Data Representation
Class 11 - computer science with python sumita arora, checkpoint 2.1.
What are the bases of decimal, octal, binary and hexadecimal systems ?
The bases are:
- Decimal — Base 10
- Octal — Base 8
- Binary — Base 2
- Hexadecimal — Base 16
What is the common property of decimal, octal, binary and hexadecimal number systems ?
Decimal, octal, binary and hexadecimal number systems are all positional-value system .
Complete the sequence of following binary numbers : 100, 101, 110, ............... , ............... , ............... .
100, 101, 110, 111 , 1000 , 1001 .
Complete the sequence of following octal numbers : 525, 526, 527, ............... , ............... , ............... .
525, 526, 527, 530 , 531 , 532 .
Complete the sequence of following hexadecimal numbers : 17, 18, 19, ............... , ............... , ............... .
17, 18, 19, 1A , 1B , 1C .
Convert the following binary numbers to decimal and hexadecimal:
(c) 101011111
(e) 10010101
(f) 11011100
Converting to decimal:
| Binary No | Power | Value | Result |
|---|---|---|---|
| 0 | 2 | 1 | 0x1=0 |
| 1 | 2 | 2 | 1x2=2 |
| 0 | 2 | 4 | 0x4=0 |
| 1 | 2 | 8 | 1x8=8 |
Equivalent decimal number = 8 + 2 = 10
Therefore, (1010) 2 = (10) 10
Converting to hexadecimal:
Grouping in bits of 4:
1010 undefined \underlinesegment{1010} 1010
| Binary Number | Equivalent Hexadecimal |
|---|---|
| 1010 | A (10) |
Therefore, (1010) 2 = (A) 16
| Binary No | Power | Value | Result |
|---|---|---|---|
| 0 | 2 | 1 | 0x1=0 |
| 1 | 2 | 2 | 1x2=2 |
| 0 | 2 | 4 | 0x4=0 |
| 1 | 2 | 8 | 1x8=8 |
| 1 | 2 | 16 | 1x16=16 |
| 1 | 2 | 32 | 1x32=32 |
Equivalent decimal number = 32 + 16 + 8 + 2 = 58
Therefore, (111010) 2 = (58) 10
0011 undefined 1010 undefined \underlinesegment{0011} \quad \underlinesegment{1010} 0011 1010
| Binary Number | Equivalent Hexadecimal |
|---|---|
| 1010 | A (10) |
| 0011 | 3 |
Therefore, (111010) 2 = (3A) 16
| Binary No | Power | Value | Result |
|---|---|---|---|
| 1 | 2 | 1 | 1x1=1 |
| 1 | 2 | 2 | 1x2=2 |
| 1 | 2 | 4 | 1x4=4 |
| 1 | 2 | 8 | 1x8=8 |
| 1 | 2 | 16 | 1x16=16 |
| 0 | 2 | 32 | 0x32=0 |
| 1 | 2 | 64 | 1x64=64 |
| 0 | 2 | 128 | 0x128=0 |
| 1 | 2 | 256 | 1x256=256 |
Equivalent decimal number = 256 + 64 + 16 + 8 + 4 + 2 + 1 = 351
Therefore, (101011111) 2 = (351) 10
0001 undefined 0101 undefined 1111 undefined \underlinesegment{0001} \quad \underlinesegment{0101} \quad \underlinesegment{1111} 0001 0101 1111
| Binary Number | Equivalent Hexadecimal |
|---|---|
| 1111 | F (15) |
| 0101 | 5 |
| 0001 | 1 |
Therefore, (101011111) 2 = (15F) 16
| Binary No | Power | Value | Result |
|---|---|---|---|
| 0 | 2 | 1 | 0x1=0 |
| 0 | 2 | 2 | 0x2=0 |
| 1 | 2 | 4 | 1x4=4 |
| 1 | 2 | 8 | 1x8=8 |
Equivalent decimal number = 8 + 4 = 12
Therefore, (1100) 2 = (12) 10
1100 undefined \underlinesegment{1100} 1100
| Binary Number | Equivalent Hexadecimal |
|---|---|
| 1100 | C (12) |
Therefore, (1100) 2 = (C) 16
| Binary No | Power | Value | Result |
|---|---|---|---|
| 1 | 2 | 1 | 1x1=1 |
| 0 | 2 | 2 | 0x2=0 |
| 1 | 2 | 4 | 1x4=4 |
| 0 | 2 | 8 | 0x8=0 |
| 1 | 2 | 16 | 1x16=16 |
| 0 | 2 | 32 | 0x32=0 |
| 0 | 2 | 64 | 0x64=0 |
| 1 | 2 | 128 | 1x128=128 |
Equivalent decimal number = 1 + 4 + 16 + 128 = 149
Therefore, (10010101) 2 = (149) 10
1001 undefined 0101 undefined \underlinesegment{1001} \quad \underlinesegment{0101} 1001 0101
| Binary Number | Equivalent Hexadecimal |
|---|---|
| 0101 | 5 |
| 1001 | 9 |
Therefore, (101011111) 2 = (95) 16
| Binary No | Power | Value | Result |
|---|---|---|---|
| 0 | 2 | 1 | 0x1=0 |
| 0 | 2 | 2 | 0x2=0 |
| 1 | 2 | 4 | 1x4=4 |
| 1 | 2 | 8 | 1x8=8 |
| 1 | 2 | 16 | 1x16=16 |
| 0 | 2 | 32 | 0x32=0 |
| 1 | 2 | 64 | 1x64=64 |
| 1 | 2 | 128 | 1x128=128 |
Equivalent decimal number = 4 + 8 + 16 + 64 + 128 = 220
Therefore, (11011100) 2 = (220) 10
1101 undefined 1100 undefined \underlinesegment{1101} \quad \underlinesegment{1100} 1101 1100
| Binary Number | Equivalent Hexadecimal |
|---|---|
| 1100 | C (12) |
| 1101 | D (13) |
Therefore, (11011100) 2 = (DC) 16
Convert the following decimal numbers to binary and octal :
Converting to binary:
| 2 | Quotient | Remainder |
|---|---|---|
| 2 | 23 | 1 (LSB) |
| 2 | 11 | 1 |
| 2 | 5 | 1 |
| 2 | 2 | 0 |
| 2 | 1 | 1 (MSB) |
| 0 |
Therefore, (23) 10 = (10111) 2
Converting to octal:
| 8 | Quotient | Remainder |
|---|---|---|
| 8 | 23 | 7 (LSB) |
| 8 | 2 | 2 (MSB) |
| 0 |
Therefore, (23) 10 = (27) 8
| 2 | Quotient | Remainder |
|---|---|---|
| 2 | 100 | 0 (LSB) |
| 2 | 50 | 0 |
| 2 | 25 | 1 |
| 2 | 12 | 0 |
| 2 | 6 | 0 |
| 2 | 3 | 1 |
| 2 | 1 | 1 (MSB) |
| 0 |
Therefore, (100) 10 = (1100100) 2
| 8 | Quotient | Remainder |
|---|---|---|
| 8 | 100 | 4 (LSB) |
| 8 | 12 | 4 |
| 8 | 1 | 1 (MSB) |
| 0 |
Therefore, (100) 10 = (144) 8
| 2 | Quotient | Remainder |
|---|---|---|
| 2 | 145 | 1 (LSB) |
| 2 | 72 | 0 |
| 2 | 36 | 0 |
| 2 | 18 | 0 |
| 2 | 9 | 1 |
| 2 | 4 | 0 |
| 2 | 2 | 0 |
| 2 | 1 | 1 (MSB) |
| 0 |
Therefore, (145) 10 = (10010001) 2
| 8 | Quotient | Remainder |
|---|---|---|
| 8 | 145 | 1 (LSB) |
| 8 | 18 | 2 |
| 8 | 2 | 2 (MSB) |
| 0 |
Therefore, (145) 10 = (221) 8
| 2 | Quotient | Remainder |
|---|---|---|
| 2 | 19 | 1 (LSB) |
| 2 | 9 | 1 |
| 2 | 4 | 0 |
| 2 | 2 | 0 |
| 2 | 1 | 1 (MSB) |
| 0 |
Therefore, (19) 10 = (10011) 2
| 8 | Quotient | Remainder |
|---|---|---|
| 8 | 19 | 3 (LSB) |
| 8 | 2 | 2 (MSB) |
| 0 |
Therefore, (19) 10 = (23) 8
| 2 | Quotient | Remainder |
|---|---|---|
| 2 | 121 | 1 (LSB) |
| 2 | 60 | 0 |
| 2 | 30 | 0 |
| 2 | 15 | 1 |
| 2 | 7 | 1 |
| 2 | 3 | 1 |
| 2 | 1 | 1 (MSB) |
| 0 |
Therefore, (121) 10 = (1111001) 2
| 8 | Quotient | Remainder |
|---|---|---|
| 8 | 121 | 1 (LSB) |
| 8 | 15 | 7 |
| 8 | 1 | 1 (MSB) |
| 0 |
Therefore, (121) 10 = (171) 8
| 2 | Quotient | Remainder |
|---|---|---|
| 2 | 161 | 1 (LSB) |
| 2 | 80 | 0 |
| 2 | 40 | 0 |
| 2 | 20 | 0 |
| 2 | 10 | 0 |
| 2 | 5 | 1 |
| 2 | 2 | 0 |
| 2 | 1 | 1 (MSB) |
| 0 |
Therefore, (161) 10 = (10100001) 2
| 8 | Quotient | Remainder |
|---|---|---|
| 8 | 161 | 1 (LSB) |
| 8 | 20 | 4 |
| 8 | 2 | 2 (MSB) |
| 0 |
Therefore, (161) 10 = (241) 8
Convert the following hexadecimal numbers to binary :
| Hexadecimal Number | Binary Equivalent |
|---|---|
| 6 | 0110 |
| A (10) | 1010 |
(A6) 16 = (10100110) 2
| Hexadecimal Number | Binary Equivalent |
|---|---|
| 7 | 0111 |
| 0 | 0000 |
| A (10) | 1010 |
(A07) 16 = (101000000111) 2
| Hexadecimal Number | Binary Equivalent |
|---|---|
| 4 | 0100 |
| B (11) | 1011 |
| A (10) | 1010 |
| 7 | 0111 |
(7AB4) 16 = (111101010110100) 2
| Hexadecimal Number | Binary Equivalent |
|---|---|
| E (14) | 1110 |
| B (11) | 1011 |
(BE) 16 = (10111110) 2
| Hexadecimal Number | Binary Equivalent |
|---|---|
| 9 | 1001 |
| C (12) | 1100 |
| B (11) | 1011 |
(BC9) 16 = (101111001001) 2
| Hexadecimal Number | Binary Equivalent |
|---|---|
| 8 | 1000 |
| C (12) | 1100 |
| B (11) | 1011 |
| 9 | 1001 |
(9BC8) 16 = (1001101111001000) 2
Convert the following binary numbers to hexadecimal and octal :
(a) 10011011101
(b) 1111011101011011
(c) 11010111010111
(d) 1010110110111
(e) 10110111011011
(f) 1111101110101111
0100 undefined 1101 undefined 1101 undefined \underlinesegment{0100} \quad \underlinesegment{1101} \quad \underlinesegment{1101} 0100 1101 1101
| Binary Number | Equivalent Hexadecimal |
|---|---|
| 1101 | D (13) |
| 1101 | D (13) |
| 0100 | 4 |
Therefore, (10011011101) 2 = (4DD) 16
Converting to Octal:
Grouping in bits of 3:
010 undefined 011 undefined 011 undefined 101 undefined \underlinesegment{010} \quad \underlinesegment{011} \quad \underlinesegment{011} \quad \underlinesegment{101} 010 011 011 101
| Binary Number | Equivalent Octal |
|---|---|
| 101 | 5 |
| 011 | 3 |
| 011 | 3 |
| 010 | 2 |
Therefore, (10011011101) 2 = (2335) 8
1111 undefined 0111 undefined 0101 undefined 1011 undefined \underlinesegment{1111} \quad \underlinesegment{0111} \quad \underlinesegment{0101} \quad \underlinesegment{1011} 1111 0111 0101 1011
| Binary Number | Equivalent Hexadecimal |
|---|---|
| 1011 | B (11) |
| 0101 | 5 |
| 0111 | 7 |
| 1111 | F (15) |
Therefore, (1111011101011011) 2 = (F75B) 16
001 undefined 111 undefined 011 undefined 101 undefined 011 undefined 011 undefined \underlinesegment{001} \quad \underlinesegment{111} \quad \underlinesegment{011} \quad \underlinesegment{101} \quad \underlinesegment{011} \quad \underlinesegment{011} 001 111 011 101 011 011
| Binary Number | Equivalent Octal |
|---|---|
| 011 | 3 |
| 011 | 3 |
| 101 | 5 |
| 011 | 3 |
| 111 | 7 |
| 001 | 1 |
Therefore, (1111011101011011) 2 = (173533) 8
0011 undefined 0101 undefined 1101 undefined 0111 undefined \underlinesegment{0011} \quad \underlinesegment{0101} \quad \underlinesegment{1101} \quad \underlinesegment{0111} 0011 0101 1101 0111
| Binary Number | Equivalent Hexadecimal |
|---|---|
| 0111 | 7 |
| 1101 | D (13) |
| 0101 | 5 |
| 0011 | 3 |
Therefore, (11010111010111) 2 = (35D7) 16
011 undefined 010 undefined 111 undefined 010 undefined 111 undefined \underlinesegment{011} \quad \underlinesegment{010} \quad \underlinesegment{111} \quad \underlinesegment{010} \quad \underlinesegment{111} 011 010 111 010 111
| Binary Number | Equivalent Octal |
|---|---|
| 111 | 7 |
| 010 | 2 |
| 111 | 7 |
| 010 | 2 |
| 011 | 3 |
Therefore, (11010111010111) 2 = (32727) 8
0001 undefined 0101 undefined 1011 undefined 0111 undefined \underlinesegment{0001} \quad \underlinesegment{0101} \quad \underlinesegment{1011} \quad \underlinesegment{0111} 0001 0101 1011 0111
| Binary Number | Equivalent Hexadecimal |
|---|---|
| 0111 | 7 |
| 1011 | B (11) |
| 0101 | 5 |
| 0001 | 1 |
Therefore, (1010110110111) 2 = (15B7) 16
001 undefined 010 undefined 110 undefined 110 undefined 111 undefined \underlinesegment{001} \quad \underlinesegment{010} \quad \underlinesegment{110} \quad \underlinesegment{110} \quad \underlinesegment{111} 001 010 110 110 111
| Binary Number | Equivalent Octal |
|---|---|
| 111 | 7 |
| 110 | 6 |
| 110 | 6 |
| 010 | 2 |
| 001 | 1 |
Therefore, (1010110110111) 2 = (12667) 8
0010 undefined 1101 undefined 1101 undefined 1011 undefined \underlinesegment{0010} \quad \underlinesegment{1101} \quad \underlinesegment{1101} \quad \underlinesegment{1011} 0010 1101 1101 1011
| Binary Number | Equivalent Hexadecimal |
|---|---|
| 1011 | B (11) |
| 1101 | D (13) |
| 1101 | D (13) |
| 0010 | 2 |
Therefore, (10110111011011) 2 = (2DDB) 16
010 undefined 110 undefined 111 undefined 011 undefined 011 undefined \underlinesegment{010} \quad \underlinesegment{110} \quad \underlinesegment{111} \quad \underlinesegment{011} \quad \underlinesegment{011} 010 110 111 011 011
| Binary Number | Equivalent Octal |
|---|---|
| 011 | 3 |
| 011 | 3 |
| 111 | 7 |
| 110 | 6 |
| 010 | 2 |
Therefore, (10110111011011) 2 = (26733) 8
1111 undefined 1011 undefined 1010 undefined 1111 undefined \underlinesegment{1111} \quad \underlinesegment{1011} \quad \underlinesegment{1010} \quad \underlinesegment{1111} 1111 1011 1010 1111
| Binary Number | Equivalent Hexadecimal |
|---|---|
| 1111 | F (15) |
| 1010 | A (10) |
| 1011 | B (11) |
| 1111 | F (15) |
Therefore, (1111101110101111) 2 = (FBAF) 16
001 undefined 111 undefined 101 undefined 110 undefined 101 undefined 111 undefined \underlinesegment{001} \quad \underlinesegment{111} \quad \underlinesegment{101} \quad \underlinesegment{110} \quad \underlinesegment{101} \quad \underlinesegment{111} 001 111 101 110 101 111
| Binary Number | Equivalent Octal |
|---|---|
| 111 | 7 |
| 101 | 5 |
| 110 | 6 |
| 101 | 5 |
| 111 | 7 |
| 001 | 1 |
Therefore, (1111101110101111) 2 = (175657) 8

