise in fighting coronavirus
- Watch the testimonial of Carmen Hurtado , researcher of the EPIORGABOLISM project.
Host switching pathogens, infectious outbreaks and zoonosis; a Marie Sklodowska-Curie Training Network
HONOURs is teaching 15 talented young researchers, including coronavirologists, to become “preparedness-experts”. The project involves 11 laboratories, all at the forefront of novel virus investigations and characterizations. HONOUR reacted in January 2020, immediately after the emergence of COVID-19, by starting work on SARS-CoV-2. A synthetic biology virus culture system was developed to swiftly evaluate therapy options, next to rapid tests to determine virus shedding on location. The quality of protective immunity was evaluated, and a search started on the most suitable animal model to battle the virus and provide therapy options. HONOURs is devoting its expert knowledge to fight this coronavirus and provide therapy options.
- HONOURs: Virus Outbreak Preparedness and COVID-19
- Visit the HONOURs website
INnate-ImmunomeTabolIsm as Antiviral TargEt
The global COVID-19 pandemic highlights an urgent need for innovation in the development of novel antiviral strategies and therapies. INITIATE has recruited 15 young PhD candidates to become experts in the field of antiviral immunometabolism, with a focus on RNA viruses – including coronaviruses. While it is clear that viral replication, metabolic pathways, and host immune responses are tightly interconnected, the host molecular pathways that impact viral pathogenesis are not well-defined. With the emergence of COVID-19, eight of the INITIATE projects have included SARS-CoV-2 in their research programs to understand coronavirus molecular virology, the role of the host immune response in driving COVID-19 immunopathogenesis and the potential of targeting host metabolism as therapeutic strategies.

Organoids for Virus Research - An innovative training-ETN programme
ORGANOVIR is contributing to COVID-19 research in a variety of ways, and several of its researchers are currently working on the development of new antivirals to combat the disease. Researchers at KU Leuven (Belgium) are studying the way in which coronaviruses evolve, and are searching out possible targets for further remedies. The project also investigating active substances – or a combination of them – in existing medicines that could be effective against SARS-CoV-2. ORGANOVIR is also conducting pre-clinical tests for a vaccine against COVID-19 using a technology based on the yellow fever vaccine.
In parallel, a group of researchers at the Jagiellonian University (Poland) is studying the infection on the single-cell and tissue level in different organs and cell types, working on virus inhibitors and collaborating with companies to create a point of care diagnostics based on different platforms. The group is also studying the course of the pandemic in Poland and monitoring the virus variability in the country.
ORGANOVIR’s coordinators have been intensively working on clinical and diagnostic tasks and set up new COVID-19 research at the Amsterdam UMC (The Netherlands). This has resulted in the launch of COVID-KIDS, a study on immunity in children, and the use of 3D culture models for COVID-19 studies.
- Read about the COVID-19 activities of ORGANOVIR partners
- Read the testimonial of Mariana Guedes , researcher for the OrganoVir project
- Read the testimonial of Thuc Do , researcher for the OrganoVir project
- Air-liquid interface cultures of nasal epithelial cells to investigate factors critical for viral entry into host cells
MECHANISMS OF INFECTION, IMMUNE REACTIONS AND HOST-PATHAGEN INTERACTION
Unravelling species barriers of coronaviruses.
COV RESTRIC targeted the precise mechanisms that allow coronaviruses to jump across species. Dr Stephanie Pfänder, researcher of COV RESTRIC, worked on various virological aspects of emerging viruses – with a focus on emerging coronaviruses. Her work has the potential to lead to novel strategies to protect cells against coronavirus infection. This is crucial to fight the COVID-19 pandemic – and to help insulate society against future coronavirus outbreaks.
- Read the testimonial of Stephanie Pfänder , researcher of the COV RESTRIC project.
- Host proteins involved in species barriers of viral infections
DIGITAL TOOLS, DATA AND MODELLING
Research and innovation staff exchange network of european data scientists.
The NeEDS consortium is currently focusing on the emerging data challenges that come with the COVID-19 pandemic. In Spain, the first cases of the COVID-19 pandemic were confirmed late February 2020 and data started to be collected daily by the different regions. Data and Data Science tools turned out to be crucial to assist decision makers in this highly uncertain context. NeEDS and the scientific collaborations they enjoy were fundamental to create a working group of data scientists from different European universities, which has developed an Artificial Intelligence tool to provide short-term predictions of the pandemic’s evolution. With this novel methodology, NeEDS as contributed to the cooperative efforts coordinated by the Spanish Commission of Mathematics to support data-driven decision making related to the COVID19 pandemic. In a recent interview , Project Coordinator Dolores Romero Morales has reflected on the potential of the NeEDS expertise and the efforts of tackling these data challenges within the team. The consortium is tackling other important Data Science questions, e.g., using spatial data to support COVID19 information apps or addressing the pressing data privacy needs.
- Read about the COVID-19 activities of NeEDS and its partners
- On Sparse Ensemble Methods: An Application to Short-Term Predictions of the Evolution of COVID-19
- Read the testimonials of Remedios Sillero, Cristina Molero and Sandra Benitez , seconded researchers for the NeEDS project.
Pan-genome Graph Algorithms and Data Integration
Researchers involved in PANGAIA are investigating how massive amounts of genome sequence data can be ordered and analysed for their use in biomedicine. Their work has important implications in areas such as bacteria and virus research, investigation of drug resistance mechanisms and vaccine development: big data technology can help to identify the characteristics of new strains of viruses such as SARS-CoV-2 and bacteria by comparing their genomes.
- Identifying large data sets to help coronavirus research
- Identifying pathogenic genes in virus strains at a glance
Modelling Infectious Diseases in Dynamic, relocated, refugee populations
In order to assist policy-makers in mitigating outbreaks, MIDIDP has created realistic models to simulate the spread of infectious diseases in under-vaccinated refugee populations in Europe and neighbouring countries. Dr Hasan Güçlü, researcher of MIDIDP, has created a model that simulates the spread of COVID-19 in populations with variable demographics.
- Read the testimonial of Hasan Güçlü , researcher of the MIDIDP project.
PUBLIC HEALTH, PREPAREDNESS AND RESPONSE
Disability and disease during the 1918 influenza pandemic: implications for preparedness policies.
As the current COVID-19 pandemic shows, people with disabilities are at increased risk for complications and death as they are often neglected in epidemic responses. Dr Jessica Dimka, researcher of DIS2 , is exploring disability as a risk factor in pandemics. Using the 1918 Spanish influenza pandemic as a model, the project seeks to promote more equitable public health plans and interventions. Dimka points out that people with disabilities must be considered in all pandemic strategies: their lives, livelihoods and rights are not expendable.
- Read the testimonial of Jessica Dimka , researcher of the DIS2 project.
MULTIDISCIPLINARY PROJECTS
Protecting human rights and public health in global pandemics.
THEMIS is an interdisciplinary research project that reacts to the increasing occurrence of global pandemics, like the caused by the present COVID-19 disease, and restrictive public health measures taken to respond to these threats. Using a rights-based approach, Dr Patrycja Dąbrowska-Kłosińska, researcher of THEMIS, intends to create a better understanding of how to prepare for, and respond to, global pandemics.
The project seeks to offer a vital reference for policy-making at national, regional and global levels – one that prioritises fair pandemic preparedness to cross-border health threats. The project has offered critical guidance during the current COVID-19 pandemic, which has required a previously unimagined scale of coordinated, public health-control measures as well as consideration of human-rights implications worldwide.
- Read the testimonial of Patrycja Dąbrowska-Kłosińska , researcher of the THEMIS project.
Martí I Franquès COFUND
Since the emergence of COVID-19, several fellows involved in the Martí Franquès Programme (MFP) have been working on solutions to the current crisis. Researchers are developing an epidemiological mathematical model that infers the status of the epidemic, thereby monitoring and estimating the impact of interventions on the spread of COVID-19.
In parallel, another group of researchers is implementing an original virtual screening protocol to reposition approved drugs. This would allow predicting which of them could inhibit the main protease of the virus (M-pro), a key target for antiviral drugs given its essential role in the virus replication.
- Read the testimonial of Benjamin Steinegger , whose research is developing a mathematical framework to monitor and estimate the impact of interventions on the COVID-19 pandemic.
Project outcomes
- Modelling the impact of interventions on the spread of COVID-19
- Prediction of novel inhibitors of the main protease of SARS-CoV-2
- See all the results relevant to COVID-19 produced by MFP fellows
The launch of a new industrial PhD programme at EPFL
Several fellows involved in the EPFLinnovators project are working on solutions to COVID-19 since the start of the crisis. Teams of researchers are developing subunit vaccines against the SARS-CoV-2 virus, investigating the potential use of cyclodextrin derivatives to prevent and treat the infections caused by SARS-CoV-2, and analysing the mechanical aspects of SARS-CoV-2 entry into cells.
- Read the testimonial of Xiaomeng Hu , researcher of the EPFLInnovators project.
- Subunit vaccines against SARS-CoV-2
- Non-toxic cyclodextrin derivative against viruses at micromolar concentration
- Variations in clathrin mediated endocytosis on a mammalian cell membrane
SOCIAL BEHAVIOUR AND IMPACT
Leading fellows.