Checkpoint 2.2
Multiple choice questions.
The value of radix in binary number system is ..........
The value of radix in octal number system is ..........
The value of radix in decimal number system is ..........
The value of radix in hexadecimal number system is ..........
Which of the following are not valid symbols in octal number system ?
Which of the following are not valid symbols in hexadecimal number system ?
Which of the following are not valid symbols in decimal number system ?
The hexadecimal digits are 1 to 0 and A to ..........
The binary equivalent of the decimal number 10 is ..........
Question 10
ASCII code is a 7 bit code for ..........
- other symbol
- all of these ✓
Question 11
How many bytes are there in 1011 1001 0110 1110 numbers?
Question 12
The binary equivalent of the octal Numbers 13.54 is.....
- 1101.1110 ✓
- None of these
Question 13
The octal equivalent of 111 010 is.....
Question 14
The input hexadecimal representation of 1110 is ..........
Question 15
Which of the following is not a binary number ?
Question 16
Convert the hexadecimal number 2C to decimal:
Question 17
UTF8 is a type of .......... encoding.
- extended ASCII
Question 18
UTF32 is a type of .......... encoding.
Question 19
Which of the following is not a valid UTF8 representation?
- 2 octet (16 bits)
- 3 octet (24 bits)
- 4 octet (32 bits)
- 8 octet (64 bits) ✓
Question 20
Which of the following is not a valid encoding scheme for characters ?
Fill in the Blanks
The Decimal number system is composed of 10 unique symbols.
The Binary number system is composed of 2 unique symbols.
The Octal number system is composed of 8 unique symbols.
The Hexadecimal number system is composed of 16 unique symbols.
The illegal digits of octal number system are 8 and 9 .
Hexadecimal number system recognizes symbols 0 to 9 and A to F .
Each octal number is replaced with 3 bits in octal to binary conversion.
Each Hexadecimal number is replaced with 4 bits in Hex to binary conversion.
ASCII is a 7 bit code while extended ASCII is a 8 bit code.
The Unicode encoding scheme can represent all symbols/characters of most languages.
The ISCII encoding scheme represents Indian Languages' characters on computers.
UTF8 can take upto 4 bytes to represent a symbol.
UTF32 takes exactly 4 bytes to represent a symbol.
Unicode value of a symbol is called code point .
True/False Questions
A computer can work with Decimal number system. False
A computer can work with Binary number system. True
The number of unique symbols in Hexadecimal number system is 15. False
Number systems can also represent characters. False
ISCII is an encoding scheme created for Indian language characters. True
Unicode is able to represent nearly all languages' characters. True
UTF8 is a fixed-length encoding scheme. False
UTF32 is a fixed-length encoding scheme. True
UTF8 is a variable-length encoding scheme and can represent characters in 1 through 4 bytes. True
UTF8 and UTF32 are the only encoding schemes supported by Unicode. False
Type A: Short Answer Questions
What are some number systems used by computers ?
The most commonly used number systems are decimal, binary, octal and hexadecimal number systems.
What is the use of Hexadecimal number system on computers ?
The Hexadecimal number system is used in computers to specify memory addresses (which are 16-bit or 32-bit long). For example, a memory address 1101011010101111 is a big binary address but with hex it is D6AF which is easier to remember. The Hexadecimal number system is also used to represent colour codes. For example, FFFFFF represents White, FF0000 represents Red, etc.
What does radix or base signify ?
The radix or base of a number system signifies how many unique symbols or digits are used in the number system to represent numbers. For example, the decimal number system has a radix or base of 10 meaning it uses 10 digits from 0 to 9 to represent numbers.
What is the use of encoding schemes ?
Encoding schemes help Computers represent and recognize letters, numbers and symbols. It provides a predetermined set of codes for each recognized letter, number and symbol. Most popular encoding schemes are ASCI, Unicode, ISCII, etc.
Discuss UTF-8 encoding scheme.
UTF-8 is a variable width encoding that can represent every character in Unicode character set. The code unit of UTF-8 is 8 bits called an octet. It uses 1 to maximum 6 octets to represent code points depending on their size i.e. sometimes it uses 8 bits to store the character, other times 16 or 24 or more bits. It is a type of multi-byte encoding.
How is UTF-8 encoding scheme different from UTF-32 encoding scheme ?
UTF-8 is a variable length encoding scheme that uses different number of bytes to represent different characters whereas UTF-32 is a fixed length encoding scheme that uses exactly 4 bytes to represent all Unicode code points.
What is the most significant bit and the least significant bit in a binary code ?
In a binary code, the leftmost bit is called the most significant bit or MSB. It carries the largest weight. The rightmost bit is called the least significant bit or LSB. It carries the smallest weight. For example:
1 M S B 0 1 1 0 1 1 0 L S B \begin{matrix} \underset{\bold{MSB}}{1} & 0 & 1 & 1 & 0 & 1 & 1 & \underset{\bold{LSB}}{0} \end{matrix} MSB 1 0 1 1 0 1 1 LSB 0
What are ASCII and extended ASCII encoding schemes ?
ASCII encoding scheme uses a 7-bit code and it represents 128 characters. Its advantages are simplicity and efficiency. Extended ASCII encoding scheme uses a 8-bit code and it represents 256 characters.
What is the utility of ISCII encoding scheme ?
ISCII or Indian Standard Code for Information Interchange can be used to represent Indian languages on the computer. It supports Indian languages that follow both Devanagari script and other scripts like Tamil, Bengali, Oriya, Assamese, etc.
What is Unicode ? What is its significance ?
Unicode is a universal character encoding scheme that can represent different sets of characters belonging to different languages by assigning a number to each of the character. It has the following significance:
- It defines all the characters needed for writing the majority of known languages in use today across the world.
- It is a superset of all other character sets.
- It is used to represent characters across different platforms and programs.
What all encoding schemes does Unicode use to represent characters ?
Unicode uses UTF-8, UTF-16 and UTF-32 encoding schemes.
What are ASCII and ISCII ? Why are these used ?
ASCII stands for American Standard Code for Information Interchange. It uses a 7-bit code and it can represent 128 characters. ASCII code is mostly used to represent the characters of English language, standard keyboard characters as well as control characters like Carriage Return and Form Feed. ISCII stands for Indian Standard Code for Information Interchange. It uses a 8-bit code and it can represent 256 characters. It retains all ASCII characters and offers coding for Indian scripts also. Majority of the Indian languages can be represented using ISCII.
What are UTF-8 and UTF-32 encoding schemes. Which one is more popular encoding scheme ?
UTF-8 is a variable length encoding scheme that uses different number of bytes to represent different characters whereas UTF-32 is a fixed length encoding scheme that uses exactly 4 bytes to represent all Unicode code points. UTF-8 is the more popular encoding scheme.
What do you understand by code point ?
Code point refers to a code from a code space that represents a single character from the character set represented by an encoding scheme. For example, 0x41 is one code point of ASCII that represents character 'A'.
What is the difference between fixed length and variable length encoding schemes ?
Variable length encoding scheme uses different number of bytes or octets (set of 8 bits) to represent different characters whereas fixed length encoding scheme uses a fixed number of bytes to represent different characters.
Type B: Application Based Questions
Convert the following binary numbers to decimal:
| Binary No | Power | Value | Result |
|---|---|---|---|
| 1 | 2 | 1 | 1x1=1 |
| 0 | 2 | 2 | 0x2=0 |
| 1 | 2 | 4 | 1x4=4 |
| 1 | 2 | 8 | 1x8=8 |
Equivalent decimal number = 1 + 4 + 8 = 13
Therefore, (1101) 2 = (13) 10
Equivalent decimal number = 2 + 8 + 16 + 32 = 58
Equivalent decimal number = 1 + 2 + 4 + 8 + 16 + 64 + 256 = 351
Convert the following binary numbers to decimal :
Equivalent decimal number = 4 + 8 = 12
(b) 10010101
(c) 11011100
Convert the following decimal numbers to binary:
| Multiply | = | Resultant | Carry |
|---|---|---|---|
| 0.25 x 2 | = | 0.5 | 0 |
| 0.5 x 2 | = | 0 | 1 |
Therefore, (0.25) 10 = (0.01) 2
| 2 | Quotient | Remainder |
|---|---|---|
| 2 | 122 | 0 (LSB) |
| 2 | 61 | 1 |
| 2 | 30 | 0 |
| 2 | 15 | 1 |
| 2 | 7 | 1 |
| 2 | 3 | 1 |
| 2 | 1 | 1 (MSB) |
| 0 |
Therefore, (122) 10 = (1111010) 2
| Multiply | = | Resultant | Carry |
|---|---|---|---|
| 0.675 x 2 | = | 0.35 | 1 |
| 0.35 x 2 | = | 0.7 | 0 |
| 0.7 x 2 | = | 0.4 | 1 |
| 0.4 x 2 | = | 0.8 | 0 |
| 0.8 x 2 | = | 0.6 | 1 |
(We stop after 5 iterations if fractional part doesn't become 0)
Therefore, (0.675) 10 = (0.10101) 2
Convert the following decimal numbers to octal:
| 8 | Quotient | Remainder |
|---|---|---|
| 8 | 122 | 2 (LSB) |
| 8 | 15 | 7 |
| 8 | 1 | 1 (MSB) |
| 0 |
Therefore, (122) 10 = (172) 8
| Multiply | = | Resultant | Carry |
|---|---|---|---|
| 0.675 x 8 | = | 0.4 | 5 |
| 0.4 x 8 | = | 0.2 | 3 |
| 0.2 x 8 | = | 0.6 | 1 |
| 0.6 x 8 | = | 0.8 | 4 |
| 0.8 x 8 | = | 0.4 | 6 |
Therefore, (0.675) 10 = (0.53146) 8
Convert the following hexadecimal numbers to binary:
| Hexadecimal Number | Binary Equivalent |
|---|---|
| D (13) | 1101 |
| 3 | 0011 |
| 2 | 0010 |
(23D) 16 = (1000111101) 2
Convert the following binary numbers to hexadecimal:
(a) 1010110110111
(b) 10110111011011
(c) 0110101100
0001 undefined 1010 undefined 1100 undefined \underlinesegment{0001} \quad \underlinesegment{1010} \quad \underlinesegment{1100} 0001 1010 1100
| Binary Number | Equivalent Hexadecimal |
|---|---|
| 1100 | C (12) |
| 1010 | A (10) |
| 0001 | 1 |
Therefore, (0110101100) 2 = (1AC) 16
Convert the following octal numbers to decimal:
| Octal No | Power | Value | Result |
|---|---|---|---|
| 7 | 8 | 1 | 7x1=7 |
| 5 | 8 | 8 | 5x8=40 |
| 2 | 8 | 64 | 2x64=128 |
Equivalent decimal number = 7 + 40 + 128 = 175
Therefore, (257) 8 = (175) 10
| Octal No | Power | Value | Result |
|---|---|---|---|
| 7 | 8 | 1 | 7x1=7 |
| 2 | 8 | 8 | 2x8=16 |
| 5 | 8 | 64 | 5x64=320 |
| 3 | 8 | 512 | 3x512=1536 |
Equivalent decimal number = 7 + 16 + 320 + 1536 = 1879
Therefore, (3527) 8 = (1879) 10
| Octal No | Power | Value | Result |
|---|---|---|---|
| 3 | 8 | 1 | 3x1=3 |
| 2 | 8 | 8 | 2x8=16 |
| 1 | 8 | 64 | 1x64=64 |
Equivalent decimal number = 3 + 16 + 64 = 83
Therefore, (123) 8 = (83) 10
Integral part
| Octal No | Power | Value | Result |
|---|---|---|---|
| 5 | 8 | 1 | 5x1=5 |
| 0 | 8 | 8 | 0x8=0 |
| 6 | 8 | 64 | 6x64=384 |
Fractional part
| Octal No | Power | Value | Result |
|---|---|---|---|
| 1 | 8 | 0.125 | 1x0.125=0.125 |
| 2 | 8 | 0.0156 | 2x0.0156=0.0312 |
Equivalent decimal number = 5 + 384 + 0.125 + 0.0312 = 389.1562
Therefore, (605.12) 8 = (389.1562) 10
Convert the following hexadecimal numbers to decimal:
| Hexadecimal Number | Power | Value | Result |
|---|---|---|---|
| 6 | 16 | 1 | 6x1=6 |
| A (10) | 16 | 16 | 10x16=160 |
Equivalent decimal number = 6 + 160 = 166
Therefore, (A6) 16 = (166) 10
| Hexadecimal Number | Power | Value | Result |
|---|---|---|---|
| B (11) | 16 | 1 | 11x1=11 |
| 3 | 16 | 16 | 3x16=48 |
| 1 | 16 | 256 | 1x256=256 |
| A (10) | 16 | 4096 | 10x4096=40960 |
Equivalent decimal number = 11 + 48 + 256 + 40960 = 41275
Therefore, (A13B) 16 = (41275) 10
| Hexadecimal Number | Power | Value | Result |
|---|---|---|---|
| 5 | 16 | 1 | 5x1=5 |
| A (10) | 16 | 16 | 10x16=160 |
| 3 | 16 | 256 | 3x256=768 |
Equivalent decimal number = 5 + 160 + 768 = 933
Therefore, (3A5) 16 = (933) 10
| Hexadecimal Number | Power | Value | Result |
|---|---|---|---|
| 9 | 16 | 1 | 9x1=9 |
| E (14) | 16 | 16 | 14x16=224 |
Equivalent decimal number = 9 + 224 = 233
Therefore, (E9) 16 = (233) 10
| Hexadecimal Number | Power | Value | Result |
|---|---|---|---|
| 3 (11) | 16 | 1 | 3x1=3 |
| A (10) | 16 | 16 | 10x16=160 |
| C (12) | 16 | 256 | 12x256=3072 |
| 7 | 16 | 4096 | 7x4096=28672 |
Equivalent decimal number = 3 + 160 + 3072 + 28672 = 31907
Therefore, (7CA3) 16 = (31907) 10
Convert the following decimal numbers to hexadecimal:
| 16 | Quotient | Remainder |
|---|---|---|
| 16 | 132 | 4 |
| 16 | 8 | 8 |
| 0 |
Therefore, (132) 10 = (84) 16
| 16 | Quotient | Remainder |
|---|---|---|
| 16 | 2352 | 0 |
| 16 | 147 | 3 |
| 16 | 9 | 9 |
| 0 |
Therefore, (2352) 10 = (930) 16
| 16 | Quotient | Remainder |
|---|---|---|
| 16 | 122 | A (10) |
| 16 | 7 | 7 |
| 0 |
Therefore, (122) 10 = (7A) 16
| Multiply | = | Resultant | Carry |
|---|---|---|---|
| 0.675 x 16 | = | 0.8 | A (10) |
| 0.8 x 16 | = | 0.8 | C (12) |
| 0.8 x 16 | = | 0.8 | C (12) |
| 0.8 x 16 | = | 0.8 | C (12) |
| 0.8 x 16 | = | 0.8 | C (12) |
Therefore, (0.675) 10 = (0.ACCCC) 16
| 16 | Quotient | Remainder |
|---|---|---|
| 16 | 206 | E (14) |
| 16 | 12 | C (12) |
| 0 |
Therefore, (206) 10 = (CE) 16
| 16 | Quotient | Remainder |
|---|---|---|
| 16 | 3619 | 3 |
| 16 | 226 | 2 |
| 16 | 14 | E (14) |
| 0 |
Therefore, (3619) 10 = (E23) 16
Convert the following hexadecimal numbers to octal:
| Hexadecimal Number | Binary Equivalent |
|---|---|
| C (12) | 1100 |
| A (10) | 1010 |
| 8 | 1000 |
| 3 | 0011 |
(38AC) 16 = (11100010101100) 2
011 undefined 100 undefined 010 undefined 101 undefined 100 undefined \underlinesegment{011}\medspace\underlinesegment{100}\medspace\underlinesegment{010}\medspace\underlinesegment{101}\medspace\underlinesegment{100} 011 100 010 101 100
| Binary Number | Equivalent Octal |
|---|---|
| 100 | 4 |
| 101 | 5 |
| 010 | 2 |
| 100 | 4 |
| 011 | 3 |
(38AC) 16 = (34254) 8
| Hexadecimal Number | Binary Equivalent |
|---|---|
| 6 | 0110 |
| D (13) | 1101 |
| F (15) | 1111 |
| 7 | 0111 |
(7FD6) 16 = (111111111010110) 2
111 undefined 111 undefined 111 undefined 010 undefined 110 undefined \underlinesegment{111}\medspace\underlinesegment{111}\medspace\underlinesegment{111}\medspace\underlinesegment{010}\medspace\underlinesegment{110} 111 111 111 010 110
| Binary Number | Equivalent Octal |
|---|---|
| 110 | 6 |
| 010 | 2 |
| 111 | 7 |
| 111 | 7 |
| 111 | 7 |
(7FD6) 16 = (77726) 8
| Hexadecimal Number | Binary Equivalent |
|---|---|
| D (13) | 1101 |
| C (12) | 1100 |
| B (11) | 1011 |
| A (10) | 1010 |
(ABCD) 16 = (1010101111001101) 2
001 undefined 010 undefined 101 undefined 111 undefined 001 undefined 101 undefined \underlinesegment{001}\medspace\underlinesegment{010}\medspace\underlinesegment{101}\medspace\underlinesegment{111}\medspace\underlinesegment{001}\medspace\underlinesegment{101} 001 010 101 111 001 101
| Binary Number | Equivalent Octal |
|---|---|
| 101 | 5 |
| 001 | 1 |
| 111 | 7 |
| 101 | 5 |
| 010 | 2 |
| 001 | 1 |
(ABCD) 16 = (125715) 8
Convert the following octal numbers to binary:
| Octal Number | Binary Equivalent |
|---|---|
| 3 | 011 |
| 2 | 010 |
| 1 | 001 |
Therefore, (123) 8 = ( 001 undefined 010 undefined 011 undefined \bold{\underlinesegment{001}}\medspace\bold{\underlinesegment{010}}\medspace\bold{\underlinesegment{011}} 001 010 011 ) 2
| Octal Number | Binary Equivalent |
|---|---|
| 7 | 111 |
| 2 | 010 |
| 5 | 101 |
| 3 | 011 |
Therefore, (3527) 8 = ( 011 undefined 101 undefined 010 undefined 111 undefined \bold{\underlinesegment{011}}\medspace\bold{\underlinesegment{101}}\medspace\bold{\underlinesegment{010}}\medspace\bold{\underlinesegment{111}} 011 101 010 111 ) 2
| Octal Number | Binary Equivalent |
|---|---|
| 5 | 101 |
| 0 | 000 |
| 7 | 111 |
Therefore, (705) 8 = ( 111 undefined 000 undefined 101 undefined \bold{\underlinesegment{111}}\medspace\bold{\underlinesegment{000}}\medspace\bold{\underlinesegment{101}} 111 000 101 ) 2
| Octal Number | Binary Equivalent |
|---|---|
| 2 | 010 |
| 4 | 100 |
| 6 | 110 |
| 7 | 111 |
Therefore, (7642) 8 = ( 111 undefined 110 undefined 100 undefined 010 undefined \bold{\underlinesegment{111}}\medspace\bold{\underlinesegment{110}}\medspace\bold{\underlinesegment{100}}\medspace\bold{\underlinesegment{010}} 111 110 100 010 ) 2
| Octal Number | Binary Equivalent |
|---|---|
| 5 | 101 |
| 1 | 001 |
| 0 | 000 |
| 7 | 111 |
Therefore, (7015) 8 = ( 111 undefined 000 undefined 001 undefined 101 undefined \bold{\underlinesegment{111}}\medspace\bold{\underlinesegment{000}}\medspace\bold{\underlinesegment{001}}\medspace\bold{\underlinesegment{101}} 111 000 001 101 ) 2
| Octal Number | Binary Equivalent |
|---|---|
| 6 | 110 |
| 7 | 111 |
| 5 | 101 |
| 3 | 011 |
Therefore, (3576) 8 = ( 011 undefined 101 undefined 111 undefined 110 undefined \bold{\underlinesegment{011}}\medspace\bold{\underlinesegment{101}}\medspace\bold{\underlinesegment{111}}\medspace\bold{\underlinesegment{110}} 011 101 111 110 ) 2
Convert the following binary numbers to octal
111 undefined 010 undefined \underlinesegment{111} \quad \underlinesegment{010} 111 010
| Binary Number | Equivalent Octal |
|---|---|
| 010 | 2 |
| 111 | 7 |
Therefore, (111010) 2 = (72) 8
(b) 110110101
110 undefined 110 undefined 101 undefined \underlinesegment{110} \quad \underlinesegment{110} \quad \underlinesegment{101} 110 110 101
| Binary Number | Equivalent Octal |
|---|---|
| 101 | 5 |
| 110 | 6 |
| 110 | 6 |
Therefore, (110110101) 2 = (665) 8
(c) 1101100001
001 undefined 101 undefined 100 undefined 001 undefined \underlinesegment{001} \quad \underlinesegment{101} \quad \underlinesegment{100} \quad \underlinesegment{001} 001 101 100 001
| Binary Number | Equivalent Octal |
|---|---|
| 001 | 1 |
| 100 | 4 |
| 101 | 5 |
| 001 | 1 |
Therefore, (1101100001) 2 = (1541) 8
011 undefined 001 undefined \underlinesegment{011} \quad \underlinesegment{001} 011 001
| Binary Number | Equivalent Octal |
|---|---|
| 001 | 1 |
| 011 | 3 |
Therefore, (11001) 2 = (31) 8
(b) 10101100
010 undefined 101 undefined 100 undefined \underlinesegment{010} \quad \underlinesegment{101} \quad \underlinesegment{100} 010 101 100
| Binary Number | Equivalent Octal |
|---|---|
| 100 | 4 |
| 101 | 5 |
| 010 | 2 |
Therefore, (10101100) 2 = (254) 8
(c) 111010111
111 undefined 010 undefined 111 undefined \underlinesegment{111} \quad \underlinesegment{010} \quad \underlinesegment{111} 111 010 111
| Binary Number | Equivalent Octal |
|---|---|
| 111 | 7 |
| 010 | 2 |
| 111 | 7 |
Therefore, (111010111) 2 = (727) 8
Add the following binary numbers:
(i) 10110111 and 1100101
1 1 0 1 1 1 0 1 1 1 1 1 1 + 1 1 0 0 1 0 1 1 0 0 0 1 1 1 0 0 \begin{matrix} & & \overset{1}{1} & \overset{1}{0} & 1 & 1 & \overset{1}{0} & \overset{1}{1} & \overset{1}{1} & 1 \\ + & & & 1 & 1 & 0 & 0 & 1 & 0 & 1 \\ \hline & \bold{1} & \bold{0} & \bold{0} & \bold{0} & \bold{1} & \bold{1} & \bold{1} & \bold{0} & \bold{0} \end{matrix} + 1 1 1 0 0 1 1 0 1 1 0 1 0 1 0 1 0 1 1 1 1 1 1 1 0 0 1 1 0
Therefore, (10110111) 2 + (1100101) 2 = (100011100) 2
(ii) 110101 and 101111
1 1 1 1 0 1 1 1 0 1 1 + 1 0 1 1 1 1 1 1 0 0 1 0 0 \begin{matrix} & & \overset{1}{1} & \overset{1}{1} & \overset{1}{0} & \overset{1}{1} & \overset{1}{0} & 1 \\ + & & 1 & 0 & 1 & 1 & 1 & 1 \\ \hline & \bold{1} & \bold{1} & \bold{0} & \bold{0} & \bold{1} & \bold{0} & \bold{0} \end{matrix} + 1 1 1 1 1 1 1 0 0 0 1 1 0 1 1 1 1 0 1 1 0 1 1 0
Therefore, (110101) 2 + (101111) 2 = (1100100) 2
(iii) 110111.110 and 11011101.010
0 1 0 1 1 1 1 1 0 1 1 1 1 1 1 1 . 1 1 1 0 + 1 1 0 1 1 1 0 1 . 0 1 0 1 0 0 0 1 0 1 0 1 . 0 0 0 \begin{matrix} & & \overset{1}{0} & \overset{1}{0} & \overset{1}{1} & \overset{1}{1} & \overset{1}{0} & \overset{1}{1} & \overset{1}{1} & \overset{1}{1} & . & \overset{1}{1} & 1 & 0 \\ + & & 1 & 1 & 0 & 1 & 1 & 1 & 0 & 1 & . & 0 & 1 & 0 \\ \hline & \bold{1} & \bold{0} & \bold{0} & \bold{0} & \bold{1} & \bold{0} & \bold{1} & \bold{0} & \bold{1} & \bold{.} & \bold{0} & \bold{0} & \bold{0} \end{matrix} + 1 0 1 1 0 0 1 1 0 1 1 0 0 1 1 1 1 0 1 1 0 1 1 1 1 1 1 0 0 1 1 1 1 . . . 1 1 0 0 1 1 0 0 0 0
Therefore, (110111.110) 2 + (11011101.010) 2 = (100010101) 2
(iv) 1110.110 and 11010.011
0 1 1 1 1 1 1 0 1 . 1 1 1 0 + 1 1 0 1 0 . 0 1 1 1 0 1 0 0 1 . 0 0 1 \begin{matrix} & & \overset{1}{0} & \overset{1}{1} & \overset{1}{1} & 1 & \overset{1}{0} & . & \overset{1}{1} & 1 & 0 \\ + & & 1 & 1 & 0 & 1 & 0 & . & 0 & 1 & 1 \\ \hline & \bold{1} & \bold{0} & \bold{1} & \bold{0} & \bold{0} & \bold{1} & \bold{.} & \bold{0} & \bold{0} & \bold{1} \end{matrix} + 1 0 1 1 0 1 1 1 1 1 1 0 0 1 1 0 0 1 0 1 . . . 1 1 0 0 1 1 0 0 1 1
Therefore, (1110.110) 2 + (11010.011) 2 = (101001.001) 2
Question 21
Given that A's code point in ASCII is 65, and a's code point is 97. What is the binary representation of 'A' in ASCII ? (and what's its hexadecimal representation). What is the binary representation of 'a' in ASCII ?
Binary representation of 'A' in ASCII will be binary representation of its code point 65.
Converting 65 to binary:
| 2 | Quotient | Remainder |
|---|---|---|
| 2 | 65 | 1 (LSB) |
| 2 | 32 | 0 |
| 2 | 16 | 0 |
| 2 | 8 | 0 |
| 2 | 4 | 0 |
| 2 | 2 | 0 |
| 2 | 1 | 1 (MSB) |
| 0 |
Therefore, binary representation of 'A' in ASCII is 1000001.
Converting 65 to Hexadecimal:
| 16 | Quotient | Remainder |
|---|---|---|
| 16 | 65 | 1 |
| 16 | 4 | 4 |
| 0 |
Therefore, hexadecimal representation of 'A' in ASCII is (41) 16 .
Similarly, converting 97 to binary:
| 2 | Quotient | Remainder |
|---|---|---|
| 2 | 97 | 1 (LSB) |
| 2 | 48 | 0 |
| 2 | 24 | 0 |
| 2 | 12 | 0 |
| 2 | 6 | 0 |
| 2 | 3 | 1 |
| 2 | 1 | 1 (MSB) |
| 0 |
Therefore, binary representation of 'a' in ASCII is 1100001.
Question 22
Convert the following binary numbers to decimal, octal and hexadecimal numbers.
(i) 100101.101
Decimal Conversion of integral part:
| Binary No | Power | Value | Result |
|---|---|---|---|
| 1 | 2 | 1 | 1x1=1 |
| 0 | 2 | 2 | 0x2=0 |
| 1 | 2 | 4 | 1x4=4 |
| 0 | 2 | 8 | 0x8=0 |
| 0 | 2 | 16 | 0x16=0 |
| 1 | 2 | 32 | 1x32=32 |
Decimal Conversion of fractional part:
| Binary No | Power | Value | Result |
|---|---|---|---|
| 1 | 2 | 0.5 | 1x0.5=0.5 |
| 0 | 2 | 0.25 | 0x0.25=0 |
| 1 | 2 | 0.125 | 1x0.125=0.125 |
Equivalent decimal number = 1 + 4 + 32 + 0.5 + 0.125 = 37.625
Therefore, (100101.101) 2 = (37.625) 10
Octal Conversion
100 undefined 101 undefined . 101 undefined \underlinesegment{100} \quad \underlinesegment{101} \quad \bold{.} \quad \underlinesegment{101} 100 101 . 101
| Binary Number | Equivalent Octal |
|---|---|
| 101 | 5 |
| 100 | 4 |
| 101 | 5 |
Therefore, (100101.101) 2 = (45.5) 8
Hexadecimal Conversion
0010 undefined 0101 undefined . 1010 undefined \underlinesegment{0010} \quad \underlinesegment{0101} \medspace . \medspace \underlinesegment{1010} 0010 0101 . 1010
| Binary Number | Equivalent Hexadecimal |
|---|---|
| 0101 | 5 |
| 0010 | 2 |
| . | |
| 1010 | A (10) |
Therefore, (100101.101) 2 = (25.A) 16
(ii) 10101100.01011
| Binary No | Power | Value | Result |
|---|---|---|---|
| 0 | 2 | 1 | 0x1=0 |
| 0 | 2 | 2 | 0x2=0 |
| 1 | 2 | 4 | 1x4=4 |
| 1 | 2 | 8 | 1x8=8 |
| 0 | 2 | 16 | 0x16=0 |
| 1 | 2 | 32 | 1x32=32 |
| 0 | 2 | 64 | 0x64=0 |
| 1 | 2 | 128 | 1x128=128 |
| Binary No | Power | Value | Result |
|---|---|---|---|
| 0 | 2 | 0.5 | 0x0.5=0 |
| 1 | 2 | 0.25 | 1x0.25=0.25 |
| 0 | 2 | 0.125 | 0x0.125=0 |
| 1 | 2 | 0.0625 | 1x0.0625=0.0625 |
| 1 | 2 | 0.03125 | 1x0.03125=0.03125 |
Equivalent decimal number = 4 + 8 + 32 + 128 + 0.25 + 0.0625 + 0.03125 = 172.34375
Therefore, (10101100.01011) 2 = (172.34375) 10
010 undefined 101 undefined 100 undefined . 010 undefined 110 undefined \underlinesegment{010} \quad \underlinesegment{101} \quad \underlinesegment{100} \quad \bold{.} \quad \underlinesegment{010} \quad \underlinesegment{110} 010 101 100 . 010 110
| Binary Number | Equivalent Octal |
|---|---|
| 100 | 4 |
| 101 | 5 |
| 010 | 2 |
| 010 | 2 |
| 110 | 6 |
Therefore, (10101100.01011) 2 = (254.26) 8
1010 undefined 1100 undefined . 0101 undefined 1000 undefined \underlinesegment{1010} \quad \underlinesegment{1100} \medspace . \medspace \underlinesegment{0101} \medspace \underlinesegment{1000} 1010 1100 . 0101 1000
| Binary Number | Equivalent Hexadecimal |
|---|---|
| 1100 | C (12) |
| 1010 | A (10) |
| . | |
| 0101 | 5 |
| 1000 | 8 |
Therefore, (10101100.01011) 2 = (AC.58) 16
Decimal Conversion:
| Binary No | Power | Value | Result |
|---|---|---|---|
| 0 | 2 | 1 | 0x1=0 |
| 1 | 2 | 2 | 1x2=2 |
| 0 | 2 | 4 | 0x4=0 |
| 1 | 2 | 8 | 1x8=8 |
Equivalent decimal number = 2 + 8 = 10
001 undefined 010 undefined \underlinesegment{001} \quad \underlinesegment{010} 001 010
| Binary Number | Equivalent Octal |
|---|---|
| 010 | 2 |
| 001 | 1 |
Therefore, (1010) 2 = (12) 8
(iv) 10101100.010111
| Binary No | Power | Value | Result |
|---|---|---|---|
| 0 | 2 | 0.5 | 0x0.5=0 |
| 1 | 2 | 0.25 | 1x0.25=0.25 |
| 0 | 2 | 0.125 | 0x0.125=0 |
| 1 | 2 | 0.0625 | 1x0.0625=0.0625 |
| 1 | 2 | 0.03125 | 1x0.03125=0.03125 |
| 1 | 2 | 0.015625 | 1x0.015625=0.015625 |
Equivalent decimal number = 4 + 8 + 32 + 128 + 0.25 + 0.0625 + 0.03125 + 0.015625 = 172.359375
Therefore, (10101100.010111) 2 = (172.359375) 10
010 undefined 101 undefined 100 undefined . 010 undefined 111 undefined \underlinesegment{010} \quad \underlinesegment{101} \quad \underlinesegment{100} \quad \bold{.} \quad \underlinesegment{010} \quad \underlinesegment{111} 010 101 100 . 010 111
| Binary Number | Equivalent Octal |
|---|---|
| 100 | 4 |
| 101 | 5 |
| 010 | 2 |
| 010 | 2 |
| 111 | 7 |
Therefore, (10101100.010111) 2 = (254.27) 8
1010 undefined 1100 undefined . 0101 undefined 1100 undefined \underlinesegment{1010} \quad \underlinesegment{1100} \medspace . \medspace \underlinesegment{0101} \medspace \underlinesegment{1100} 1010 1100 . 0101 1100
| Binary Number | Equivalent Hexadecimal |
|---|---|
| 1100 | C (12) |
| 1010 | A (10) |
| . | |
| 0101 | 5 |
| 1100 | C (12) |
Therefore, (10101100.010111) 2 = (AC.5C) 16
Data Representation in Computer MCQ [PDF] 40 Top Question
Data representation in computer MCQ . Questions and answers with PDF for all Computer Related Entrance & Competitive Exams Preparation. Helpful for Class 11, GATE, IBPS, SBI (Bank PO & Clerk), SSC, Railway etc.
Data Representation in Computer MCQ
1. To perform calculation on stored data computer, uses ……… number system. [SBI Clerk 2009]
(1) decimal
(2) hexadecimal
2. The number system based on ‘0’ and ‘1’ only, is known as
(1) binary system
(2) barter system
(3) number system
(4) hexadecimal system
3. Decimal number system is the group of ………… numbers.
(4) 0 to 9 and A to F
4. A hexadecimal number is represented by
(1) three digits
(2) four binary digits
(3) four digits
(4) All of these
5. A hexadigit can be represented by [IBPS Clerk 2012]
(1) three binary (consecutive) bits
(2) four binary (consecutive) bits
(3) eight binary (consecutive) bits
(4) sixteen binary (consecutive) bits
(5) None of the above
6. What type of information system would be recognised by digital circuits?
(1) Hexadecimal system
(2) Binary system
(3) Both ‘1’ and ‘2’
(4) Only roman system
7. The binary equivalent of decimal number 98 is [IBPS Clerk 2012]
(1) 1110001
(2) 1110100
(3) 1100010
(4) 1111001
(5) None of these
8. What is the value of the binary number 101?
9. The binary number 10101 is equivalent to decimal number ………….
10. To convert binary number to decimal, multiply the all binary digits by power of
11. Which of the following is a hexadecimal number equal to 3431 octal number?
12. LSD stands for
(1) Long Significant Digit
(2) Least Significant Digit
(3) Large Significant Digit
(4) Longer Significant Decimal
13. How many values can be represented by a single byte?
14. Which of the following is not a computer code?
(4) UNICODE
15. MSD refers as
(1) Most Significant Digit
(2) Many Significant Digit
(3) Multiple Significant Digit
(4) Most Significant Decimal
16. The most widely used code that represents each character as a unique 8-bit code is [IBPS Clerk 2011]
(2) UNICODE
17. Today’s mostly used coding system is/are
(4) Both ‘1’ and ‘2’
18. In EBCDIC code, maximum possible characters set size is
19. Code ‘EBCDIC’ that is used in computing stands for
(1) Extension BCD Information Code
(2) Extended BCD Information Code
(3) Extension BCD Interchange Conduct
(4) Extended BCD Interchange Conduct
20. Most commonly used codes for representing bits are
21. The coding system allows non-english characters and special characters to be represented
22. Which of the following is invalid hexadecimal number?
23. Gate having output 1 only when one of its input is 1 is called
24. Gate is also known as inverter.
25. The only function of NOT gate is to ……..
(1) Stop signal
(2) Invert input signal
(3) Act as a universal gate
(4) Double input signal
26. Which of following are known as universal gates?
(1) NAND & NOR
(2) AND & OR
(3) XOR & OR
27. Gate whose output is 0 only when inputs are different is called
28. In the binary language, each letter of the alphabet, each number and each special character is made up of a unique combination of [BOB Clerk 2010]
c) 8 character
29. Decimal equivalent of (1111) 2 is [IBPS Clerk 2012]
30. ASCII code for letter A is
a) 1100011
b) 1000001
c) 1111111
31. Which of the following is not a binary number? [IBPS Clerk 2011]
32. Which of the following is an example of binary number? [IBPS Clerk 2011]
33. Numbers that are written with base 10 are classified as
(1) decimal number
(2) whole number
(3) hexadecimal number
(4) exponential integers
34. The octal system [IBPS Clerk 2011]
(1) needs less digits to represent a number than in the binary system
(2) needs more digits to represent a number than in the binary system
(3) needs the same number of digits to represent a number as in the binary system
(4) needs the same number of digits to represent a number as in the decimal system
35. Hexadecimal number system has ………. base.
36. ASCII stands for [IBPS Clerk 2011,2014]
(1) American Special Computer for Information Interaction
(2) American Standard Computer for Information Interchange
(3) American Special Code for Information Interchange
(4) American Special Computer for Information Interchange
(5) American Standard Code for Information Interchange
| [PDF Download] |
- Show all results for " "
🎧 New: AI-Generated Podcasts Turn your study notes into engaging audio conversations. Learn more

Data Representation in Computer Science Quiz
More actions.
- PDF Questions
- Make a copy
Podcast Beta
Questions and answers, what are binary digits and how are they represented.
Binary digits, also known as bits, are represented by the digits 1 and 0, to signify the states 'on' and 'off' respectively.
What are transistors and how do they function in computers?
Transistors are electronic devices that control the flow of electricity and act as switches which can be turned on or off by an electrical signal. In computers, millions or even billions of transistors are organized in intricate patterns on integrated circuits to perform calculations, store data, and execute various tasks.
How do computers manipulate complex information using binary switches?
By combining simple binary switches (bits), computers can represent and manipulate complex information such as numbers, text, images, and videos.
What is the foundation of digital computing and the operation of modern computers?
<p>The binary system, which uses binary digits to represent information, forms the foundation of digital computing and is fundamental to the operation of modern computers.</p> Signup and view all the answers
What is a numeral system (or system of numeration) and how does it relate to number representation?
<p>A numeral system is a system of representing numbers. In the context of computers, the binary system is a numeral system that uses 0 and 1 to represent numbers, forming the basis for number representation in digital computing.</p> Signup and view all the answers
Study Notes
Binary representation in computers.
- Computers use binary digits, also known as bits, to represent information.
- Each bit can be in one of two states: "on" or "off", represented by the digits 1 and 0, respectively.
- These individual switches are called transistors, which are electronic devices controlling the flow of electricity.
Transistors in Computers
- Transistors act as switches that can be turned on or off by an electrical signal.
- Computers contain millions, or even billions, of transistors organized in intricate patterns on integrated circuits.
- These transistors work together to perform calculations, store data, and execute various tasks.
Binary System in Digital Computing
- By combining these simple binary switches, computers can represent and manipulate complex information such as numbers, text, images, and videos.
- The binary system forms the foundation of digital computing and is fundamental to the operation of modern computers.
Number Systems
- A numeral system (or system of numeration) is a way of representing numbers.
Studying That Suits You
Use AI to generate personalized quizzes and flashcards to suit your learning preferences.
Description
Test your knowledge of data representation in computer science with this quiz. Explore the fundamentals of binary digits, bits, transistors, and their role in representing and processing information in computers.
More Quizzes Like This