Over the last decade, the reliance on online products and services has steadily increased, but since the beginning of the COVID-19 pandemic it has escalated to an unprecedented level. Dr Matthew Dennis, researcher of the LEaDing Fellows COFUND project at TU Delft (the Netherlands), examines the ethical implications and value trade-offs as societies attempt to transition across the digital divide. His project highlights that an ethical reflection on this digital transition is urgently needed, as digital solutions to problems generated by COVID-19 may create winners and losers – likely disproportionately affecting vulnerable users. By addressing these issues, the pandemic may foster the kind of social and political interconnectedness that was envisioned at the start of the crisis.
- Read the testimonial of Matthew James Dennis , researcher of the LEaDing Fellows project.
MSCA on social media
The MSCA social media are continuously updated with testimonials of MSCA fellows, supervisors, coordinators and projects working to find solutions to challenges posed by COVID-19 and other infectious diseases.
- MSCA on Twitter
- MSCA Facebook page
Related content
Related information, thanks for your feedback.
We are happy to see that your experience was positive. Don't forget to share the pages you like with your friends and colleagues.
If you need to ask a question, please contact Europe direct .
- Frontiers in Immunology
- Vaccines and Molecular Therapeutics
- Research Topics
mRNA Design, Manufacturing, Delivery, and Applications in Medicine
Total Downloads
Total Views and Downloads
About this Research Topic
In vitro transcribed mRNA has lived up to its potential as a game-changing disruptive vaccine modality during the COVID-19 pandemic. As a result, research into mRNA therapeutics, which extends beyond the realm of vaccines to fight infectious diseases, has gained increasing attention both in academia and industry. Indeed, mRNA therapeutics have been explored beyond vaccines and other immune modulating strategies, typically used in context of infectious diseases and cancer, entering indications such as genetic disorders and heart failure. A number of challenges need to be overcome for mRNA to fulfil its role as a well-established therapeutic modality applicable to broad relevance to a variety of diseases. Technology advances to adapt the mRNA design to the intended application, to facilitate mRNA manufacturing, to improve mRNA delivery to specific cell types in situ and to avoid unintended reactogenicity coinciding with an in-depth understanding of how mRNA interacts with different cellular components mediating recognition, stability versus degradation and translation are some of the research areas that merit attention. We welcome research articles as well as comprehensive reviews that cover significant research findings in key areas of research, including but not limited to: 1. Engineering of the mRNA molecular structure to improve protein expression versus antigen processing and presentation, including alternative approaches to mRNA such as chemical, circular and self-amplifying RNA for therapeutic agents; 2. Development of technology platforms to synthetize, purify and analyse mRNA; 3. Development of novel mRNA delivery systems, including tissue and cell-targeted mRNA delivery; 4. Reducing the reactogenicity/immunogenicity of mRNA; 5. Application of mRNA to resolve diseases, including infectious disease, cancer, genetic disorders, etc.
Keywords : RNA engineering, stability, translation, fidelity, RNA synthesis, purification, quality control methods, RNA formulations, intracellular transport, RNA reactogenicity/immunogenicity, Applications of RNA, infectious diseases, cancer
Important Note : All contributions to this Research Topic must be within the scope of the section and journal to which they are submitted, as defined in their mission statements. Frontiers reserves the right to guide an out-of-scope manuscript to a more suitable section or journal at any stage of peer review.
Topic Editors
Topic coordinators, submission deadlines, participating journals.
Manuscripts can be submitted to this Research Topic via the following journals:
total views
- Demographics
No records found
total views article views downloads topic views
Top countries
Top referring sites, about frontiers research topics.
With their unique mixes of varied contributions from Original Research to Review Articles, Research Topics unify the most influential researchers, the latest key findings and historical advances in a hot research area! Find out more on how to host your own Frontiers Research Topic or contribute to one as an author.
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- Medicine (Baltimore)
- v.99(43); 2020 Oct 23
The COVID-19 research landscape
Junhui wang.
a Institute of Medical Information, Chinese Academy of Medical Sciences
b Digital China Health Technologies Co. Ltd., Beijing, China.
Objectives:
The Coronavirus Disease 2019 (COVID-19) caused heavy burdens and brought tremendous challenges to global public health. This study aimed to investigate collaboration relationships, research topics, and research trends on COVID-19 using scientific literature.
COVID-19-related articles published from January 1 to July 1, 2020 were retrieved from PubMed database. A total of 27,370 articles were included. Excel 2010, Medical Text Indexer (MTI), VOSviewer, and D3.js were used to summarize bibliometric features.
The number of the COVID-19 research publications has been continuously increasing after its break. United States was the most productive and active country for COVID-19 research, with the largest number of publications and collaboration relationships. Huazhong University of Science and Technology from China was the most productive institute on the number of publications, and University of Toronto from Canada ranked as Top 1 institute for global research collaboration. Four key research topics were identified, of which the topic of epidemiology and public health interventions has gathered highest attentions. Topic of virus infection and immunity has been more focused during the early stage of COVID-19 outbreak compared with later stage. The topic popularity of clinical symptoms and diagnosis has been steady.
Conclusions:
Our topic analysis results revealed that the study of drug treatment was insufficient. To achieve critical breakthroughs of this research area, more interdisciplinary, multi-institutional, and global research collaborations are needed.
1. Introduction
A novel coronavirus emerged and caused a rapid spread of phenomena in Wuhan, China, at the end of 2019. In February 11, 2020, the World Health Organization named this disease Coronavirus Disease 2019 (COVID-19). [ 1 ] With the global spread of COVID-19, it threatened human lives, caused heavy burdens, and brought tremendous challenges to social development. To support the public health decision-making and scientific countermeasures implementation, researchers around the world were racing to study on the disease transmission, diagnostic tests, treatments, vaccines, among others. With the joint efforts of researchers and clinicians around the world, more and more COVID-19-related articles have been published and the outputs of scientific research are constantly emerging. As of July 1, 2020, PubMed has included 27,370 published articles on COVID-19.
State of the art literature review about COVID-19 demonstrated that most available literature-based studies could be basically divided into 2 kinds. The first kind is systematic reviews or meta-analyses. Most of them focused on a certain specific subfields of COVID-19 research, such as drug therapy, diagnostic methods, or clinical symptoms. For example, Alzghari et al [ 2 ] performed a systematic review to investigate the effect of Tocilizumab on COVID-19, and Zhu et al [ 3 ] systematically reviewed the CT imaging features of COVID-19 to provide reference for clinical practice. The second kind is the bibliometric analysis which uses quantitative analysis methods to describe literature in a particular research domain. However, some of the bibliometric analysis were targeting at coronavirus, not just severe acute respiratory syndrome coronavirus 2 (SARS-COV-2), for the purpose of providing reference for COVID-19 research, and the time window was usually set for a long retrospective duration. [ 4 – 7 ] For example, Mao et al [ 7 ] analyzed coronavirus articles published from 2003 to 2020. Up to the investigation time of this study, there were limited number of bibliometric studies specific to COVID-19 and most of them were found and implemented at early stage of COVID-19 outbreak. [ 8 , 9 ] For example, Lou et al [ 8 ] executed a query in PubMed using keyword “COVID-19” and analyzed 183 related articles. Most of these previous literature-based studies of COVID-19 provided a specific review for COVID-19 research progresses or clinical observations; however, the description of a whole picture of COVID-19 scientific research using systematical methods was still insufficient.
Therefore, to answer who, what, where, and when questions of COVID-19 studies, we adopted a hybrid method that integrated multi-approaches, including bibliometrics, topic analysis, collaboration analysis, trends analysis, and visualization, to give a timely and systematic review of COVID-19 literatures. The analysis objectives include countries/regions, institutes, collaboration relationships, research topics, and research trends of COVID-19 studies.
2. Materials and methods
2.1. data source.
The data scope of this study is COVID-19-related articles published from January 1, to July 1, 2020. Since PubMed has served as the primary database for retrieving biomedical literature, it was selected as the only data source. [ 10 ] Ethical approval was not required because no human and animal subjects were enrolled.
2.2. Search strategy
The advanced search option was adopted, and the query “((novel coronavirus[Title/Abstract] OR COVID-19[Title/Abstract] OR 2019-nCov[Title/Abstract] OR SARS-Cov-2[Title/Abstract] OR COVID19[Title/Abstract] OR coronavirus disease 2019[Title/Abstract] OR coronavirus disease-19[Title/Abstract]) OR COVID-19[Supplementary Concept]) AND (“2020/01/01”: “2020/07/01”[dp])”was executed on July 1, 2020. In total, 27,370 COVID-19 articles were collected.
2.3. Data collection
All of the retrieved articles were downloaded and saved with PubMed default format. Microsoft Excel 2010 was used to pre-process the data and, in conjunction with Visual Basic for Applications (VBA), to extract analysis objects such as country/region names and institute names. The number of publications of a country is derived by counting the number of publications that contain at least one author's affiliation belongs to this country, and the first affiliation will be selected when an author has more than one affiliations.
2.4. Bibliometric and visualized analysis
MTI (National Library of Medicine, Bethesda, MD), [ 11 ] VOSviewer 1.6.15 (Leiden University, Leiden, Netherlands) [ 12 ] and D3.js (Mike Bostock, Observable, Inc., San Francisco, CA) [ 13 ] were used to carry out bibliometric and visual analysis of the publications. Since Medical Subject Headings (MeSH) represent much richer semantics that author-selected keywords, they were chosen as the object of topic analysis. MTI was used to extract MeSH terms from title and abstract of articles because newly created articles in PubMed will not be indexed with MeSH terms immediately. VOSviewer was used to generate collaborative network of countries/regions/institutes and co-occurrence network of MeSH terms. Finally, D3.js was used to visualize the internal hierarchy and the popularity trend of topics, which identified by MeSH terms co-occurrence clustering.
2.5. Analytical methods
Topic popularity was calculated by proportional frequency equation and tracked in a certain period of time window (10 days window) to identify the research trends. The equation of proportional frequency is as follows:

Where Dpro_t is the proportional frequency of the term in the t time window, D_t is the document frequency of the term, that is, the number of publications containing the term. DAll_t is the total number of publications and DAvg is the average number of publications on each time window. Topic popularity is measured by adding up proportional frequency of all the terms in this topic.
3.1. The Scale of COVID-19 publications
The number of COVID-19 research publications has been continuously increasing after its break. According to the growth trend from the view of global to country level, as shown in Figure Figure1, 1 , United States overtook China Mainland as the largest contributor in publishing COVID-19-related articles in early May 2020. As of July 1, 2020, United States had published 5949 (21.7% of the total) articles, and China Mainland had published 4080 (14.9% of the total) articles in total that are much higher than any of the other countries. The following Italy (10.7%) and UK (8.4%) were also prolific among the top 10 countries (Table (Table1). 1 ). In addition, China Mainland had the highest rate of domestic collaboration (79.4%), whereas Australia had the lowest (34.8%) among the top 10 productive countries.

The growth trend on number of publications about COVID-19 research.
The top 10 productive countries/regions that published COVID-19 research.

3.2. The collaborative network of countries/regions
Collaboration activities on country/region level were measured based on co-author analysis. As shown in Figure Figure2, 2 , there were 76 countries/regions involved in COVID-19 research collaboration which divided into 3 clusters.

The collaboration network on COVID-19 research across countries/regions.
Cluster 1 (blue color) mainly included United States, China Mainland, Canada, and Australia, which were all ranked as Top 10 productive countries. When measuring the collaboration activities, our study further disclosed that United States and China Mainland played the leading role of the COVID-19 research. These two countries had strong internal co-authorship relations, and at the same time had strong external co-authorship relations with other countries/regions. Cluster 2 (green color) was composed with 27 European countries that included UK, Italy, Germany, and France, among others. There were frequent internal collaboration activities among these European countries. In addition, Cluster 3 (red color) included India, Brazil, and other countries of Asia, Africa, and South America with a relatively low frequency of internal collaboration.
Furthermore, total link strength analysis showed that United States was the most active country with the highest number of collaboration relationships with other countries/regions. United States and China Mainland had the largest number of link strength compared with other countries, with a total of 439 collaboration papers. However, Chinese researchers had mostly co-authored with their domestic collaborators, only 20.6% of the studies were collaborated with international researchers outside China Mainland (Table (Table1 1 ).
3.3. The collaborative network of research institutes
The most productive institutes were located at United States, China Mainland, and Europe. There were 307 institutes that had published >10 articles. Table Table2 2 lists the number of publications and internal collaboration publications for top 10 productive institutes. Huazhong University of Science and Technology (523), Wuhan University (340), and University of California (300) were ranked as Top 3 productive institutes by number of publications. Besides, the BMJ editors published 193 latest news and comments about COVID-19 research with the highest rate of internal collaboration of 100%.
The top 10 productive institutes that published COVID-19 research.

Collaboration network among productive institutes was generated based on co-author analysis. Institutes were clearly separated into 5 clusters as shown in Figure Figure3. 3 . Cluster 1 (red color) included 96 institutes which were mostly universities and hospitals of United States, as well as 10 universities from Canada, among which University of Toronto ranked as Top 1 institute for global research collaboration with the largest number of total link strength. Besides, University of California and University of Washington were also the collaboration centers with large number of co-authored articles. The universities, hospitals, and research institutes came from China composed Cluster 2 (blue color), from which Huazhong University of Science and Technology and Wuhan University had the largest number of link strength compared with other institutes, with a total of 60 collaboration papers. Furthermore, >100 institutes from Europe composed Cluster 3 (green color) and Cluster 4 (yellow color), of which universities and hospitals from Italy composed Cluster 4 and the remaining institutes composed Cluster 3. According to co-author analysis on these 2 clusters, University College London and University of Oxford were most active on research collaboration with other institutes. In addition, it was interesting to observe that Cluster 5 (purple color) contributed a relatively small volume of publications but was a self-centered research community mainly composed with 8 universities from Iran.

The collaboration network on COVID-19 research across institutes.
3.4. The identified COVID-19 research topics
To achieve better understanding of what are the researcher's focuses and research progress of COVID-19 with its break timeline, MeSH terms of each article were selected as the observation objects to measure the research topics and topic trends. On the analysis of selected 2000 MeSH terms with their frequency above 10, a MeSH terms co-occurrence network with 584 high-frequency terms were generated, as shown in Figure Figure4. 4 . The network center nodes are COVID-19, severe acute respiratory syndrome coronavirus 2, and Coronavirus Infections. Four topics about COVID-19 research were obviously identified: epidemiology and public health interventions, virus infection and immunity, clinical symptoms and diagnosis, drug treatments, and clinical studies, as shown in Figure Figure5 5 .

The MeSH terms co-occurrence network on COVID-19 research.

The hierarchy of four identified COVID-19 topics.
3.4.1. Topic I: epidemiology and public health interventions
The research topic of epidemiology and public health interventions had gathered great attentions. It contained 281 of the 584 MeSH terms, indicating that the prevention and control of COVID-19 was the most concerned issue at all the stages of disease break. It mainly contained epidemic transmission dynamics, prevention and control measures and effect analysis at different regional levels (global, national, and urban), [ 14 , 15 ] epidemiological investigation, modeling, and trend prediction from the perspective of public health, [ 16 , 17 ] as well as various personal protective measures (Disinfection, Hand Hygiene, Masks, Personal Protective Equipment, Protective Devices), [ 18 , 19 ] and social prevention and control measures (Airway Management, Mass Screening, Social Distance, Social Isolation). [ 20 ] In addition, high attention had been paid to the psychological and mental state (Anxiety, Anxiety Disorders, Depression, Fear, Mental Disorders, Mental Health) of the general public, infected people, and medical workers. [ 21 ]
3.4.2. Topic II: virus infection and immunity
A total of 168 MeSH terms were included in this topic, which was mainly for the molecular biology and immunology studies of SARS-CoV-2 for the purpose of detection and prevention. Three subtopics of Topic II were identified based on content analysis. The first subtopic was the research on the pathogenesis of COVID-19 that included the replication process and infection mechanism of SARS-CoV-2 in human cells, with emphasis on the interaction between SARS-CoV-2 and biological enzymes (RNA-directed DNA polymerase, angiotensin-converting enzyme [ACE2], serine endopeptidases). [ 22 , 23 ] The second subtopic was the studies on the etiological detection methods of SARS-CoV-2 and the most important methods involved were real-time polymerase chain reaction and reverse transcriptase polymerase chain reaction (PCR). [ 24 , 25 ] In addition, COVID-19 vaccine development with the aim of inducing immune response composed the third subtopic. [ 26 , 27 ]
3.4.3. Topic III: clinical symptoms and diagnosis
A total of 111 MeSH terms were included in Topic III, which mainly covered clinical symptoms of COVID-19 patients and various testing methods used for diagnosis. The clinical symptoms (or complications) of COVID-19 mentioned in the literature mainly included: abdominal pain, cough, diarrhea, dyspnea, fatigue, fever, headache, leukopenia, lymphopenia, myalgia, nausea, pharyngitis, pleural effusion, pneumonia, pulmonary embolism, respiratory distress syndrome, respiratory insufficiency, vomiting, among others. [ 28 , 29 ] The diagnostic methods, mostly discussed in the literature, were routine blood tests (alanine transaminase, aspartate aminotransferases, biomarkers, C-reactive protein, leukocyte count, l -lactate dehydrogenase, lymphocyte count, neutrophils, platelet count) and imaging examinations (radiography, tomography, x-rays). [ 30 ]
3.4.4. Topic IV: drug treatments and clinical studies
Topic IV contained 24 MeSH terms, which was the smallest topic. The research content in this topic was mainly in vivo and in vitro trials of multiple drugs and their combinations for the purpose of treating COVID-19. The studied drugs involved antibacterial/antiviral drugs (azithromycin, favipiravir, lopinavir, remdesivir, ribavirin, ritonavir), antimalarials, and rheumatoid arthritis drugs (chloroquine, hydroxychloroquine, tocilizumab) among others. Because of the difference of clinical endpoint and experimental design, the trials results obtained so far are not consistent. For example, some researchers conclude that remdesivir can be used as potent drugs against COVID-19 [ 31 ] ; however, some studies show that remdesivir cannot significantly improve the symptoms of patients with severe COVID-19. [ 32 ] Chloroquine and hydroxychloroquine are in a similar situation to remdesivir. [ 33 , 34 ] Therefore, there is still no widely accepted standard on specific drugs or the best drug treatment options of COVID-19. [ 35 – 37 ]
3.5. Topic popularities and evolvements about COVID-19 research
Topic popularity of the above 4 COVID-19 topics was measured by using proportional frequency equation in Section 2, and the measured results, as shown in Figure Figure6, 6 , were consistent with manually validation results by reviewing literature. According to trend analysis, the topic of epidemiology and public health interventions has gathered great attentions and continuously with high popularity. The characteristics of SARS-CoV-2, such as biological structure, genetic sequence, and infection mechanism, have been well studied, and beyond this, consensus has been reached on COVID-19 clinical symptoms and diagnostic methods.