The Binary World
Data Representation Quiz
Data Representation in Computers: Chapter Four 4.1
Share this lesson
Upgrade to continue
Today's Special Offer
Save an additional 20% with coupon: SAVE20
Upgrade to a paid plan to continue
Trusted by top students and educators worldwide
We are constantly improving Quizgecko and would love to hear your feedback. You can also submit feature requests here: feature requests.
Create your free account
By continuing, you agree to Quizgecko's Terms of Service and Privacy Policy .
Data representation 1: Introduction
This course investigates how systems software works: what makes programs work fast or slow, and how properties of the machines we program impact the programs we write. We discuss both general ideas and specific tools, and take an experimental approach.
Textbook readings
- How do computers represent different kinds of information?
- How do data representation choices impact performance and correctness?
- What kind of language is understood by computer processors?
- How is code you write translated to code a processor runs?
- How do hardware and software defend against bugs and attacks?
- How are operating systems interfaces implemented?
- What kinds of computer data storage are available, and how do they perform?
- How can we improve the performance of a system that stores data?
- How can programs on the same computer cooperate and interact?
- What kinds of operating systems interfaces are useful?
- How can a single program safely use multiple processors?
- How can multiple computers safely interact over a network?
- Six problem sets
- Midterm and final
- Starting mid-next week
- Attendance checked for simultaneously-enrolled students
- Rough breakdown: >50% assignments, <35% tests, 15% participation
- Course grading: A means mastery
Collaboration
Discussion, collaboration, and the exchange of ideas are essential to doing academic work, and to engineering. You are encouraged to consult with your classmates as you work on problem sets. You are welcome to discuss general strategies for solutions as well as specific bugs and code structure questions, and to use Internet resources for general information.
However, the work you turn in must be your own—the result of your own efforts. You should understand your code well enough that you could replicate your solution from scratch, without collaboration.
In addition, you must cite any books, articles, online resources, and so forth that helped you with your work, using appropriate citation practices; and you must list the names of students with whom you have collaborated on problem sets and briefly describe how you collaborated. (You do not need to list course staff.)
On our programming language
We use the C++ programming language in this class.
C++ is a boring, old, and unsafe programming language, but boring languages are underrated . C++ offers several important advantages for this class, including ubiquitous availability, good tooling, the ability to demonstrate impactful kinds of errors that you should understand, and a good standard library of data structures.
Pset 0 links to several C++ tutorials and references, and to a textbook.
Each program runs in a private data storage space. This is called its memory . The memory “remembers” the data it stores.
Programs work by manipulating values . Different programming languages have different conceptions of value; in C++, the primitive values are integers, like 12 or -100; floating-point numbers, like 1.02; and pointers , which are references to other objects.
An object is a region of memory that contains a value. (The C++ standard specifically says “a region of data storage in the execution environment, the contents of which can represent values”.)
Objects, values, and variables
Which are the objects? Which are the values?
Variables generally correspond to objects, and here there are three objects, one for each variable i1 , i2 , and i3 . The compiler and operating system associate the names with their corresponding objects. There are three values, too, one used to initialize each object: 61 , 62 , and 63 . However, there are other values—for instance, each argument to the printf calls is a value.
What does the program print?
i1: 61 i2: 62 i3: 63
C and C++ pointer types allow programs to access objects indirectly. A pointer value is the address of another object. For instance, in this program, the variable i4 holds a pointer to the object named by i3 :
There are four objects, corresponding to variables i1 through i4 . Note that the i4 object holds a pointer value, not an integer. There are also four values: 61 , 62 , 63 , and the expression &i3 (the address of i3 ). Note that there are three integer values, but four values overall.
What does this program print?
i1: 61 i2: 62 i3: 63 value pointed to by i4: 63
Here, the expressions i3 and *i4 refer to exactly the same object. Any modification to i3 can be observed through *i4 and vice versa. We say that i3 and *i4 are aliases : different names for the same object.
We now use hexdump_object , a helper function declared in our hexdump.hh helper file , to examine both the contents and the addresses of these objects.
Exactly what is printed will vary between operating systems and compilers. In Docker in class, on my Apple-silicon Macbook, we saw:
But on an Intel-based Amazon EC2 native Linux machine:
The data bytes look similar—identical for i1 through i3 —but the addresses vary.
But on Intel Mac OS X: 103c63020 3d 00 00 00 |=...| 103c5ef60 3e 00 00 00 |>...| 7ffeebfa4abc 3f 00 00 00 |?...| 7ffeebfa4ab0 bc 4a fa eb fe 7f 00 00 |.J......| And on Docker on an Intel Mac: 56499f239010 3d 00 00 00 |=...| 56499f23701c 3e 00 00 00 |>...| 7fffebf8b19c 3f 00 00 00 |?...| 7fffebf8b1a0 9c b1 f8 eb ff 7f 00 00 |........|
A hexdump printout shows the following information on each line.
- An address , like 4000004010 . This is a hexadecimal (base-16) number indicating the value of the address of the object. A line contains one to sixteen bytes of memory starting at this address.
- The contents of memory starting at the given address, such as 3d 00 00 00 . Memory is printed as a sequence of bytes , which are 8-bit numbers between 0 and 255. All modern computers organize their memory in units of 8-bit bytes.
- A textual representation of the memory contents, such as |=...| . This is useful when examining memory that contains textual data, and random garbage otherwise.
Dynamic allocation
Must every data object be given a name? No! In C++, the new operator allocates a brand-new object with no variable name. (In C, the malloc function does the same thing.) The C++ expression new T returns a pointer to a brand-new, never-before-seen object of type T . For instance:
This prints something like
The new int{64} expression allocates a fresh object with no name of its own, though it can be located by following the i4 pointer.
What do you notice about the addresses of these different objects?
- i3 and i4 , which are objects corresponding to variables declared local to main , are located very close to one another. In fact they are just 4 bytes part: i3 directly abuts i4 . Their addresses are quite high. In native Linux, in fact, their addresses are close to 2 47 !
- i1 and i2 are at much lower addresses, and they do not abut. i2 ’s location is below i1 , and about 0x2000 bytes away.
- The anonymous storage allocated by new int is located between i1 / i2 and i3 / i4 .
Although the values may differ on other operating systems, you’ll see qualitatively similar results wherever you run ./objects .
What’s happening is that the operating system and compiler have located different kinds of object in different broad regions of memory. These regions are called segments , and they are important because objects’ different storage characteristics benefit from different treatment.
i2 , the const int global object, has the smallest address. It is in the code or text segment, which is also used for read-only global data. The operating system and hardware ensure that data in this segment is not changed during the lifetime of the program. Any attempt to modify data in the code segment will cause a crash.
i1 , the int global object, has the next highest address. It is in the data segment, which holds modifiable global data. This segment keeps the same size as the program runs.
After a jump, the anonymous new int object pointed to by i4 has the next highest address. This is the heap segment, which holds dynamically allocated data. This segment can grow as the program runs; it typically grows towards higher addresses.
After a larger jump, the i3 and i4 objects have the highest addresses. They are in the stack segment, which holds local variables. This segment can also grow as the program runs, especially as functions call other functions; in most processors it grows down , from higher addresses to lower addresses.
Experimenting with the stack
How can we tell that the stack grows down? Do all functions share a single stack? This program uses a recursive function to test. Try running it; what do you see?

Snapsolve any problem by taking a picture. Try it in the Numerade app?
The Essentials Of Computer Organization And Architecture
Linda null, julia lobur, data representation in computer systems - all with video answers.

Chapter Questions
Perform the following base conversions using subtraction or division-remainder: a) $458_{10}=$________ 3 b) $677_{10}=$________ 5 c) $1518_{10}=$_______ 7 d) $4401_{10}=$_______ 9

Perform the following base conversions using subtraction or division-remainder: a) $588_{10}=$_________ 3 b) $2254_{10}=$________ 5 c) $652_{10}=$________ 7 d) $3104_{10}=$________ 9

Convert the following decimal fractions to binary with a maximum of six places to the right of the binary point: a) 26.78125 b) 194.03125 c) 298.796875 d) 16.1240234375
Convert the following decimal fractions to binary with a maximum of six places to the right of the binary point: a) 25.84375 b) 57.55 c) 80.90625 d) 84.874023
Represent the following decimal numbers in binary using 8 -bit signed magnitude, one's complement, and two's complement: a) 77 b) -42 c) 119 d) -107

Using a "word" of 3 bits, list all of the possible signed binary numbers and their decimal equivalents that are representable in: a) Signed magnitude b) One's complement c) Two's complement

Using a "word" of 4 bits, list all of the possible signed binary numbers and their decimal equivalents that are representable in: a) Signed magnitude b) One's complement c) Two's complement
From the results of the previous two questions, generalize the range of values (in decimal) that can be represented in any given $x$ number of bits using: a) Signed magnitude b) One's complement c) Two's complement
Given a (very) tiny computer that has a word size of 6 bits, what are the smallest negative numbers and the largest positive numbers that this computer can represent in each of the following representations? a) One's complement b) Two's complement

You have stumbled on an unknown civilization while sailing around the world. The people, who call themselves Zebronians, do math using 40 separate characters (probably because there are 40 stripes on a zebra). They would very much like to use computers, but would need a computer to do Zebronian math, which would mean a computer that could represent all 40 characters. You are a computer designer and decide to help them. You decide the best thing is to use $\mathrm{BCZ}$, Binary-Coded Zebronian (which is like $\mathrm{BCD}$ except it codes Zebronian, not Decimal). How many bits will you need to represent each character if you want to use the minimum number of bits?

Perform the following binary multiplications: a) 1100 $\times 101$ b) 10101 $\times 111$ c) 11010 $\times 1100$
Perform the following binary multiplications: a) 1011 $\times 101$ b) 10011 $\times 1011$ c) 11010 $\times 101$
Perform the following binary divisions: a) $101101 \div 101$ b) $10000001 \div 101$ c) $1001010010 \div 1011$
Perform the following binary divisions: a) $11111101 \div 1011$ b) $110010101 \div 1001$ c) $1001111100 \div 1100$
Use the double-dabble method to convert $10212_{3}$ directly to decimal. (Hint: you have to change the multiplier.)