Trends of topic popularity.
On the topic tracking analysis of epidemiology and public health interventions, we found that most of the early studies and reports were mainly focus on China's epidemic prevention and control. [ 38 , 39 ] By implementing a series of preventive control and medical treatment measures, the pandemic in China had been effectively contained, but the number of confirmed cases outside China continued to increase, as did the corresponding research on epidemiology and public health interventions, which was consistent with the continuously high popularity trending curve of this topic (blue curve), as displayed in Figure Figure6 6 .
For virus infection and immunity study, the topic popularity decreased since early of February 2020. As studying the etiological characteristics of a novel virus, such as biological structure, genetic sequence, and infection mechanism, is the key to pandemic prevention and control, the trend curve of Topic II was in the highest position in the pre-outbreak period (January 2020). With the joint efforts of scientists around the world, substantial progress had been achieved in the understanding of SARS-CoV-2. For example, the genetic sequencing of SARS-CoV-2 was performed by Chinese scientists on January 7, 2020 and the results were timely shared with the WHO on January 12, 2020. Furthermore, the infection mechanism of SARS-CoV-2, especially its relationship with ACE2 was identified, and specific diagnostic PCR tests were produced. [ 40 , 41 ] The above achievements were mainly completed in January and February 2020, starting from February, the trend curve of Topic II gradually declined. However, the curve will remain at a high level because more and more attentions have been paid to vaccine-related research. According to literature reports, there are more than 100 candidate vaccine projects targeting COVID-19 worldwide, and some of them have entered clinical trials. [ 42 , 43 ]
With the continuous increase of confirmed and treated cases, clinicians achieved deeper understanding about COVID-19. Since March 2020, there has been a global consensus on the symptoms and diagnostic criteria for COVID-19. [ 28 , 44 ] In addition, the seventh and final edition of “Diagnosis and Treatment Protocol of COVID-19,” issued by the National Health Commission of the PRC, was also released on March 3, 2020. [ 45 ] As a result, the trend curve of Topic III starts to smooth out since March 2020 (Fig. (Fig.6 6 ).
Although lopinavir/ritonavir was recommended as antiviral drug by the first edition of “Diagnosis and Treatment Protocol of COVID-19” on January 16, 2020 at the beginning of the pandemic, the widespread interest in using antiviral drugs to treat COVID-19 began with a report of the first diagnosed patient who benefit from remdesivir in United States, which was published in NEJM on January 31, 2020. [ 46 ] Therefore, the trend curve of Topic IV in Figure Figure6 6 has risen slightly since February 2020. However, the minimal topic size and low trend curve suggest that drug therapy remains the weak point in the response to COVID-19.
4. Discussion and conclusion
The number of COVID-19 publications has been growing dramatically since March 2020. According to our search strategy, as of the submission of this manuscript (July 13, 2020), the number of COVID-19 publications has exceeded 30,000. Given that COVID-19 pandemic has not been well contained at the global level, relevant research will continue to be carried out and the number of publications will increase accordingly. The methodology in this study can be easily implemented to analyze the future research status of COVID-19, or even applied to other fields.
Although United States and China were the most productive countries, they were not in the identical situation. Since the initial outbreak was in China, Chinese scholars quickly carried out a series of studies and published numerous articles in the early stages of the epidemic. However, Chinese scholars tend to collaborate with domestic scholars rather than aboard. Unlike China, United States has seen a significant increase in the number of publications since April 2020, and has quickly occupied the highest level of participation in global collaboration due to its strong scientific research strength and influence.
Collaboration at the institutional level has obvious geographical characteristics, especially the frequent internal collaborations among institutes located in China, as well as United States. For example, Huazhong University of Science and Technology and Wuhan University, which ranked first and second by the number of publications, co-authored a total of 60 articles, making up the most productive institute pair. Both universities are located in Wuhan and their affiliated hospitals, such as Tongji Hospital, Union Hospital, and Renmin Hospital, are major hospitals for treating COVID-19 patients. The front-line clinical medical workers in those hospitals have conducted a lot of research on virus detection, clinical diagnosis and treatment while fighting against the epidemic.
COVID-19 research topics are continuously evolving with their publication timeline, measuring these changes will help researchers and scientific policy makers understanding the status of COVID-19 research. As indicated by the trend curves of topic popularity, the prevention and control of COVID-19 remains the most important issue at present, and drug therapy remains the weak point in the response to COVID-19. In addition, more support should be given to vaccine research and development, because vaccines are the ultimate solution to the epidemic. [ 5 ]
This study provided an overall investigation of COVID-19 scientific progresses using multiple qualitative and quantitative analysis methods. The collaboration status of COVID-19 research at national and institutional levels was disclosed and 4 topics (epidemiology and public health interventions, virus infection and immunity, clinical symptoms and diagnosis, drug treatments, and clinical studies) were identified and interpreted. Our topic analysis results revealed that the study of drug treatment was insufficient. To achieve critical breakthroughs of this research area, more interdisciplinary, multi-institutional, and global research collaborations are needed.
4.1. Strengths and limitations
Publications on COVID-19 research were retrieved from PubMed, and the collaboration status and research trends of COVID-19 were measured via bibliometric and visualized analysis, which was considered to be relatively objective and comprehensive. Moreover, well curated MeSH terms were used as the object of topic analysis in this study, compared with author-selected keywords which were usually chosen by existing COVID-19-related bibliometric analysis. [ 4 – 7 ] Due to the limited number and randomness of author-selected keywords, the derived results, especially the co-occurrence analysis results, cannot reflect the real status of the COVID-19 research. Our MeSH terms-based methodology could better disclose the research topics and trends of COVID-19. However, limitations also exist in our research. On the one hand, PubMed was selected as the only data source, so some articles only indexed in other databases such as Web of Science and Scopus might be left out. On the other hand, for the sparisity reason of citation network of published COVID-19 articles, citation analysis has not been adopted in this study. In the future, studies based on citation analysis, such as identification of influential authors and highly-cited articles, will be conducted and included in our further analysis.
Author contributions
Conceptualization, N.H.; Data curation, J.W.; Software, J.W. and N.H.; Visualization, J.W. and N.H.; Writing—original draft, J.W. and N.H.; Writing—review & editing, J.W. and N.H. All authors have read and agreed to the published version of the manuscript.
Conceptualization: Na Hong.
Data curation: Junhui Wang.
Software: Junhui Wang, Na Hong.
Visualization: Junhui Wang, Na Hong.
Writing – original draft: Junhui Wang, Na Hong.
Writing – review & editing: Junhui Wang, Na Hong.
Abbreviations: ACE2 = Angiotensin Converting Enzyme 2, COVID-19 = Coronavirus Disease 2019, MeSH = Medical Subject Headings, MTI = Medical Text Indexer, SARS-COV-2 = severe acute respiratory syndrome coronavirus 2, VBA = Visual Basic for Applications.
How to cite this article: Wang J, Hong N. The COVID-19 research landscape: Measuring topics and collaborations using scientific literature. Medicine . 2020;99:43(e22849).
The authors report no conflicts of interest.
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- My Account Login
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Open access
- Published: 06 May 2024
Vaccine effectiveness against emerging COVID-19 variants using digital health data
- Tanner J. Varrelman ORCID: orcid.org/0000-0002-8766-0129 1 ,
- Benjamin Rader 1 , 2 ,
- Christopher Remmel 1 ,
- Gaurav Tuli 1 ,
- Aimee R. Han ORCID: orcid.org/0000-0001-8927-3432 1 ,
- Christina M. Astley ORCID: orcid.org/0000-0002-5063-8470 1 , 3 , 4 , 5 &
- John S. Brownstein ORCID: orcid.org/0000-0001-8568-5317 1 , 4
Communications Medicine volume 4 , Article number: 81 ( 2024 ) Cite this article
514 Accesses
3 Altmetric
Metrics details
- Computational biology and bioinformatics
Participatory surveillance of self-reported symptoms and vaccination status can be used to supplement traditional public health surveillance and provide insights into vaccine effectiveness and changes in the symptoms produced by an infectious disease. The University of Maryland COVID Trends and Impact Survey provides an example of participatory surveillance that leveraged Facebook’s active user base to provide self-reported symptom and vaccination data in near real-time.
Here, we develop a methodology for identifying changes in vaccine effectiveness and COVID-19 symptomatology using the University of Maryland COVID Trends and Impact Survey data from three middle-income countries (Guatemala, Mexico, and South Africa). We implement conditional logistic regression to develop estimates of vaccine effectiveness conditioned on the prevalence of various definitions of self-reported COVID-like illness in lieu of confirmed diagnostic test results.
We highlight a reduction in vaccine effectiveness during Omicron-dominated waves of infections when compared to periods dominated by the Delta variant (median change across COVID-like illness definitions: −0.40, IQR[−0.45, −0.35]. Further, we identify a shift in COVID-19 symptomatology towards upper respiratory type symptoms (i.e., cough and sore throat) during Omicron periods of infections. Stratifying COVID-like illness by the National Institutes of Health’s (NIH) description of mild and severe COVID-19 symptoms reveals a similar level of vaccine protection across different levels of COVID-19 severity during the Omicron period.
Conclusions
Participatory surveillance data alongside methodologies described in this study are particularly useful for resource-constrained settings where diagnostic testing results may be delayed or limited.
Plain language summary
Surveys that are sent out to users of social media can be used to supplement traditional methods to monitor the spread of infectious diseases. This has the potential to be particularly useful in areas where other data is unavailable, such as areas with less surveillance of infectious disease prevalence and access to infectious disease diagnostics. We used data from a survey available to users of the social media platform Facebook to collect information about any potential symptoms of COVID-19 infection and vaccines received during the COVID-19 pandemic. We found a potential reduction in vaccine effectiveness and change in symptoms when the Omicron variant was known to be circulating compared to the earlier Delta variant. This method could be adapted to monitor the spread of COVID-19 and other infectious diseases in the future, which might enable the impact of infectious diseases to be recognized more quickly.
Similar content being viewed by others

Long COVID: major findings, mechanisms and recommendations

A guide to vaccinology: from basic principles to new developments
Exploring the reported adverse effects of COVID-19 vaccines among vaccinated Arab populations: a multi-national survey study
Introduction.
Timely identification of alterations in vaccine effectiveness (VE) with the emergence of novel COVID-19 variants, such as Omicron, is important for informing the global public health response. The attributable risk proportion of vaccine-preventable diseases is often estimated using relative risk measures obtained from cohort studies or odds ratios determined through case-control designs, which typically rely on gold-standard diagnostic testing 1 , 2 . These studies are conducted retrospectively, leading to a lag between variant emergence and VE estimates. In an effort to provide timely VE insights, monitoring systems have been developed that leverage digital health data 3 , 4 . However, even these real-time methodologies are bounded by some form of diagnostic testing data, whether it be self-reported or through other means of collection. While resource-rich locales across the world have managed to scale up diagnostic testing to inform pandemic response efforts, many low-and middle-income countries (LMICs) have struggled to establish widespread testing 5 , 6 , therefore limiting the applicability of current VE monitoring systems. Alternatively, digital health surveys of self-reported symptoms and vaccination status provide a data source that may be used in place of limited/delayed testing data 7 , 8 , 9 .
In this study, we use data from the University of Maryland Global COVID Trends and Impact Survey (UMD-CTIS) to develop a methodology to simultaneously characterize potential changes in VE and COVID-19 symptomatology for Delta and Omicron-dominated periods of infections. UMD-CTIS is a digital health survey that leveraged Facebook’s active user base, providing cross-sectional survey data in near real-time from 114 countries, starting in 2020 and ending in 2022. Our analyses utilize aggregate data from three MICs that were selected based on the quality of UMD-CTIS data and the presence of distinct Delta and Omicron periods of infections. The selected countries include Guatemala, Mexico, and South Africa. Our analyses of this data reveal reduced vaccine effectiveness against suspected COVID-19 infection during the Omicron period compared to Delta, as well as a shift towards more upper respiratory-type symptoms like cough and sore throat.
Syndromic surveillance data
The University of Maryland Global COVID Trends and Impact Survey (UMD-CTIS), in partnership with Facebook, is a cross-sectional survey that sampled Facebook’s active user base on a daily basis. Facebook users were presented an invitation at the top of their news feed, inviting them to participate in the survey. It is important to note that survey invitations did not include any type of incentive, and participation was driven purely by individuals’ willingness to contribute to digital health. If an individual decided to accept the invitation, they were navigated off of the Facebook platform to the digital health survey hosted by Qualtrics, with data collection being performed by the Joint Program in Survey Methodology at the University of Maryland. On the Qualtrics survey itself, respondents were shown the consent page explaining the purpose of the research to gain a better public understanding of where and how the coronavirus pandemic is spreading, that the survey would take 3–5 min, and that their responses would remain confidential and anonymous. After providing informed consent and confirmation of being at least 18 years of age, respondents could proceed with the survey. Survey respondents and non-respondents were entered back into the sampling pool after a duration of a few weeks or months, depending on the sample size for a given area. Survey data included self-reported information such as demographics, recent symptoms, and COVID-19 vaccination status. While Facebook acts as the survey sampling frame, the company cannot access individually identified respondent answers. Further, to work with these data, institutions must have a signed Data Use Agreement (data access and survey questions available https://covidmap.umd.edu ) 7 , 10 , which our institution signed in order to access and analyze the UMD-CTIS data. Boston Children’s Hospital Institutional Review Board (P00023700) approved this study using UMD-CTIS data. Additional details on the survey design, methodology, and validation can be found in Astley et al. (2021) 7 .
To select the study locations, we began by focusing on countries that met three criteria: they are included in the UMD-CTIS sample, have encountered distinct waves of COVID-19 infections primarily driven by the Delta and Omicron variants, and are considered a low or middle-income country as described by the Organization for Economic Co-operation and Development (OECD). Next, we visualized the time-series symptom data and ruled out countries where the UMD-CTIS data was noticeably erratic.
Using peak detection (Python (3.8.2), scipy.signal.argrelextrema (1.7.1), order parameter = 70) for all CLI time series (April 2021–February 2022), we infer 2-week consensus variant periods prior to each peak, for Delta and Omicron, respectively, for Guatemala (peak date September 13, 2021 [survey No. 4137] and peak date February 2, 2022 [survey No. 2387]), South Africa (July 22, 2021 [survey No. 7371] and December 19, 2021 [survey No. 5320]), and Mexico (August 22, 2021 [survey No. 52775] and January 26, 2022 [survey No. 71990]), that coincided with >80% variant share per public reports 11 .
Statistics and reproducibility
We utilize conditional logistic regression to estimate the attributable risk proportion (ARP) for illness in 2-dose vaccinated individuals (clogit function with method=’approximate’, R (4.1.1), survival library (3.2-13)). VE is given by VE = ARP ≈ 1−OR. We consider exposure as the vaccination status of a respondent (unvaccinated vs. 2-dose vaccinated), and the outcome as to whether a respondent reported CLI in past 14 days, with missing symptoms assumed absent. We also include strata for dichotomized age (>44 years), gender (male/female), and country of the survey respondent to limit potential confounding and differences in country-level sampling. Importantly, UMD-CTIS does not collect data on vaccine formulation. Consequently, we cannot definitively determine whether a single dose of any specific vaccine within our dataset consistently provides full protection, as seen with the Janssen COVID-19 vaccine formulation. Therefore, we have chosen not to include individuals who have received only one dose in this study. Age and gender were dichotomized in order to maintain sufficient sample sizes per stratum. We do not filter the individual vaccine effectiveness estimates by p -value, as we are interested in the group behavior of the CLI definitions and not the hypothesis of whether a single definition of CLI produces a statistically significant vaccine effectiveness estimate. Moreover, to maintain the same number of data points for each of our comparisons, we do not remove outlier data from the analyses in this study.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
To estimate VE, we adapted case-control methods 1 for prevalent COVID-like illness (CLI) as a proxy for confirmed COVID-19 cases. Therefore, our estimates of VE measure a vaccine’s ability to prevent suspected symptomatic infections defined by CLI. To allow for changes in variant-specific symptomatology, we iterate across all possible CLI defined by 66 pair-wise combinations of 12 self-reported symptoms (fever, cough, difficulty breathing, fatigue, stuffy or runny nose, aches or muscle pain, sore throat, chest pain, nausea, loss of smell or taste, headache, chills). We then cluster the vaccine effectiveness estimates according to a single symptom of interest and evaluate the median vaccine effectiveness across all CLI definitions in the cluster. As an example, using a COVID-19-specific symptom (loss of smell or taste) as an anchor symptom, we evaluate VE estimates for all CLI definitions inclusive of this symptom during Delta and Omicron waves of infections, resulting in VE estimates for 11 pairwise combinations of symptoms. Consistent with previous estimates of VE that used PCR test data as the outcome 2 , our analyses reveal a median VE Delta of 0.77, IQR[0.76, 0.80] (Fig. 1 a, triangle). In comparison, analyzing the data from the Omicron period reveals a median VE Omicron of 0.47, IQR[0.41, 0.53] (Fig. 1 a, circle). Further expanding the approach to all CLI definitions reveals a median VE Delta of 0.71, IQR[0.65, 0.75] (Fig. 1 b). In contrast, the VE Omicron estimate is even lower (median 0.29, IQR[0.20, 0.38]). Notably, our findings align with those from a recent meta-analysis study focused on real-world vaccine effectiveness for fully vaccinated individuals. This study reported a VE of 70.9% (95% CI, 68.9–72.7) against Delta infections and a VE of 23.5% (95% CI, 17.0–29.5) against Omicron variant infections 12 . To understand how VE estimates for each CLI definition vary by wave, we take the difference between the two VE period estimates (VE Omicron −VE Delta ) for each CLI definition. Doing so reveals a median within-CLI definition change of −0.40, IQR[−0.45, −0.35] (Fig. 2 a), suggesting lower VE Omicron regardless of the CLI definition that is used. Additionally, we find that the pattern of change in VE across CLI definitions is similar when evaluating individual country estimates (see Supplementary Fig. 1 ).