Using signed-magnitude representation, complete the following operations: \[ \begin{aligned} +0+(-0) &=\\ (-0)+0 &=\\ 0+0 &=\\ (-0)+(-0) &= \end{aligned} \]

Suppose a computer uses 4 -bit one's complement numbers. Ignoring overflows, what value will be stored in the variable $j$ after the following pseudocode routine terminates? \[ \begin{array}{ll} 0 \rightarrow j & \text { // Store } 0 \text { in } j \text { . } \\ -3 \rightarrow k & \text { // store }-3 \text { in } k \text { . } \end{array} \] while $k \neq 0$ \[ j=j+1 \] \[ k=k-1 \] end while
If the floating-point number storage on a certain system has a sign bit, a 3 -bit exponent, and a 4-bit significand: a) What is the largest positive and the smallest negative number that can be stored on this system if the storage is normalized? (Assume no bits are implied, there is no biasing, exponents use two's complement notation, and exponents of all zeros and all ones are allowed.) b) What bias should be used in the exponent if we prefer all exponents to be nonnegative? Why would you choose this bias?
Using the model in the previous question, including your chosen bias, add the following floating-point numbers and express your answer using the same notation as the addend and augend:
$$\begin{array}{|llllllll|} \hline 0 & 1 & 1 & 1 & 1 & 0 & 0 & 0 \\ 0 & 1 & 0 & 1 & 1 & 0 & 0 & 1 \\ \hline \end{array}$$ Calculate the relative error, if any, in your answer to the previous question.

Assume we are using the simple model for floating-point representation as given in this book (the representation uses a 14 -bit format, 5 bits for the exponent with a bias of $16, \text { a normalized mantissa of } 8 \text { bits, and a single sign bit for the number })$ a) Show how the computer would represent the numbers 100.0 and 0.25 using this floating-point format. b) Show how the computer would add the two floating-point numbers in part a by changing one of the numbers so they are both expressed using the same power of 2. c) Show how the computer would represent the sum in part b using the given floating-point representation. What decimal value for the sum is the computer actually storing? Explain.
What causes divide underflow and what can be done about it?
Why do we usually store floating-point numbers in normalized form? What is the advantage of using a bias as opposed to adding a sign bit to the exponent?

Let $a=1.0 \times 2^{9}, b=-1.0 \times 2^{9}$ and $c=1.0 \times 2^{1} .$ Using the floating-point model described in the text (the representation uses a 14 -bit format, 5 bits for the exponent with a bias of $16,$ a normalized mantissa of 8 bits, and a single sign bit for the number), perform the following calculations, paying close attention to the order of operations. What can you say about the algebraic properties of floating-point arithmetic in our finite model? Do you think this algebraic anomaly holds under multiplication as well as addition? $$\begin{array}{l} b+(a+c)= \\ (b+a)+c= \end{array}$$

a) Given that the ASCII code for A is 1000001 , what is the ASCII code for $\mathrm{J} ?$ b) Given that the EBCDIC code for A is 11000001 , what is the EBCDIC code for J?

Assume a 24 -bit word on a computer. In these 24 bits, we wish to represent the value 295 a) If our computer uses even parity, how would the computer represent the decimal value $295 ?$ b) If our computer uses 8 -bit ASCII and even parity, how would the computer represent the string $295 ?$ c) If our computer uses packed $\mathrm{BCD}$, how would the computer represent the number $+295 ?$
Decode the following ASCII message, assuming 7 -bit ASCII characters and no parity: 1001010100111110010001001110010000010001001000101
Why would a system designer wish to make Unicode the default character set for their new system? What reason(s) could you give for not using Unicode as a default?

Write the 7 -bit ASCII code for the character 4 using the following encoding: a) Non-return-to-zero b) Non-return-to-zero-invert c) Manchester code d) Frequency modulation e) Modified frequency modulation f) Run length limited (Assume 1 is "high," and 0 is "'low.")
Why is NRZ coding seldom used for recording data on magnetic media?

Assume we wish to create a code using 3 information bits, 1 parity bit (appended to the end of the information), and odd parity. List all legal code words in this code. What is the Hamming distance of your code?

Are the error-correcting Hamming codes systematic? Explain.

Compute the Hamming distance of the following code: 0011010010111100 0000011110001111 0010010110101101 0001011010011110

Compute the Hamming distance of the following code: 0000000101111111 0000001010111111 0000010011011111 0000100011101111 0001000011110111 0010000011111011 01000000111111101 1000000011111110
Suppose we want an error-correcting code that will allow all single-bit errors to be corrected for memory words of length 10. a) How many parity bits are necessary? b) Assuming we are using the Hamming algorithm presented in this chapter to design our error-correcting code, find the code word to represent the 10 -bit information word: 1001100110.
Suppose we are working with an error-correcting code that will allow all single-bit errors to be corrected for memory words of length $7 .$ We have already calculated that we need 4 check bits, and the length of all code words will be $11 .$ Code words are created according to the Hamming algorithm presented in the text. We now receive the following code word: 10101011110 Assuming even parity, is this a legal code word? If not, according to our error-correcting code, where is the error?
Repeat exercise 35 using the following code word: 01111010101

Name two ways in which Reed-Soloman coding differs from Hamming coding.
When would you choose a CRC code over a Hamming code? A Hamming code over a CRC?
Find the quotients and remainders for the following division problems modulo 2 a) $1010111_{2} \div 1101_{2}$ b) $1011111_{2} \div 11101_{2}$ c) $1011001101_{2} \div 10101_{2}$ d) $111010111_{2} \div 10111_{2}$

Find the quotients and remainders for the following division problems modulo 2 a) $1111010_{2} \div 1011_{2}$ b) $1010101_{2} \div 1100_{2}$ c) $1101101011_{2} \div 10101_{2}$ d) $1111101011_{2} \div 101101_{2}$
Using the CRC polynomial 1011 , compute the CRC code word for the information word, 1011001 . Check the division performed at the receiver.
Using the CRC polynomial 1101 , compute the CRC code word for the information word, $01001101 .$ Check the division performed at the receiver.

Pick an architecture (such as 80486 , Pentium, Pentium IV, SPARC, Alpha, or MIPS). Do research to find out how your architecture approaches the concepts introduced in this chapter. For example, what representation does it use for negative values? What character codes does it support?
Data Representation 5.3. Numbers
Data representation.
- 5.1. What's the big picture?
- 5.2. Getting started
Understanding the base 10 number system
Representing whole numbers in binary, shorthand for binary numbers - hexadecimal, computers representing numbers in practice, how many bits are used in practice, representing negative numbers in practice.
- 5.5. Images and Colours
- 5.6. Program Instructions
- 5.7. The whole story!
- 5.8. Further reading
In this section, we will look at how computers represent numbers. To begin with, we'll revise how the base 10 number system that we use every day works, and then look at binary , which is base 2. After that, we'll look at some other charactertistics of numbers that computers must deal with, such as negative numbers and numbers with decimal points.
The number system that humans normally use is in base 10 (also known as decimal). It's worth revising quickly, because binary numbers use the same ideas as decimal numbers, just with fewer digits!
In decimal, the value of each digit in a number depends on its place in the number. For example, in $123, the 3 represents $3, whereas the 1 represents $100. Each place value in a number is worth 10 times more than the place value to its right, i.e. there are the "ones", the "tens", the "hundreds", the "thousands" the "ten thousands", the "hundred thousands", the "millions", and so on. Also, there are 10 different digits (0,1,2,3,4,5,6,7,8,9) that can be at each of those place values.
If you were only able to use one digit to represent a number, then the largest number would be 9. After that, you need a second digit, which goes to the left, giving you the next ten numbers (10, 11, 12... 19). It's because we have 10 digits that each one is worth 10 times as much as the one to its right.
You may have encountered different ways of expressing numbers using "expanded form". For example, if you want to write the number 90328 in expanded form you might have written it as:
A more sophisticated way of writing it is:
If you've learnt about exponents, you could write it as:
The key ideas to notice from this are:
- Decimal has 10 digits – 0, 1, 2, 3, 4, 5, 6, 7, 8, 9.
- A place is the place in the number that a digit is, i.e. ones, tens, hundreds, thousands, and so on. For example, in the number 90328, 3 is in the "hundreds" place, 2 is in the "tens" place, and 9 is in the "ten thousands" place.
- Numbers are made with a sequence of digits.
- The right-most digit is the one that's worth the least (in the "ones" place).
- The left-most digit is the one that's worth the most.
- Because we have 10 digits, the digit at each place is worth 10 times as much as the one immediately to the right of it.
All this probably sounds really obvious, but it is worth thinking about consciously, because binary numbers have the same properties.
As discussed earlier, computers can only store information using bits, which have 2 possible states. This means that they cannot represent base 10 numbers using digits 0 to 9, the way we write down numbers in decimal. Instead, they must represent numbers using just 2 digits – 0 and 1.
Binary works in a very similar way to decimal, even though it might not initially seem that way. Because there are only 2 digits, this means that each digit is 2 times the value of the one immediately to the right.
The base 10 (decimal) system is sometimes called denary, which is more consistent with the name binary for the base 2 system. The word "denary" also refers to the Roman denarius coin, which was worth ten asses (an "as" was a copper or bronze coin). The term "denary" seems to be used mainly in the UK; in the US, Australia and New Zealand the term "decimal" is more common.
The interactive below illustrates how this binary number system represents numbers. Have a play around with it to see what patterns you can see.
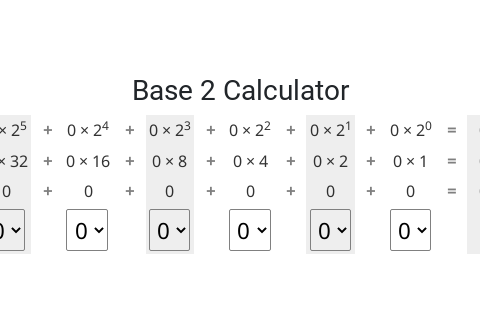
Base Calculator
Find the representations of 4, 7, 12, and 57 using the interactive.
What is the largest number you can make with the interactive? What is the smallest? Is there any integer value in between the biggest and the smallest that you can’t make? Are there any numbers with more than one representation? Why/ why not?
- 000000 in binary, 0 in decimal is the smallest number.
- 111111 in binary, 63 in decimal is the largest number.
- All the integer values (0, 1, 2... 63) in the range can be represented (and there is a unique representation for each one). This is exactly the same as decimal!
You have probably noticed from the interactive that when set to 1, the leftmost bit (the "most significant bit") adds 32 to the total, the next adds 16, and then the rest add 8, 4, 2, and 1 respectively. When set to 0, a bit does not add anything to the total. So the idea is to make numbers by adding some or all of 32, 16, 8, 4, 2, and 1 together, and each of those numbers can only be included once.