a VE estimates for symptoms paired with the loss of smell or taste for the Delta (triangle) and Omicron (circle) periods. 95% confidence intervals are calculated for each VE estimate, with Delta and Omicron period estimates derived from 64,283 and 79,697 survey responses, respectively. b Box and whisker plot of VE estimates across all 66 possible CLI defined by pairwise combinations of symptoms for Delta and Omicron periods. The box represents the interquartile range (IQR) of estimates, with the horizontal line inside the box indicating the median. The whiskers extend to the largest/smallest values up to 1.5 times the IQR. Outlier values are represented as points. The sample size for each VE estimate is consistent with the sample sizes described in panel ( a ).
a Distribution of within-CLI change (VE Omicron −VE Delta ) across all CLI definitions. b Distributions of VE Omicron −VE Delta among CLI definitions within each anchor symptom. Each box-plot contains estimates for an anchor symptom paired with the 11 other symptoms. Box-plots are ordered according to the magnitude of the median change, with the median across all VE indicated by the gray dashed line. Each box represents the interquartile range (IQR) of estimates, with the horizontal line inside the box indicating the median. The whiskers extend to the largest/smallest values up to 1.5 times the IQR. Outlier values are represented as points. Each VE estimate from the Delta and Omicron periods is derived from 64,283 and 79,697 survey responses, respectively.
To identify potential alterations in COVID-19 symptomatology, we evaluate the change in VE estimates for CLI definitions with a single anchor symptom, like loss of smell and taste. We reason that if symptoms are similar across variants, the within-anchor median change in VE will be similar across anchor symptoms. Our analyses provide evidence for a potential change in COVID-19 symptomatology from the Delta period to the Omicron period, as we note that some symptoms have more or less decline in VE (Fig. 2 b). Specifically, we find that CLI definitions that include loss of smell or taste have the smallest median change in VE (median: −0.31, IQR[−0.34, −0.28]), while definitions with the largest median change include a cough, or sore throat (cough median: −0.49, IQR[−0.52, −0.45]; sore throat median: −0.47, IQR[−0.49, −0.45]). The observed pattern of change in VE across anchor symptoms is similar when evaluating VE estimates from individual countries (see Supplementary Fig. 2) , however, with increased uncertainty in estimates as measured by the span of anchor symptom distributions (see Supplementary Results ). Similarly, a survey-based study that used PCR testing data as the outcome demonstrated a shift away from symptomatology that includes loss of smell or taste and towards upper-respiratory type symptoms (i.e., sore throat) during the Omicron period 13 . Furthermore, a study conducted in Jalisco, Mexico, analyzed reported symptoms for confirmed infections with wild-type SARS-CoV-2, Delta, and Omicron variants, revealing that Omicron infections were linked to a higher incidence of runny nose and sore throat, aligning with the findings of our country-level analysis for Mexico (see Supplementary Fig. 3) 14 . These results corroborate our overall findings, which also identified increased reporting of sore throat during a wave of COVID-19 infections dominated by the Omicron variant. Collectively, these findings suggest a shift in symptomatology associated with the Omicron variant towards more upper respiratory-type symptoms.
In addition to providing insights into changes in COVID-19 symptomatology, the VE estimates also include information about a vaccine’s ability to protect against COVID-19 illness presenting at different levels of severity as defined by pairwise combinations of symptoms. Importantly, we do not have information about the true severity of each respondent’s reported illness, and we instead infer severity based on the presence and absence of key symptoms. For instance, all CLI definitions that include at least a fever, cough, aches or muscle pain, sore throat, nausea, loss of smell or taste, or a headache in the absence of difficulty breathing or chest pain are considered mild syndromes. However, according to the NIH, CLI definitions that include difficulty breathing or chest pain are considered more severe forms of illness 15 . To understand potential changes in VE against mild and severe COVID-19 syndromes, we partition our CLI-informed VE estimates according to the above classifications. As a result, we end up with 42 mild and 21 severe definitions of CLI. We find that severe definitions of illness were more protected than mild definitions during the Delta period (median severe VE: 0.74, IQR[0.70, 0.79], median mild VE: 0.54, IQR[0.45, 0.64]) (Fig. 3 ). However, protection against mild and severe illness was similar during Omicron (median severe VE: 0.30, IQR[0.25, 0.38], median mild VE: 0.22, IQR[0.16, 0.33]). Importantly, VE against severe illness may appear higher, as vaccines are producing milder illness when an individual is infected with COVID-19 16 , making it seem as if VE against mild illness is less effective. During the Delta wave of infections, we observed a total of 13,220 reports of mild illness and 5316 reports of severe illness. In contrast, during the Omicron wave of infections, there were 24,408 reports of mild illness and 10,234 reports of severe illness.
VE estimates for pairwise combinations of symptoms that include a fever, cough, aches or muscle pain, sore throat, nausea, loss of smell or taste, or a headache in the absence of difficulty breathing or chest pain (mild illness), and pairwise combinations of symptoms that include difficulty breathing or chest pain (severe illness). Each box represents the interquartile range (IQR) of estimates, with the horizontal line inside the box indicating the median. The whiskers extend to the largest/smallest values up to 1.5 times the IQR. Outlier values are represented as points. Each VE estimate from the Delta and Omicron periods is derived from 64,283 and 79,697 survey responses, respectively.
It is critical to note that our estimates of VE measure the preventable syndrome attributed to receiving 2-doses of vaccine and represent only one of many components that contribute to true vaccine effectiveness. For instance, we are unable to account for asymptomatic breakthrough infections, and we do not have information on natural immunity among the unvaccinated nor on vaccine formulation or timing for the vaccinated. Therefore, we do not have enough information to distill whether changes in VE are caused by waning vaccine immunity, or increased penetration of an emerging variant. To this end, we would suggest that future digital health surveys include information on vaccine formulation, the general timing of vaccination, as well as information on booster doses that have been administered. While quickly adapting a digital health survey is a monumental task, it would enhance the capabilities of methods such as those described in this study. Furthermore, our VE estimates are solely derived from self-reported survey data and are thus vulnerable to a range of biases 17 . For instance, self-report bias is likely influenced by the perception around COVID-19 vaccination at a given time for a given locale. Even so, a U.S.-based survey that incorporated viral testing demonstrated that self-reported vaccination is a strong predictor for true vaccination status 18 , thus providing support for self-reported measures. Further, our estimates rely on the assumption that the range of self-reported CLI definitions defined in this study is a valid proxy for incident COVID-19 infection. Consequently, our VE estimates may be an underestimation if CLI is capturing non-COVID illness. We limit this assumption by selecting time periods reflective of when COVID-19 is circulating within the unvaccinated population of survey respondents for each country.
Although the assumptions mentioned above limit the interpretation of our VE estimates, the methodology still demonstrates notable strengths that should not be discounted. For example, simple surveys that collect self-reported symptoms and vaccination status can be collected rapidly and at a fraction of the cost of traditional surveillance measures 19 . Moreover, while we performed the retrospective analysis with knowledge of specific COVID-19 variants, CLI-informed VE estimates can be derived during suspected variant spread, with careful contextualization of a country’s epidemiological situation (i.e., absence of co-circulating pathogens and sufficient geographic coverage of surveys). In the case of UMD-CTIS, there was a two-week delay between survey completion and its availability for our modeling, allowing us to use it as a valuable near-real-time dataset for VE analyses. It is critical to note that UMD-CTIS collected a substantial number of survey samples from numerous countries, enabling meaningful insights into COVID-19. However, some countries within the UMD-CTIS sample exhibited noisy data, characterized by high variability in the number of reported CLI instances between time steps, which limited the utility of these specific datasets. While UMD-CTIS has yielded valuable data from a wide range of countries, it’s important to acknowledge that the determination of survey sampling intensity, size, and other attributes of sampling can impact the reliability and applicability of findings. To truly understand the minimum number of samples required for robust statistical analyses, further research, and investigation into these sampling parameters are essential. Such efforts will not only enhance the effectiveness of syndromic surveillance but also contribute to more accurate and comprehensive insights into COVID-19 dynamics.
Historically, understanding the impact of infectious diseases, including the effectiveness of vaccination, has relied on detailed clinical data, often gathered through sentinel surveillance networks 20 . For example, the CDC’s U.S. Outpatient Influenza-like Illness Surveillance Network (ILINet) provides information about symptom prevalence for suspected flu cases across the United States over time. While an invaluable resource, ILINet is limited to individuals seeking medical care due to its reliance on sentinel providers for data collection. Therefore, individuals who lack access to such sentinel providers or those who do not seek care will not be represented in these data. Consequently, epidemiological parameters derived from these data may not be entirely representative of the population of interest. Participatory digital surveillance systems like Flu Near You, the ZOE App, and UMD-CTIS enable broader symptom tracking by collecting data directly from the public 3 , 21 . These community-based data sources can provide complementary signals to those derived through clinical data-dependent systems like ILINet 22 . Our analysis of self-reported symptoms from UMD-CTIS demonstrates how digital health data can also be rapidly utilized to infer symptomatic shifts across populations, with the advantage of timeliness and scope beyond only those seeking care. While this application does not provide the same level of clinical confirmation as traditional studies, combining evidence from both clinical and digital participatory data sources allows for earlier response guidance while gold-standard data are collected. For instance, applying our methodology of detecting potential changes in symptomatology could help direct early public health mitigation strategies.
The COVID-19 pandemic exposed vulnerabilities in health infrastructure, particularly for LMICs that struggled to establish testing facilities 8 , needed to support real-time epidemiological parameter estimation that depends on diagnostic testing results. Leveraging the power of global participatory epidemiology in the form of digital health surveys 23 has the potential to supplement these critical testing gaps. Thus, our methods of using self-reported symptom data to understand VE and changes in symptomatology is a powerful rapid response tool, that can provide the medical community with timely insights into emerging variants. Due to our agnostic approach in defining a syndrome (i.e., all pairwise symptoms), the utility of our methods goes beyond COVID-19 and can be applied to other upper-respiratory illnesses and/or locations to support response to emerging threats.
Data availability
To access the raw data used in this manuscript, a request must be submitted to the Facebook Data for Good website: https://dataforgood.facebook.com/dfg/docs/covid-19-trends-and-impact-survey-request-for-data-access . The Global UMD-CTIS Open Data API, Microdata Repository, and contingency tables are available from The University of Maryland Social Data Science Center Global COVID-19 Trends and Impact Survey website ( https://covidmap.umd.edu ). The results of the conditional logistic regression can be found in Supplemental Data 1 and Supplemental Data 2 .
Code availability
The R and Python code used to perform the analyses in this study is available at https://doi.org/10.5281/zenodo.10775701 24 .
Collie, S., Champion, J., Moultrie, H., Bekker, L.-G. & Gray, G. Effectiveness of bnt162b2 vaccine against omicron variant in South Africa. N. Engl. J. Med. 386 , 494–496 (2022).
Article PubMed Google Scholar
Andrews, N. et al. Covid-19 vaccine effectiveness against the omicron (b.1.1.529) variant. N. Engl. J. Med. 386 , 1532–1546 (2022).
Article CAS PubMed Google Scholar
Menni, C. et al. Covid-19 vaccine waning and effectiveness and side-effects of boosters: a prospective community study from the Zoe Covid study. Lancet Infect. Dis. 22 , 1002–1010 (2022).
Bitzegeio, J., Hemmers, L., Bartel, A. & Werber, D. Timely monitoring COVID-19 vaccine protection, Berlin, Germany, April 15 to December 15, 2021. Int. J. Public Health 67 , 1604633 (2022).
Batista, C. et al. The silent and dangerous inequity around access to COVID-19 testing: a call to action. EClinicalMedicine 43 , 101230 (2022).
Aziz, A. B. et al. Integrated control of COVID-19 in resource-poor countries. Int. J. Infect. Dis. 101 , 98–101 (2020).
Article CAS PubMed PubMed Central Google Scholar
Astley, C. M. et al. Global monitoring of the impact of the COVID-19 pandemic through online surveys sampled from the Facebook user base. Proc. Natl Acad. Sci. USA 118 , https://www.pnas.org/content/118/51/e2111455118 (2021).
Fulcher, I. R. et al. Syndromic surveillance using monthly aggregate health systems information data: methods with application to COVID-19 in Liberia. Int. J. Epidemiol. 50 , 1091–1102 (2021).
Article PubMed PubMed Central Google Scholar
Eames, K. et al. Rapid assessment of influenza vaccine effectiveness: analysis of an internet-based cohort. Epidemiol. Infect. 140 , 1309–1315 (2012).
Kreuter, F. Partnering with a global platform to inform research and public policy making. Survey Res. Methods 14 , 159–163 (2020).
Google Scholar
Hodcroft, E. B. Covariants: SARS-CoV-2 Mutations and Variants of Interest (accessed 30 March 2022). CoVariants https://covariants.org/ (2022).
Zeng, B., Gao, L., Zhou, Q., Yu, K. & Sun, F. Effectiveness of COVID-19 vaccines against SARS-CoV-2 variants of concern: a systematic review and meta-analysis. BMC Med. 20 , 200 (2022).
Vihta, K.-D. et al. Omicron-associated changes in severe acute respiratory syndrome coronavirus 2 (sars-cov-2) symptoms in the United Kingdom. Clin. Infect. Dis. 76 , e133–e141 (2022).
Peña Rodríguez, M. et al. Prevalence of symptoms, comorbidities, and reinfections in individuals infected with wild-type sars-cov-2, delta, or omicron variants: a comparative study in western Mexico. Front. Public Health 11 , 1149795 (2023).
National Institutes of Health. Clinical Spectrum of SARS-COV-2 infection (accessed 12 June 2022). National Institutes of Health https://www.covid19treatmentguidelines.nih.gov/overview/clinical-spectrum/ (2022).
Centers for Disease Control and Prevention. Covid-19 After Vaccination: Possible Breakthrough Infection (accessed 12 June 2022). Centers for Disease Control and Prevention https://www.cdc.gov/coronavirus/2019-ncov/vaccines/effectiveness/why-measure-effectiveness/breakthrough-cases.html (2022).
Bradley, V. C. et al. Unrepresentative big surveys significantly overestimated us vaccine uptake. Nature https://doi.org/10.1038/s41586-021-04198-4 (2021).
Siegler, A. J. et al. Trajectory of COVID-19 vaccine hesitancy over time and association of initial vaccine hesitancy with subsequent vaccination. JAMA Netw. Open 4 , e2126882–e2126882 (2021).
Aiello, A. E., Renson, A. & Zivich, P. Social media-and Internet-based disease surveillance for public health. Annu. Rev. Public Health 41 , 101 (2020).
Torner, N. et al. Influenza vaccine effectiveness assessment through sentinel virological data in three post-pandemic seasons. Hum. Vaccines Immunother. 11 , 225–230 (2014).
Article Google Scholar
Chunara, R., Aman, S., Smolinski, M. S. & Brownstein, J. S. Flu near you: an online self-reported influenza surveillance system in the USA. Online J. Public Health Inform. 5 , https://api.semanticscholar.org/CorpusID:31097654 (2013).
Wójcik, O., Brownstein, J., Chunara, R. & Johansson, M. Public health for the people: participatory infectious disease surveillance in the digital age. Emerg. Themes Epidemiol. 11 , 7 (2014).
Chan, A. T. & Brownstein, J. S. Putting the public back in public health—surveying symptoms of COVID-19. N. Engl. J. Med. 383 , e45 (2020).
Varrelman, T. Code from: Vaccine Effectiveness Against Emerging COVID-19 Variants using Digital Health Data https://doi.org/10.5281/zenodo.10775701 https://doi.org/10.5281/zenodo.10775701 (2024).
Download references
Acknowledgements
This work was supported by a Facebook Sponsored Research Agreement (T.J.V., B.R., C.R., G.T., A.H., C.M.A., J.S.B., INB1116217). Authors report research grant funding from the Massachusetts Consortium on Pathogen Readiness (J.S.B.), the Rockefeller Foundation (J.S.B.), and the National Institutes of Health (CMA, K23 DK120899) during the conduct of the study.
Author information
Authors and affiliations.
Computational Epidemiology Lab, Boston Children’s Hospital, Boston, MA, 02115, USA
Tanner J. Varrelman, Benjamin Rader, Christopher Remmel, Gaurav Tuli, Aimee R. Han, Christina M. Astley & John S. Brownstein
Department of Epidemiology, Boston University, Boston, MA, 02118, USA
Benjamin Rader
Division of Endocrinology, Boston Children’s Hospital, Boston, MA, 02115, USA
Christina M. Astley
Harvard Medical School, Boston, MA, 02115, USA
Christina M. Astley & John S. Brownstein
Broad Institute of Harvard and MIT, Cambridge, MA, 02142, USA
You can also search for this author in PubMed Google Scholar
Contributions
T.J.V., B.R., C.M.A., and J.B. conceived the study. T.J.V. and G.T. analyzed the data. T.J.V, B.R., C.M.A., and J.B. drafted the manuscript. T.J.V., B.R., G.T., C.R., A.H., C.M.A., and J.B. reviewed and edited the manuscript. All authors approved of the final manuscript.