Choose a number less than 61 (perhaps your house number, your age, a friend's age, or the day of the month you were born on), set all the binary digits to zero, and then start with the left-most digit (32), trying out if it should be zero or one. See if you can find a method for converting the number without too much trial and error. Try different numbers until you find a quick way of doing this.
Figure out the binary representation for 23 without using the interactive? What about 4, 0, and 32? Check all your answers using the interactive to verify they are correct.
Can you figure out a systematic approach to counting in binary? i.e. start with the number 0, then increment it to 1, then 2, then 3, and so on, all the way up to the highest number that can be made with the 7 bits. Try counting from 0 to 16, and see if you can detect a pattern. Hint: Think about how you add 1 to a number in base 10. e.g. how do you work out 7 + 1, 38 + 1, 19 + 1, 99 + 1, 230899999 + 1, etc? Can you apply that same idea to binary?
Using your new knowledge of the binary number system, can you figure out a way to count to higher than 10 using your 10 fingers? What is the highest number you can represent using your 10 fingers? What if you included your 10 toes as well (so you have 20 fingers and toes to count with).
A binary number can be incremented by starting at the right and flipping all consecutive bits until a 1 comes up (which will be on the very first bit half of the time).
Counting on fingers in binary means that you can count to 31 on 5 fingers, and 1023 on 10 fingers. There are a number of videos on YouTube of people counting in binary on their fingers. One twist is to wear white gloves with the numbers 16, 8, 4, 2, 1 on the 5 fingers respectively, which makes it easy to work out the value of having certain fingers raised.
The interactive used exactly 6 bits. In practice, we can use as many or as few bits as we need, just like we do with decimal. For example, with 5 bits, the place values would be 16, 8, 4, 2 and 1, so the largest value is 11111 in binary, or 31 in decimal. Representing 14 with 5 bits would give 01110.
Write representations for the following. If it is not possible to do the representation, put "Impossible".
- Represent 101 with 7 bits
- Represent 28 with 10 bits
- Represent 7 with 3 bits
- Represent 18 with 4 bits
- Represent 28232 with 16 bits
The answers are (spaces are added to make the answers easier to read, but are not required).
- 101 with 7 bits is: 110 0101
- 28 with 10 bits is: 00 0001 1100
- 7 with 3 bits is: 111
- 18 with 4 bits is: Impossible (not enough bits to represent value)
- 28232 with 16 bits is: 0110 1110 0100 1000
An important concept with binary numbers is the range of values that can be represented using a given number of bits. When we have 8 bits the binary numbers start to get useful – they can represent values from 0 to 255, so it is enough to store someone's age, the day of the month, and so on.
Groups of 8 bits are so useful that they have their own name: a byte . Computer memory and disk space are usually divided up into bytes, and bigger values are stored using more than one byte. For example, two bytes (16 bits) are enough to store numbers from 0 to 65,535. Four bytes (32 bits) can store numbers up to 4,294,967,295. You can check these numbers by working out the place values of the bits. Every bit that's added will double the range of the number.
In practice, computers store numbers with either 16, 32, or 64 bits. This is because these are full numbers of bytes (a byte is 8 bits), and makes it easier for computers to know where each number starts and stops.
Candles on birthday cakes use the base 1 numbering system, where each place is worth 1 more than the one to its right. For example, the number 3 is 111, and 10 is 1111111111. This can cause problems as you get older – if you've ever seen a cake with 100 candles on it, you'll be aware that it's a serious fire hazard.
Luckily it's possible to use binary notation for birthday candles – each candle is either lit or not lit. For example, if you are 18, the binary notation is 10010, and you need 5 candles (with only two of them lit).
There's a video on using binary notation for counting up to 1023 on your hands, as well as using it for birthday cakes .
Most of the time binary numbers are stored electronically, and we don't need to worry about making sense of them. But sometimes it's useful to be able to write down and share numbers, such as the unique identifier assigned to each digital device (MAC address), or the colours specified in an HTML page.
Writing out long binary numbers is tedious – for example, suppose you need to copy down the 16-bit number 0101001110010001. A widely used shortcut is to break the number up into 4-bit groups (in this case, 0101 0011 1001 0001), and then write down the digit that each group represents (giving 5391). There's just one small problem: each group of 4 bits can go up to 1111, which is 15, and the digits only go up to 9.
The solution is simple: we introduce symbols for the digits from 1010 (10) to 1111 (15), which are just the letters A to F. So, for example, the 16-bit binary number 1011 1000 1110 0001 can be written more concisely as B8E1. The "B" represents the binary 1011, which is the decimal number 11, and the E represents binary 1110, which is decimal 14.
Because we now have 16 digits, this representation is base 16, and known as hexadecimal (or hex for short). Converting between binary and hexadecimal is very simple, and that's why hexadecimal is a very common way of writing down large binary numbers.
Here's a full table of all the 4-bit numbers and their hexadecimal digit equivalent:
| 0000 | 0 |
| 0001 | 1 |
| 0010 | 2 |
| 0011 | 3 |
| 0100 | 4 |
| 0101 | 5 |
| 0110 | 6 |
| 0111 | 7 |
| 1000 | 8 |
| 1001 | 9 |
| 1010 | A |
| 1011 | B |
| 1100 | C |
| 1101 | D |
| 1110 | E |
| 1111 | F |
For example, the largest 8-bit binary number is 11111111. This can be written as FF in hexadecimal. Both of those representations mean 255 in our conventional decimal system (you can check that by converting the binary number to decimal).
Which notation you use will depend on the situation; binary numbers represent what is actually stored, but can be confusing to read and write; hexadecimal numbers are a good shorthand of the binary; and decimal numbers are used if you're trying to understand the meaning of the number or doing normal math. All three are widely used in computer science.
It is important to remember though, that computers only represent numbers using binary. They cannot represent numbers directly in decimal or hexadecimal.
A common place that numbers are stored on computers is in spreadsheets or databases. These can be entered either through a spreadsheet program or database program, through a program you or somebody else wrote, or through additional hardware such as sensors, collecting data such as temperatures, air pressure, or ground shaking.
Some of the things that we might think of as numbers, such as the telephone number (03) 555-1234, aren't actually stored as numbers, as they contain important characters (like dashes and spaces) as well as the leading 0 which would be lost if it was stored as a number (the above number would come out as 35551234, which isn't quite right). These are stored as text , which is discussed in the next section.
On the other hand, things that don't look like a number (such as "30 January 2014") are often stored using a value that is converted to a format that is meaningful to the reader (try typing two dates into Excel, and then subtract one from the other – the result is a useful number). In the underlying representation, a number is used. Program code is used to translate the underlying representation into a meaningful date on the user interface.
The difference between two dates in Excel is the number of days between them; the date itself (as in many systems) is stored as the amount of time elapsed since a fixed date (such as 1 January 1900). You can test this by typing a date like "1 January 1850" – chances are that it won't be formatted as a normal date. Likewise, a date sufficiently in the future may behave strangely due to the limited number of bits available to store the date.
Numbers are used to store things as diverse as dates, student marks, prices, statistics, scientific readings, sizes and dimensions of graphics.
The following issues need to be considered when storing numbers on a computer:
- What range of numbers should be able to be represented?
- How do we handle negative numbers?
- How do we handle decimal points or fractions?
In practice, we need to allocate a fixed number of bits to a number, before we know how big the number is. This is often 32 bits or 64 bits, although can be set to 16 bits, or even 128 bits, if needed. This is because a computer has no way of knowing where a number starts and ends, otherwise.
Any system that stores numbers needs to make a compromise between the number of bits allocated to store the number, and the range of values that can be stored.
In some systems (like the Java and C programming languages and databases) it's possible to specify how accurately numbers should be stored; in others it is fixed in advance (such as in spreadsheets).
Some are able to work with arbitrarily large numbers by increasing the space used to store them as necessary (e.g. integers in the Python programming language). However, it is likely that these are still working with a multiple of 32 bits (e.g. 64 bits, 96 bits, 128 bits, 160 bits, etc). Once the number is too big to fit in 32 bits, the computer would reallocate it to have up to 64 bits.
In some programming languages there isn't a check for when a number gets too big (overflows). For example, if you have an 8-bit number using two's complement, then 01111111 is the largest number (127), and if you add one without checking, it will change to 10000000, which happens to be the number -128. (Don't worry about two's complement too much, it's covered later in this section.) This can cause serious problems if not checked for, and is behind a variant of the Y2K problem, called the Year 2038 problem , involving a 32-bit number overflowing for dates on Tuesday, 19 January 2038.
On tiny computers, such as those embedded inside your car, washing machine, or a tiny sensor that is barely larger than a grain of sand, we might need to specify more precisely how big a number needs to be. While computers prefer to work with chunks of 32 bits, we could write a program (as an example for an earthquake sensor) that knows the first 7 bits are the lattitude, the next 7 bits are the longitude, the next 10 bits are the depth, and the last 8 bits are the amount of force.
Even on standard computers, it is important to think carefully about the number of bits you will need. For example, if you have a field in your database that could be either "0", "1", "2", or "3" (perhaps representing the four bases that can occur in a DNA sequence), and you used a 64 bit number for every one, that will add up as your database grows. If you have 10,000,000 items in your database, you will have wasted 62 bits for each one (only 2 bits is needed to represent the 4 numbers in the example), a total of 620,000,000 bits, which is around 74 MB. If you are doing this a lot in your database, that will really add up – human DNA has about 3 billion base pairs in it, so it's incredibly wasteful to use more than 2 bits for each one.
And for applications such as Google Maps, which are storing an astronomical amount of data, wasting space is not an option at all!
It is really useful to know roughly how many bits you will need to represent a certain value. Have a think about the following scenarios, and choose the best number of bits out of the options given. You want to ensure that the largest possible number will fit within the number of bits, but you also want to ensure that you are not wasting space.
- Storing the day of the week - a) 1 bit - b) 4 bits - c) 8 bits - d) 32 bits
- Storing the number of people in the world - a) 16 bits - b) 32 bits - c) 64 bits - d) 128 bits
- Storing the number of roads in New Zealand - a) 16 bits - b) 32 bits - c) 64 bits - d) 128 bits
- Storing the number of stars in the universe - a) 16 bits - b) 32 bits - c) 64 bits - d) 128 bits
- b (actually, 3 bits is enough as it gives 8 values, but amounts that fit evenly into 8-bit bytes are easier to work with)
- c (32 bits is slightly too small, so you will need 64 bits)
- b (This is a challenging question, but one a database designer would have to think about. There's about 94,000 km of roads in New Zealand, so if the average length of a road was 1km, there would be too many roads for 16 bits. Either way, 32 bits would be a safe bet.)
- d (Even 64 bits is not enough, but 128 bits is plenty! Remember that 128 bits isn't twice the range of 64 bits.)
The binary number representation we have looked at so far allows us to represent positive numbers only. In practice, we will want to be able to represent negative numbers as well, such as when the balance of an account goes to a negative amount, or the temperature falls below zero. In our normal representation of base 10 numbers, we represent negative numbers by putting a minus sign in front of the number. But in binary, is it this simple?
We will look at two possible approaches: Adding a simple sign bit, much like we do for decimal, and then a more useful system called two's complement.
Using a simple sign bit
On a computer we don’t have minus signs for numbers (it doesn't work very well to use the text based one when representing a number because you can't do arithmetic on characters), but we can do it by allocating one extra bit, called a sign bit, to represent the minus sign. Just like with decimal numbers, we put the negative indicator on the left of the number — when the sign bit is set to "0", that means the number is positive and when the sign bit is set to "1", the number is negative (just as if there were a minus sign in front of it).
For example, if we wanted to represent the number 41 using 7 bits along with an additional bit that is the sign bit (to give a total of 8 bits), we would represent it by 00101001 . The first bit is a 0, meaning the number is positive, then the remaining 7 bits give 41 , meaning the number is +41 . If we wanted to make -59 , this would be 10111011 . The first bit is a 1, meaning the number is negative, and then the remaining 7 bits represent 59 , meaning the number is -59 .
Using 8 bits as described above (one for the sign, and 7 for the actual number), what would be the binary representations for 1, -1, -8, 34, -37, -88, and 102?
The spaces are not necessary, but are added to make reading the binary numbers easier
- 1 is 0000 0001
- -1 is 1000 0001
- -8 is 1000 1000
- 34 is 0010 0010
- -37 is 1010 0101
- -88 is 1101 1000
- 102 is 0110 0110
Going the other way is just as easy. If we have the binary number 10010111 , we know it is negative because the first digit is a 1. The number part is the next 7 bits 0010111 , which is 23 . This means the number is -23 .
What would the decimal values be for the following, assuming that the first bit is a sign bit?
- 00010011 is 19
- 10000110 is -6
- 10100011 is -35
- 01111111 is 127
- 11111111 is -127
But what about 10000000? That converts to -0 . And 00000000 is +0 . Since -0 and +0 are both just 0, it is very strange to have two different representations for the same number.
This is one of the reasons that we don't use a simple sign bit in practice. Instead, computers usually use a more sophisticated representation for negative binary numbers called two's complement .
Two's complement
There's an alternative representation called two's complement , which avoids having two representations for 0, and more importantly, makes it easier to do arithmetic with negative numbers.
Representing positive numbers with two's complement
Representing positive numbers is the same as the method you have already learnt. Using 8 bits ,the leftmost bit is a zero and the other 7 bits are the usual binary representation of the number; for example, 1 would be 00000001 , and 50 would be 00110010 .
Representing negative numbers with two's complement
This is where things get more interesting. In order to convert a negative number to its two's complement representation, use the following process. 1. Convert the number to binary (don't use a sign bit, and pretend it is a positive number). 2. Invert all the digits (i.e. change 0's to 1's and 1's to 0's). 3. Add 1 to the result (Adding 1 is easy in binary; you could do it by converting to decimal first, but think carefully about what happens when a binary number is incremented by 1 by trying a few; there are more hints in the panel below).
For example, assume we want to convert -118 to its two's complement representation. We would use the process as follows. 1. The binary number for 118 is 01110110 . 2. 01110110 with the digits inverted is 10001001 . 3. 10001001 + 1 is 10001010 .
Therefore, the two's complement representation for -118 is 10001010 .
The rule for adding one to a binary number is pretty simple, so we'll let you figure it out for yourself. First, if a binary number ends with a 0 (e.g. 1101010), how would the number change if you replace the last 0 with a 1? Now, if it ends with 01, how much would it increase if you change the 01 to 10? What about ending with 011? 011111?
The method for adding is so simple that it's easy to build computer hardware to do it very quickly.
What would be the two's complement representation for the following numbers, using 8 bits ? Follow the process given in this section, and remember that you do not need to do anything special for positive numbers.
- 19 in binary is 0001 0011 , which is the two's complement for a positive number.
- For -19, we take the binary of the positive, which is 0001 0011 (above), invert it to 1110 1100, and add 1, giving a representation of 1110 1101 .
- 107 in binary is 0110 1011 , which is the two's complement for a positive number.
- For -107, we take the binary of the positive, which is 0110 1011 (above), invert it to 1001 0100, and add 1, giving a representation of 1001 0101 .
- For -92, we take the binary of the positive, which is 0101 1100, invert it to 1010 0011, and add 1, giving a representation of 1010 0100 . (If you have this incorrect, double check that you incremented by 1 correctly).
Converting a two's complement number back to decimal
In order to reverse the process, we need to know whether the number we are looking at is positive or negative. For positive numbers, we can simply convert the binary number back to decimal. But for negative numbers, we first need to convert it back to a normal binary number.
So how do we know if the number is positive or negative? It turns out (for reasons you will understand later in this section) that two's complement numbers that are negative always start in a 1, and positive numbers always start in a 0. Have a look back at the previous examples to double check this.
So, if the number starts with a 1, use the following process to convert the number back to a negative decimal number.
- Subtract 1 from the number.
- Invert all the digits.
- Convert the resulting binary number to decimal.
- Add a minus sign in front of it.
So if we needed to convert 11100010 back to decimal, we would do the following.
- Subtract 1 from 11100010 , giving 11100001 .
- Invert all the digits, giving 00011110 .
- Convert 00011110 to a binary number, giving 30 .
- Add a negative sign, giving -30 .
Convert the following two's complement numbers to decimal.
- 10001100 -> (-1) 10001011 -> (inverted) 01110100 -> (to decimal) 116 -> (negative sign added) -116
- 10111111 -> (-1) 10111110 -> (inverted) 01000001 -> (to decimal) 65 -> (negative sign added) -65
How many numbers can be represented using two's complement?
While it might initially seem that there is no bit allocated as the sign bit, the left-most bit behaves like one. With 8 bits, you can still only make 256 possible patterns of 0's and 1's. If you attempted to use 8 bits to represent positive numbers up to 255, and negative numbers down to -255, you would quickly realise that some numbers were mapped onto the same pattern of bits. Obviously, this will make it impossible to know what number is actually being represented!
In practice, numbers within the following ranges can be represented. Unsigned Range is how many numbers you can represent if you only allow positive numbers (no sign is needed), and two's complement Range is how many numbers you can represent if you require both positive and negative numbers. You can work these out because the range of 8-bit values if they are stored using unsigned numbers will be from 00000000 to 11111111 (i.e. 0 to 255 in decimal), while the signed two's complement range is from 10000000 (the lowest number, -128 in decimal) to 01111111 (the highest number, 127 in decimal). This might seem a bit weird, but it works out really well because normal binary addition can be used if you use this representation even if you're adding a negative number.
| 8 bit | 0 to 255 | -128 to 127 |
| 16 bit | 0 to 65,535 | -32,768 to 32,767 |
| 32 bit | 0 to 4,294,967,295 | −2,147,483,648 to 2,147,483,647 |
| 64 bit | 0 to 18,446,744,073,709,551,615 | −9,223,372,036,854,775,808 to 9,223,372,036,854,775,807 |
Adding negative binary numbers
Before adding negative binary numbers, we'll look at adding positive numbers. It's basically the same as the addition methods used on decimal numbers, except the rules are way simpler because there are only two different digits that you might add!
You've probably learnt about column addition. For example, the following column addition would be used to do 128 + 255 .
When you go to add 5 + 8, the result is higher than 9, so you put the 3 in the one's column, and carry the 1 to the 10's column. Binary addition works in exactly the same way.
Adding positive binary numbers
If you wanted to add two positive binary numbers, such as 00001111 and 11001110 , you would follow a similar process to the column addition. You only need to know 0+0, 0+1, 1+0, and 1+1, and 1+1+1. The first three are just what you might expect. Adding 1+1 causes a carry digit, since in binary 1+1 = 10, which translates to "0, carry 1" when doing column addition. The last one, 1+1+1 adds up to 11 in binary, which we can express as "1, carry 1". For our two example numbers, the addition works like this:
Remember that the digits can be only 1 or 0. So you will need to carry a 1 to the next column if the total you get for a column is (decimal) 2 or 3.
Adding negative numbers with a simple sign bit
With negative numbers using sign bits like we did before, this does not work. If you wanted to add +11 (01011) and -7 (10111) , you would expect to get an answer of +4 (00100) .
Which is -2 .
One way we could solve the problem is to use column subtraction instead. But this would require giving the computer a hardware circuit which could do this. Luckily this is unnecessary, because addition with negative numbers works automatically using two's complement!
Adding negative numbers with two's complement
For the above addition (+11 + -7), we can start by converting the numbers to their 5-bit two's complement form. Because 01011 (+11) is a positive number, it does not need to be changed. But for the negative number, 00111 (-7) (sign bit from before removed as we don't use it for two's complement), we need to invert the digits and then add 1, giving 11001 .
Adding these two numbers works like this:
Any extra bits to the left (beyond what we are using, in this case 5 bits) have been truncated. This leaves 00100 , which is 4 , like we were expecting.
We can also use this for subtraction. If we are subtracting a positive number from a positive number, we would need to convert the number we are subtracting to a negative number. Then we should add the two numbers. This is the same as for decimal numbers, for example 5 - 2 = 3 is the same as 5 + (-2) = 3.
This property of two's complement is very useful. It means that positive numbers and negative numbers can be handled by the same computer circuit, and addition and subtraction can be treated as the same operation.
The idea of using a "complementary" number to change subtraction to addition can be seen by doing the same in decimal. The complement of a decimal digit is the digit that adds up to 10; for example, the complement of 4 is 6, and the complement of 8 is 2. (The word "complement" comes from the root "complete" – it completes it to a nice round number.)
Subtracting 2 from 6 is the same as adding the complement, and ignoring the extra 1 digit on the left. The complement of 2 is 8, so we add 8 to 6, giving (1)4.
For larger numbers (such as subtracting the two 3-digit numbers 255 - 128), the complement is the number that adds up to the next power of 10 i.e. 1000-128 = 872. Check that adding 872 to 255 produces (almost) the same result as subtracting 128.
Working out complements in binary is way easier because there are only two digits to work with, but working them out in decimal may help you to understand what is going on.
Using sign bits vs using two's complement
We have now looked at two different ways of representing negative numbers on a computer. In practice, a simple sign bit is rarely used, because of having two different representations of zero, and requiring a different computer circuit to handle negative and positive numbers, and to do addition and subtraction.
Two's complement is widely used, because it only has one representation for zero, and it allows positive numbers and negative numbers to be treated in the same way, and addition and subtraction to be treated as one operation.
There are other systems such as "One's Complement" and "Excess-k", but two's complement is by far the most widely used in practice.

- Number System and Arithmetic
- Trigonometry
- Probability
- Mensuration
- Linear Algebra
- CBSE Class 8 Maths Formulas
- CBSE Class 9 Maths Formulas
- CBSE Class 10 Maths Formulas
- CBSE Class 11 Maths Formulas
What are the different ways of Data Representation?
The process of collecting the data and analyzing that data in large quantity is known as statistics. It is a branch of mathematics trading with the collection, analysis, interpretation, and presentation of numeral facts and figures.
It is a numerical statement that helps us to collect and analyze the data in large quantity the statistics are based on two of its concepts:
- Statistical Data
- Statistical Science
Statistics must be expressed numerically and should be collected systematically.
Data Representation
The word data refers to constituting people, things, events, ideas. It can be a title, an integer, or anycast. After collecting data the investigator has to condense them in tabular form to study their salient features. Such an arrangement is known as the presentation of data.
It refers to the process of condensing the collected data in a tabular form or graphically. This arrangement of data is known as Data Representation.
The row can be placed in different orders like it can be presented in ascending orders, descending order, or can be presented in alphabetical order.
Example: Let the marks obtained by 10 students of class V in a class test, out of 50 according to their roll numbers, be: 39, 44, 49, 40, 22, 10, 45, 38, 15, 50 The data in the given form is known as raw data. The above given data can be placed in the serial order as shown below: Roll No. Marks 1 39 2 44 3 49 4 40 5 22 6 10 7 45 8 38 9 14 10 50 Now, if you want to analyse the standard of achievement of the students. If you arrange them in ascending or descending order, it will give you a better picture. Ascending order: 10, 15, 22, 38, 39, 40, 44. 45, 49, 50 Descending order: 50, 49, 45, 44, 40, 39, 38, 22, 15, 10 When the row is placed in ascending or descending order is known as arrayed data.
Types of Graphical Data Representation
Bar chart helps us to represent the collected data visually. The collected data can be visualized horizontally or vertically in a bar chart like amounts and frequency. It can be grouped or single. It helps us in comparing different items. By looking at all the bars, it is easy to say which types in a group of data influence the other.
Now let us understand bar chart by taking this example Let the marks obtained by 5 students of class V in a class test, out of 10 according to their names, be: 7,8,4,9,6 The data in the given form is known as raw data. The above given data can be placed in the bar chart as shown below: Name Marks Akshay 7 Maya 8 Dhanvi 4 Jaslen 9 Muskan 6
A histogram is the graphical representation of data. It is similar to the appearance of a bar graph but there is a lot of difference between histogram and bar graph because a bar graph helps to measure the frequency of categorical data. A categorical data means it is based on two or more categories like gender, months, etc. Whereas histogram is used for quantitative data.
For example:
The graph which uses lines and points to present the change in time is known as a line graph. Line graphs can be based on the number of animals left on earth, the increasing population of the world day by day, or the increasing or decreasing the number of bitcoins day by day, etc. The line graphs tell us about the changes occurring across the world over time. In a line graph, we can tell about two or more types of changes occurring around the world.
For Example:
Pie chart is a type of graph that involves a structural graphic representation of numerical proportion. It can be replaced in most cases by other plots like a bar chart, box plot, dot plot, etc. As per the research, it is shown that it is difficult to compare the different sections of a given pie chart, or if it is to compare data across different pie charts.
Frequency Distribution Table
A frequency distribution table is a chart that helps us to summarise the value and the frequency of the chart. This frequency distribution table has two columns, The first column consist of the list of the various outcome in the data, While the second column list the frequency of each outcome of the data. By putting this kind of data into a table it helps us to make it easier to understand and analyze the data.
For Example: To create a frequency distribution table, we would first need to list all the outcomes in the data. In this example, the results are 0 runs, 1 run, 2 runs, and 3 runs. We would list these numerals in numerical ranking in the foremost queue. Subsequently, we ought to calculate how many times per result happened. They scored 0 runs in the 1st, 4th, 7th, and 8th innings, 1 run in the 2nd, 5th, and the 9th innings, 2 runs in the 6th inning, and 3 runs in the 3rd inning. We set the frequency of each result in the double queue. You can notice that the table is a vastly more useful method to show this data. Baseball Team Runs Per Inning Number of Runs Frequency 0 4 1 3 2 1 3 1
Sample Questions
Question 1: Considering the school fee submission of 10 students of class 10th is given below:
| Muskan | Paid |
| Kritika | Not paid |
| Anmol | Not paid |
| Raghav | Paid |
| Nitin | Paid |
| Dhanvi | Paid |
| Jasleen | Paid |
| Manas | Not paid |
| Anshul | Not paid |
| Sahil | Paid |
In order to draw the bar graph for the data above, we prepare the frequency table as given below. Fee submission No. of Students Paid 6 Not paid 4 Now we have to represent the data by using the bar graph. It can be drawn by following the steps given below: Step 1: firstly we have to draw the two axis of the graph X-axis and the Y-axis. The varieties of the data must be put on the X-axis (the horizontal line) and the frequencies of the data must be put on the Y-axis (the vertical line) of the graph. Step 2: After drawing both the axis now we have to give the numeric scale to the Y-axis (the vertical line) of the graph It should be started from zero and ends up with the highest value of the data. Step 3: After the decision of the range at the Y-axis now we have to give it a suitable difference of the numeric scale. Like it can be 0,1,2,3…….or 0,10,20,30 either we can give it a numeric scale like 0,20,40,60… Step 4: Now on the X-axis we have to label it appropriately. Step 5: Now we have to draw the bars according to the data but we have to keep in mind that all the bars should be of the same length and there should be the same distance between each graph
Question 2: Watch the subsequent pie chart that denotes the money spent by Megha at the funfair. The suggested colour indicates the quantity paid for each variety. The total value of the data is 15 and the amount paid on each variety is diagnosed as follows:
Chocolates – 3
Wafers – 3
Toys – 2
Rides – 7
To convert this into pie chart percentage, we apply the formula: (Frequency/Total Frequency) × 100 Let us convert the above data into a percentage: Amount paid on rides: (7/15) × 100 = 47% Amount paid on toys: (2/15) × 100 = 13% Amount paid on wafers: (3/15) × 100 = 20% Amount paid on chocolates: (3/15) × 100 = 20 %
Question 3: The line graph given below shows how Devdas’s height changes as he grows.
Given below is a line graph showing the height changes in Devdas’s as he grows. Observe the graph and answer the questions below.
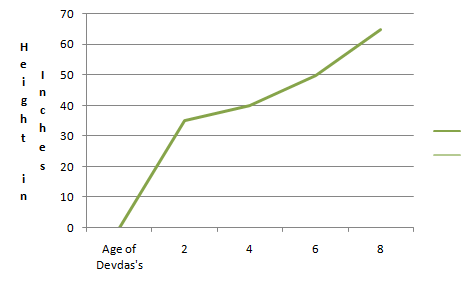
(i) What was the height of Devdas’s at 8 years? Answer: 65 inches (ii) What was the height of Devdas’s at 6 years? Answer: 50 inches (iii) What was the height of Devdas’s at 2 years? Answer: 35 inches (iv) How much has Devdas’s grown from 2 to 8 years? Answer: 30 inches (v) When was Devdas’s 35 inches tall? Answer: 2 years.
Similar Reads
- Mathematics
- School Learning
Please Login to comment...
- Top Language Learning Apps in 2024
- Top 20 Free VPN for iPhone in 2024: October Top Picks
- How to Underline in Discord
- How to Block Someone on Discord
- GeeksforGeeks Practice - Leading Online Coding Platform
Improve your Coding Skills with Practice
What kind of Experience do you want to share?
Talk to our experts
1800-120-456-456
- Introduction to Data Representation
- Computer Science

About Data Representation
Data can be anything, including a number, a name, musical notes, or the colour of an image. The way that we stored, processed, and transmitted data is referred to as data representation. We can use any device, including computers, smartphones, and iPads, to store data in digital format. The stored data is handled by electronic circuitry. A bit is a 0 or 1 used in digital data representation.
Data Representation Techniques
Classification of Computers
Computer scans are classified broadly based on their speed and computing power.
1. Microcomputers or PCs (Personal Computers): It is a single-user computer system with a medium-power microprocessor. It is referred to as a computer with a microprocessor as its central processing unit.

Microcomputer
2. Mini-Computer: It is a multi-user computer system that can support hundreds of users at the same time.

Types of Mini Computers
3. Mainframe Computer: It is a multi-user computer system that can support hundreds of users at the same time. Software technology is distinct from minicomputer technology.

Mainframe Computer
4. Super-Computer: With the ability to process hundreds of millions of instructions per second, it is a very quick computer. They are used for specialised applications requiring enormous amounts of mathematical computations, but they are very expensive.

Supercomputer
Types of Computer Number System
Every value saved to or obtained from computer memory uses a specific number system, which is the method used to represent numbers in the computer system architecture. One needs to be familiar with number systems in order to read computer language or interact with the system.
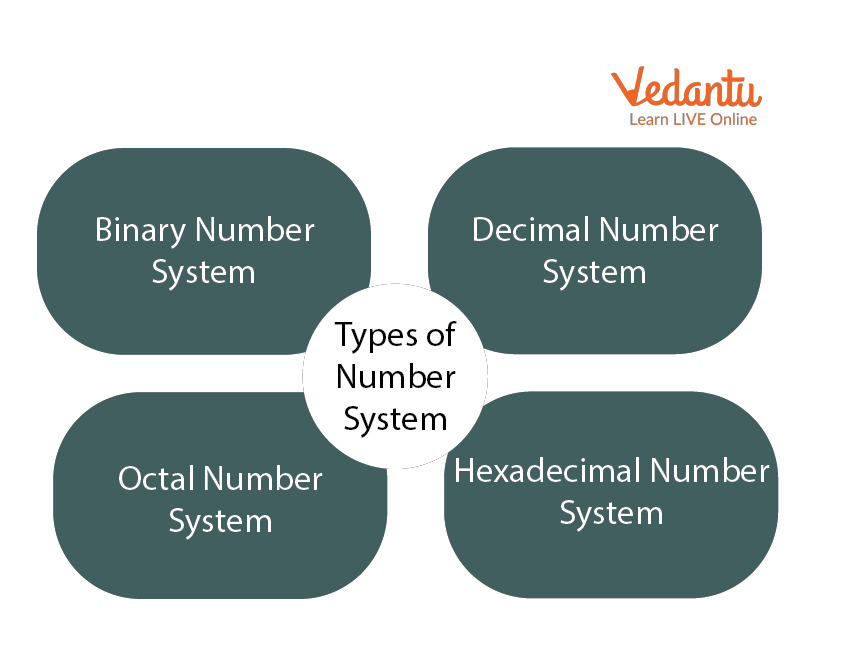
Types of Number System
1. Binary Number System
There are only two digits in a binary number system: 0 and 1. In this number system, 0 and 1 stand in for every number (value). Because the binary number system only has two digits, its base is 2.
A bit is another name for each binary digit. The binary number system is also a positional value system, where each digit's value is expressed in powers of 2.
Characteristics of Binary Number System
The following are the primary characteristics of the binary system:
It only has two digits, zero and one.
Depending on its position, each digit has a different value.
Each position has the same value as a base power of two.
Because computers work with internal voltage drops, it is used in all types of computers.

Binary Number System
2. Decimal Number System
The decimal number system is a base ten number system with ten digits ranging from 0 to 9. This means that these ten digits can represent any numerical quantity. A positional value system is also a decimal number system. This means that the value of digits will be determined by their position.
Characteristics of Decimal Number System
Ten units of a given order equal one unit of the higher order, making it a decimal system.
The number 10 serves as the foundation for the decimal number system.
The value of each digit or number will depend on where it is located within the numeric figure because it is a positional system.
The value of this number results from multiplying all the digits by each power.

Decimal Number System
Decimal Binary Conversion Table
Decimal | Binary |
0 | 0000 |
1 | 0001 |
2 | 0010 |
3 | 0011 |
4 | 0100 |
5 | 0101 |
6 | 0110 |
7 | 0111 |
8 | 1000 |
9 | 1001 |
10 | 1010 |
11 | 1011 |
12 | 1100 |
13 | 1101 |
14 | 1110 |
15 | 1111 |
3. Octal Number System
There are only eight (8) digits in the octal number system, from 0 to 7. In this number system, each number (value) is represented by the digits 0, 1, 2, 3,4,5,6, and 7. Since the octal number system only has 8 digits, its base is 8.
Characteristics of Octal Number System:
Contains eight digits: 0,1,2,3,4,5,6,7.
Also known as the base 8 number system.
Each octal number position represents a 0 power of the base (8).
An octal number's last position corresponds to an x power of the base (8).

Octal Number System
4. Hexadecimal Number System
There are sixteen (16) alphanumeric values in the hexadecimal number system, ranging from 0 to 9 and A to F. In this number system, each number (value) is represented by 0, 1, 2, 3, 5, 6, 7, 8, 9, A, B, C, D, E, and F. Because the hexadecimal number system has 16 alphanumeric values, its base is 16. Here, the numbers are A = 10, B = 11, C = 12, D = 13, E = 14, and F = 15.
Characteristics of Hexadecimal Number System:
A system of positional numbers.
Has 16 symbols or digits overall (0, 1, 2, 3, 4, 5, 6, 7, 8, 9, A, B, C, D, E, F). Its base is, therefore, 16.
Decimal values 10, 11, 12, 13, 14, and 15 are represented by the letters A, B, C, D, E, and F, respectively.
A single digit may have a maximum value of 15.
Each digit position corresponds to a different base power (16).
Since there are only 16 digits, any hexadecimal number can be represented in binary with 4 bits.
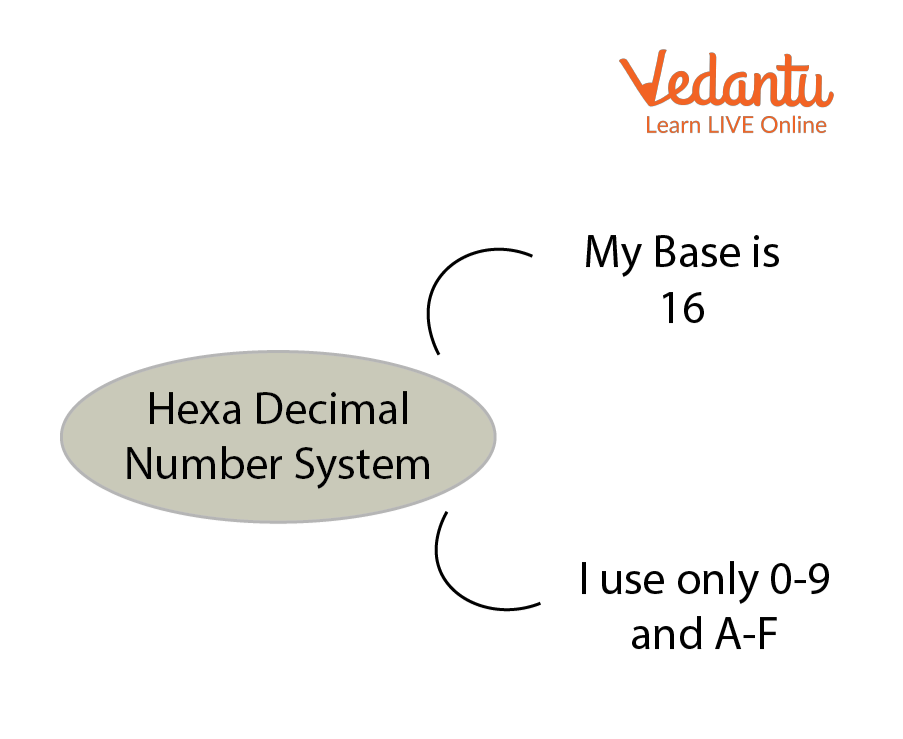
Hexadecimal Number System
So, we've seen how to convert decimals and use the Number System to communicate with a computer. The full character set of the English language, which includes all alphabets, punctuation marks, mathematical operators, special symbols, etc., must be supported by the computer in addition to numerical data.
Learning By Doing
Choose the correct answer:.
1. Which computer is the largest in terms of size?
Minicomputer
Micro Computer
2. The binary number 11011001 is converted to what decimal value?
Solved Questions
1. Give some examples where Supercomputers are used.
Ans: Weather Prediction, Scientific simulations, graphics, fluid dynamic calculations, Nuclear energy research, electronic engineering and analysis of geological data.
2. Which of these is the most costly?
Mainframe computer
Ans: C) Supercomputer


FAQs on Introduction to Data Representation
1. What is the distinction between the Hexadecimal and Octal Number System?
The octal number system is a base-8 number system in which the digits 0 through 7 are used to represent numbers. The hexadecimal number system is a base-16 number system that employs the digits 0 through 9 as well as the letters A through F to represent numbers.
2. What is the smallest data representation?
The smallest data storage unit in a computer's memory is called a BYTE, which comprises 8 BITS.
3. What is the largest data unit?
The largest commonly available data storage unit is a terabyte or TB. A terabyte equals 1,000 gigabytes, while a tebibyte equals 1,024 gibibytes.
By Gkseries see more questions
Data representation | computer system organization objective questions with answers.
- Computer Science /
- data representation
Sports GK Questions and Answers 2024 (Latest Updated)
Awards & honours gk questions 2024 (latest updated).
Computer Organization multiple choice questions and answers set contain 5 mcqs on number system and data representation in computer science. Each quiz objective question has 4 options as possible answers. Choose your option and check it with the given correct answer.
View Answer Comment
Answer: Option [C]
23D -> 0010 0011 1101 and 9AA -> 1001 1010 1010
So the sum of two hexadecimal numbers is 1011 1110 0111 i.e. BE7
Answer: Option [D]
37 H means 00110111
17 H means 00010111
On division of 37 H by 17 H the remainder is 09 H
| Article and Schedule Quiz |
DOWNLOAD CURRENT AFFAIRS PDF FROM APP
Answer: Option [A]
31 can be represented by 5 bits and the 1 bit needed for sign bit.
2 n -1 is the largest integer in 2's complement representation using n bits.
Answer: Option [B]
-2 15 is the least negative value for the two 8-bit 2's complement numbers.
Random GK Questions
Adre 2.0 free mock tests, adre 2.0 full length mock test, take mock tests.
| Missiles Mock Test | |
| SSC MTS Mock Test | |
| IBPS CLERK MOCK TEST | |
| SSC MTS 2022 JULY 26 Shift 1 (ENGLISH) | |
| SSC GD Previous Year Paper 2021 Nov 17 Shift - I (Hindi) | |
| SSC CGL Tier - 1 PYP 2022 April 21 Shift- 1 (ENGLISH) | |
| MPSC PAPER I MOCK TEST 1 (ENGLISH) | |
| IB Security Assistant Mock test 1 (english) | |
| UP POLICE CONSTABLE MOCK TEST 1 | |
| DELHI POLICE CONSTABLE MOCK TEST 1 (HINDI) |
- Digital Logic Circuits
- Digital Components
- Data Representation
- Register Transfer and Microoperations
- Assembly Language Programming
- Central Processing Unit
- Pipeline and Vector Processing
- Computer Arithmetic
- Input-Output Organization
- Memory Organization