Corresponding author
Correspondence to Tanner J. Varrelman .
Ethics declarations
Competing interests.
The authors declare no competing interests.
Peer review
Peer review information.
Communications Medicine thanks Ifeolu David, Farrokh Habibzadeh and Nick Tsinoremas for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplemental material, description of additional supplementary files, supplemental data 1, supplemental data 2, reporting summary, rights and permissions.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ .
Reprints and permissions
About this article
Cite this article.
Varrelman, T.J., Rader, B., Remmel, C. et al. Vaccine effectiveness against emerging COVID-19 variants using digital health data. Commun Med 4 , 81 (2024). https://doi.org/10.1038/s43856-024-00508-9
Download citation
Received : 07 June 2023
Accepted : 24 April 2024
Published : 06 May 2024
DOI : https://doi.org/10.1038/s43856-024-00508-9
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing newsletter — what matters in science, free to your inbox daily.

IMAGES
VIDEO
COMMENTS
3.5 Topic popularities and evolvements about COVID-19 research. Topic popularity of the above 4 COVID-19 topics was measured by using proportional frequency equation in Section 2, and the measured results, as shown in Figure 6, were consistent with manually validation results by reviewing literature. According to trend analysis, the topic of ...
DOI: 10.1056/NEJMe2402224. Nirmatrelvir-ritonavir (Paxlovid [Pfizer]) is used as first-line therapy for nonhospitalized persons with Covid-19 1 on the basis of the results of the Evaluation of ...
Covid-19 has affected humanity in a major way. An extremely dangerous virus, hitherto unknown to humanity, had to be studied and contained in order to overcome the pandemic. Research on Covid-19 had surged in the early days with an unprecedented surge in the publications on that specific topic.
Here the authors show that, in convalescent COVID-19 patients, memory T cell responses are detectable up to 317 days post-symptom onset, in which the presence of stem cell-like memory T cells ...
Scroll down the page to view all COVID-19 articles, stories, and resources from across NIH. You can also select a topic from the list to view resources on that topic. - Any -. Aging. Cancer. Children. Clinical Trials. Immune Responses. Long COVID.
The spread of the "Severe Acute Respiratory Coronavirus 2" (SARS-CoV-2), the causal agent of COVID-19, was characterized as a pandemic by the World Health Organization (WHO) in March 2020 and has triggered an international public health emergency [].The numbers of confirmed cases and deaths due to COVID-19 are rapidly escalating, counting in millions [], causing massive economic strain ...
What are the best treatments for covid-19? Written in cooperation with the World Health Organization, The BMJ's living systematic review is a meta-analysis comparing the effects of treatments for covid-19,1 using data from more than 400 randomised clinical trials worldwide. At the time of writing, it states that systemic corticosteroids (particularly dexamethasone), interleukin-6 receptor ...
The most urgent and important question for COVID-19 in Africa is how to gain access to vaccines. Without a deliberate commitment globally to scale up vaccination in Africa, its people will be ...
To the Editor—The COVID-19 pandemic has produced devastating effects worldwide, with the causative coronavirus SARS-CoV-2 infecting over 170 million patients and causing more than 3.5 million ...
The WHO Covid-19 Research Database was maintained by the WHO Library & Digital Information Networks and was funded by COVID-19 emergency funds. The database was built by BIREME, the Specialized Center of PAHO/AMRO. Its content spanned the time period March 2020 to June 2023. It has now been archived, and no longer searchable since January 2024.
Despite widespread suffering from Covid-19, ... Medicine frequently segments patients by demographic or socioeconomic traits, ... (e.g., distrust of medical research by some people of color) that ...
COVID-19 Research. Stanford Medicine scientists have launched dozens of research projects as part of the global response to COVID-19. Some aim to prevent, diagnose and treat the disease; others aim to understand how it spreads and how people's immune systems respond to it. Below is a curated selection, including summaries, of the projects.
By the end of November 2021, scientists estimate that mRNA COVID-19 vaccines had prevented at least 1 million deaths, 10 million hospitalizations, and 36 million SARS-CoV-2 infections in the United States. Sometimes people who are fully vaccinated get a breakthrough infection, meaning that they test positive for SARS-CoV-2 or become ill with ...
COVID-19: Emergence, Spread, Possible Treatments, and Global Burden. The Coronavirus (CoV) is a large family of viruses known to cause illnesses ranging from the common cold to acute respiratory tract infection. The severity of the infection may be visible as pneumonia, acute respiratory syndrome, and even death.
Abstract. Background: Since the start of the COVID-19 outbreak, a large number of COVID-19-related papers have been published. However, concerns about the risk of expedited science have been raised. We aimed at reviewing and categorizing COVID-19-related medical research and to critically appraise peer-reviewed original articles.
This SHEA white paper identifies knowledge gaps and challenges in healthcare epidemiology research related to coronavirus disease 2019 (COVID-19) with a focus on core principles of healthcare epidemiology. These gaps, revealed during the worst phases of the COVID-19 pandemic, are described in 10 sections: epidemiology, outbreak investigation ...
Introduction. SARS-CoV-2 is responsible for a large number of global COVID-19 cases. It is a highly transmissible virus among humans that has become a significant public health issue. 1 Symptoms include fever, dry cough, fatigue, shortness of breath, chills, muscle pain, headache, gastric disorders and weight loss, often leading to death. 2 ...
Although Covid is new, the ethical issues prompted by it are not, and they needn't be addressed ab initio. There is a wealth of knowledge about the appropriate values and principles to guide ...
The COVID-19 research landscape: Measuring topics and collaborations using scientific literature Medicine (Baltimore). 2020 Oct 23;99(43): e22849. doi ... The number of the COVID-19 research publications has been continuously increasing after its break. United States was the most productive and active country for COVID-19 research, with the ...
So far, COVID-19 has caused about 4.3 million deaths. "COVID is one of many pandemics," Sudlow says. "We have been sitting on pandemics for years and not using these kinds of resources ...
EPIORGABOLISM is studying how SARS-Co-V2, the coronavirus responsible for the 2019 novel coronavirus disease (COVID-19), interacts with and infects kidney cells. Together with the lung, the kidney is one of the main organs affected by the COVID-19 disease. Dr Carmen Hurtado, researcher of EPIORGABOLISM, is currently generating human kidney ...
In vitro transcribed mRNA has lived up to its potential as a game-changing disruptive vaccine modality during the COVID-19 pandemic. As a result, research into mRNA therapeutics, which extends beyond the realm of vaccines to fight infectious diseases, has gained increasing attention both in academia and industry. Indeed, mRNA therapeutics have been explored beyond vaccines and other immune ...
Objectives: We aimed to assess the risk of autoimmune- and inflammatory post-acute COVID-19 conditions. Design: Descriptive network cohort study. Setting: Electronic health records from UK and Dutch primary care, Norwegian linked health registry, hospital records of specialist centres in Spain, France, and Korea, and healthcare claims from Estonia and the US. Participants: We followed ...
Tuberculosis (TB) is the leading infectious disease cause of death worldwide. In recent years, stringent measures to contain the spread of SARS-CoV-2 have led to considerable disruptions of healthcare services for TB in many countries. The extent to which these measures have affected TB testing, treatment initiation and outcomes has not been comprehensively assessed.
CAR T-cell immunotherapy changes these collected T cells to produce chimeric antigen receptors (or CARs) that adhere to tumors to destroy them. A recent study published in the New England Journal of Medicine, suggests CAR T-cell therapy could become a highly effective treatment for SLE patients who do not respond to current lupus therapeutics ...
Orthopaedic surgeon Cassandra Lee, center, was one of the first physicians to place new shock absorber implant for knee pain. The MISHA Knee System was designed to be an option for people like Barron who are experiencing a prohibitive amount of pain and haven't found relief from other treatments. It's meant to prolong the need to seek a ...
The topics covered include the occupational health system and policy, universal health coverage, vulnerable groups of workers, mental health at work, climate change and workers' health, with a particular focus on the health, safety and well-being of health workers.During the COVID-19 pandemic, the centre coordinated the project "Actions to ...
Here, using an agnostic, hybrid-capture sequencing approach, we report the detection of H5N1 in wastewater in nine Texas cities, with a total catchment area population in the millions, over a two-month period from March 4th to April 25th, 2024. Sequencing reads uniquely aligning to H5N1 covered all eight genome segments, with best alignments to ...
The COVID-19 research landscape - PMC. Journal List. Medicine (Baltimore) v.99 (43); 2020 Oct 23. PMC7581087. As a library, NLM provides access to scientific literature. Inclusion in an NLM database does not imply endorsement of, or agreement with, the contents by NLM or the National Institutes of Health.
To estimate VE, we adapted case-control methods 1 for prevalent COVID-like illness (CLI) as a proxy for confirmed COVID-19 cases. Therefore, our estimates of VE measure a vaccine's ability to ...