Computer Science Topics
- Digital Components Data Representation
- Introduction to Programming
- C Programming Basics
- Algorithms for Problem Solving
- Conditional Statements & Loops
- Storage Classes
- Memory Allocation
- Structures and Unions
- Linked Lists
- Fundamental Programming Structures in Java
- Java Datatypes Variables & Arrays
- Java Operators
- Control Statements
- Objects and Classes
- Principles of JAVA
- Packages & Interfaces
- Exception Handling
- Java Threads
- Software Engineering Fundamentals
- Software Requirements Analysis & Specification
- Software Design
- Coding and Software Testing
- User Interface Design
- Software Configuration Management
- Software Implementation & Maintenance
- Object-Oriented SE
- Database Architecture and Modeling
- Entity Relationship Model
- Relational DBMS
- Database Normalization
- Relational Algebra and Relational Calculus
- Backup and Recovery
- Database Security and Integrity
- Fundamentals of Data Communication
- Data Modulation
- Multichannel Data Communication
- Communication Network Fundamentals
- Introduction To PHP
- Introduction To ASP.NET
- ASP.NET Server Controls
- ASP.NET Validation Controls
- Data Structures
- Operating Systems
- Computer Fundamentals
- Management Information System
- Microsoft Office
- Computer Awareness Quiz
- Cloud Computing
- Visual Basic Programming
- Automata Theory
- Computer Based Optimisation
- C Sharp Programming
- Digital Computer Fundamentals
- Discrete mathematics
- Electronics
- COA Tutorial
- Evolution of Computing Devices
- Functional units of Digital System
- Basic Operational Concepts
Basic CO and Design
- General System Architecture
- Store Program Control Concept
- Flynn's Classification of Computers
- Computer Registers
- Computer Instructions
- Design of Control Unit
- Instruction Cycle
- Control Logic Gates
Digital Logic Circuits
- Digital Computers
- Logic Gates
- Boolean Algebra
- Examples of Boolean algebra simplification
- Laws of Boolean Algebra
- Simplification using Boolean Algebra
- Map Simplification
- Combinational Circuits
- Half - Adder
- Full - Adder
Flip - Flops
- S-R Flip-Flop
- D Flip-Flop
- JK Flip-Flop
- T Flip-Flop
Digital Components
- Integrated Circuits
- Multiplexers
- De - multiplexers
- Shift Registers
- Register Transfer
- Register Transfer Language
- Bus and Memory Transfer
Micro-Operations
- Arithmetic Micro-Operations
- Binary Adder
- Binary Adder-Subtractor
- Binary Incrementer
Memory Organization
- Memory Hierarchy
- Main Memory
- Auxiliary Memory
- Associative Memory
- Cache Memory
- Parallel Processing
- Arithmetic Pipeline
- Instruction Pipeline
- Booth's Multiplication Algorithm
- Branch Instruction in Computer Organization
- Data Representation in Computer Organization
- ALU and Data Path in Computer Organization
- External memory in Computer Organization
- Structured Computer Organization
- Types of Register in Computer Organization
- Secondary Storage Devices in Computer Organization
- Types of Operands in Computer Organization
- Serial Communication in Computer organization
- Addressing Sequencing in Computer Organization
- Simplified Instructional Computer
- Arithmetic Instructions in AVR microcontroller
- Conventional Computing VS Quantum Computing
- Instruction set used in Simplified Instructional Computer
- Branch Instruction in AVR microcontroller
- Conditional Branch instruction in AVR Microcontroller
- Data transfer instruction in AVR microcontroller
- Memory-based vs Register-based addressing modes
- 1's complement Representation vs 2's complement Representation
- CALL Instructions and Stack in AVR Microcontroller
- Difference between Call and Jump Instructions
- Overflow in Arithmetic Addition in Binary number System
- Horizontal Micro-programmed Vs. Vertical Micro-programmed Control Unit
- Hardwired vs Micro-programmed Control Unit
- Non-Restoring Division Algorithm for Unsigned Integer
- Restoring Division Algorithm for Unsigned Integer
- Debugging a Machine-level Program
- Dependencies and Data Hazard in pipeline in Computer Organization
- Execution, Stages and Throughput in Pipeline
- Types of Pipeline Delay and Stalling
- Timing Diagram of MOV Instruction
- Advantages and Disadvantages of Flash Memory
- Importance/Need of negative feedback in amplifiers
- Anti-Aliasing - Computer Graphics
- Bus Arbitration in Computer Organization
- Convert a number from Base 2 (Binary) to Base 6
- Cache Coherence
- Cache Memory and Virtual Memory
- Electrical Potential and Potential Difference
- RAM and Cache
- SIM and RIM instructions in 8085 processor
- Clusters in Computer Organisation
- Data Types and Addressing Modes of 80386/80386DX Microprocessor
| In computer organization, data refers to the symbols that are used to represent events, people, things and ideas. The data can be represented in the following ways:
can be anything like a number, a name, notes in a musical composition, or the color in a photograph. Data representation can be referred to as the form in which we stored the data, processed it and transmitted it. In order to store the data in digital format, we can use any device like computers, smartphones, and iPads. Electronic circuitry is used to handle the stored data.
is a type of process in which we convert information like photos, music, number, text into digital data. Electronic devices are used to manipulate these types of data. The digital revolution has evolved with the help of 4 phases, starting with the big, expensive standalone computers and progressing to today's digital world. All around the world, small and inexpensive devices are spreading everywhere.
The or bits are used to show the digital data, which is represented by 0 and 1. The binary digits can be called the smallest unit of information in a computer. The main use of binary digit is that it can store the information or data in the form of 0s and 1s. It contains a value that can be on/off or true/false. On or true will be represented by the 1, and off or false will be represented by the 0. The digital file is a simple file, which is used to collect data contained by the storage medium like the flash drive, CD, hard disk, or DVD. The number can be represented in the following way:
is used to contain numbers, which helps us to perform arithmetic operations. The digital devices use a binary number system so that they can represent numeric data. The binary number system can only be represented by two digits 0 and 1. There can't be any other digits like 2 in the system. If we want to represent number 2 in binary, then we will write it as 10. The text can be represented in the following ways:
can be formed with the help of symbols, letters, and numerals, but they can?t be used in calculations. Using the character data, we can form our address, hair colour, name, etc. Character data normally takes the data in the form of text. With the help of the text, we can describe many things like our father name, mother name, etc.
Several types of codes are employed by the to represent character data, including Unicode, ASCII, and other types of variants. The full form of ASCII is American Standard Code for Information Interchange. It is a type of character encoding standard, which is used for electronic communication. With the help of telecommunication equipment, computers and many other devices, ASCII code can represent the text. The ASCII code needs 7 bits for each character, where the unique character is represented by every single bit. For the uppercase letter A, the ASCII code is represented as 1000001.
can be described as a superset of ASCII. The ASCII set uses 7 bits to represent every character, but the Extended ASCII uses 8 bits to represent each character. The extended ASCII contains 7 bits of ASCII characters and 1 bit for additional characters. Using the 7 bits, the ASCII code provides code for 128 unique symbols or characters, but Extended ASCII provides code for 256 unique symbols or characters. For the uppercase letter A, the Extended ASCII code is represented as 01000001.
is also known as the universal character encoding standard. Unicode provides a way through which an individual character can be represented in the form of web pages, text files, and other documents. Using ASCII, we can only represent the basic English characters, but with the help of Unicode, we can represent characters from all languages around the World. ASCII code provides code for 128 characters, while Unicode provide code for roughly 65,000 characters with the help of 16 bits. In order to represent each character, ASCII code only uses 1 bit, while Unicode supports up to 4 bytes. The Unicode encoding has several different types, but UTF-8 and UTF-16 are the most commonly used. UTF-8 is a type of variable length coding scheme. It has also become the standard character encoding, which is used on the web. Many software programs also set UTF-8 as their default encoding. can be used for numerals like phone numbers and social security numbers. ASCII text contains plain and unformatted text. This type of file will be saved in a text file format, which contains a name ending with .txt. These files are labelled differently on different systems, like Windows operating system labelled these files as "Text document" and Apple devices labelled these files as "Plain Text". There will have no formatting in the ASCII text files. If we want to make the documents with styles and formats, then we have to embed formatting codes in the text.
Microsoft word is used to create formatted text and documents. It uses the to do this. If we create a new document using the Microsoft Word 2007 or later version, then it always uses DOCX as the default file format. use to produce the documents. As compared to Microsoft Word, it is simpler to create and edit documents using page format. uses the to create the documents. The files that saved in the PDF format cannot be modified. But we can easily print and share these files. If we save our document in PDF format, then we cannot change that file into the Microsoft Office file or any other file without specified software. is the hypertext markup language. It is used for document designing, which will be displayed in a web browser. It uses to design the documents. In HTML, hypertext is a type of text in any document containing links through which we can go to other places in the document or in other documents also. The markup language can be called as a computer language. In order to define the element within a document, this language uses tags. The bits and bytes can be represented in the following ways:
In the field of digital communication or computers, bits are the most basic unit of information or smallest unit of data. It is short of binary digit, which means it can contain only one value, either 0 or 1. So bits can be represented by 0 or 1, - or +, false or true, off or on, or no or yes. Many technologies are based on bits and bytes, which is extensively useful to describe the network access speed and storage capacity. The bit is usually abbreviated as a lowercase b. In order to execute the instructions and store the data, the bits are grouped into multiple bits, which are known as bytes. Bytes can be defined as a group of eight bits, and it is usually abbreviated as an uppercase B. If we have four bytes, it will equal 32 bits (4*8 = 32), and 10 bytes will equal 80 bits (8*10 = 80).
Bits are used for data rates like speeds while movie download, speed while internet connection, etc. Bytes are used to get the storage capacity and file sizes. When we are reading something related to digital devices, it will be frequently encountered references like 90 kilobits per second, 1.44 megabytes, 2.8 gigahertz, and 2 terabytes. To quantify digital data, we have many options such as Kilo, Mega, Giga, Tera and many more similar terms, which are described as follows: Kb is also called a kilobyte or Kbyte. It is mostly used while referring to the size of small computer files. Kbps is also called kilobit, Kbit or Kb. The 56 kbps means 56 kilobits per second which are used to show the slow data rates. If our internet speed is 56 kbps, we have to face difficulty while connecting more than one device, buffering while streaming videos, slow downloading, and many other internet connectivity problems. Mbps is also called Megabit, MB or Mbit. The 50 Mbps means 50 Megabit per second, which are used to show the faster data rates. If our internet speed is 50 Mbps, we will experience online activity without any buffering, such as online gaming, downloading music, streaming HD, web browsing, etc. 50 Mbps or more than that will be known as fast internet speed. With the help of fast speed, we can easily handle more than one online activity for more than one user at a time without major interruptions in services. 3.2 MB is also called Megabyte, MB or MByte. It is used when we are referring to the size of files, which contains videos and photos. 100 Gbit is also called Gigabit or GB. It is used to show the really fast network speeds. 16 GB is also called Gigabyte, GB or GByte. It is used to show the storage capacity. The digital data is compressed to reduce transmission times and file size. Data compression is the process of reducing the number of bits used to represent data. Data compression typically uses encoding techniques to compress the data. The compressed data will help us to save storage capacity, reduce costs for storage hardware, increase file transfer speed. Compression uses some programs, which also uses algorithms and functions to find out the way to reduce the data size. Compression can be referred "zipping". The process of reconstructing files will be known as unzipping or extracting. The compressed files will contain .gz, or.tar.gz, .pkg, or .zip at the end of the files. Compression can be divided into two techniques: Lossless compression and Lossy compression. As the name implies, lossless compression is the process of compressing the data without any loss of information or data. If we compressed the data with the help of lossless compression, then we can exactly recover the original data from the compressed data. That means all the information can be completely restored by lossless compression. Many applications want to use data loss compression. For example, lossless compression can be used in the format of ZIP files and in the GNU tool gzip. The lossless data compression can also be used as a component within the technologies of lossy data compression. It is generally used for discrete data like word processing files, database records, some images, and information of the video.
Lossy compression is the process of compressing the data, but that data cannot be recovered 100% of original data. This compression is able to provide a high degree of compression, and the result of this compression will be in smaller compressed files. But in this process, some number of video frames, sound waves and original pixels are removed forever. If the compression is greater, then the size of files will be smaller. Business data and text, which needs a full restoration, will never use lossy compression. Nobody likes to lose the information, but there are a lot of files that are very large, and we don't have enough space to maintain all of the original data or many times, we don't require all the original data in the first place. For example, videos, photos and audio recording files to capture the beauty of our world. In this case, we use lossy compression. |
Latest Courses

We provides tutorials and interview questions of all technology like java tutorial, android, java frameworks
Contact info
G-13, 2nd Floor, Sec-3, Noida, UP, 201301, India
[email protected] .

Latest Post
PRIVACY POLICY
Interview Questions
Online compiler.
Advertisement
Automatic sleep stage classification using deep learning: signals, data representation, and neural networks
- Open access
- Published: 23 September 2024
- Volume 57 , article number 301 , ( 2024 )
Cite this article
You have full access to this open access article

- Peng Liu 1 ,
- Wei Qian 1 ,
- Hua Zhang 1 ,
- Yabin Zhu 2 ,
- Qi Hong 3 ,
- Qiang Li 3 &
- Yudong Yao 4
227 Accesses
Explore all metrics
In clinical practice, sleep stage classification (SSC) is a crucial step for physicians in sleep assessment and sleep disorder diagnosis. However, traditional sleep stage classification relies on manual work by sleep experts, which is time-consuming and labor-intensive. Faced with this obstacle, computer-aided diagnosis (CAD) has the potential to become an intelligent assistant tool for sleep experts, aiding doctors in the assessment and decision-making process. In fact, in recent years, CAD supported by artificial intelligence, especially deep learning (DL) techniques, has been widely applied in SSC. DL offers higher accuracy and lower costs, making a significant impact. In this paper, we will systematically review SSC research based on DL methods (DL-SSC). We explores DL-SSC from several important perspectives, including signal and data representation, data preprocessing, deep learning models, and performance evaluation. Specifically, this paper addresses three main questions: (1) What signals can DL-SSC use? (2) What are the various methods to represent these signals? (3) What are the effective DL models? Through addressing on these questions, this paper will provide a comprehensive overview of DL-SSC.
Similar content being viewed by others

A Deep Learning-Based Method for Sleep Stage Classification Using Physiological Signal

Sensitive deep learning application on sleep stage scoring by using all PSG data
SleepExpertNet: high-performance and class-balanced deep learning approach inspired from the expert neurologists for sleep stage classification
Explore related subjects.
- Artificial Intelligence
Avoid common mistakes on your manuscript.
1 Introduction
Sleep is the most fundamental biological process, occupying approximately one-third of human life and playing a vital role in human existence (Siegel 2009 ). Unfortunately, sleep disorders are prevalent in modern society. A global study involving nearly 500,000 people in 2022 indicated that the insomnia rate among the public reached as high as 40.5% during the COVID-19 pandemic (Jahrami et al. 2022 ). Sleep disorders are closely associated with various neurologic and psychiatric disorders (Van Someren 2021 ). For instance, research by Zhang et al. demonstrated a correlation between reduced deep sleep proportion in Alzheimer’s disease patients and the severity of dementia (Zhang et al. 2022b ). Additionally, insomnia was found to double the risk of depression in people without depressive symptoms, as stated in Baglioni et al. ( 2011 ). Timely and effective treatment of insomnia was able to serve as a primary preventive measure for depression (Clarke and Harvey 2012 ). In summary, sleep issues have a significant impact on both physiological and psychological well-being, necessitating timely diagnosis. The essential step in clinical sleep disorder diagnosis and assessment is referred to as sleep stage classification (SSC) (Wulff et al. 2010 ), also known as sleep staging or sleep scoring.
In clinical practice, the gold standard for classifying sleep stages is the polysomnogram (PSG), which includes a set of nocturnal sleep signals such as electroencephalogram (EEG), electrooculogram (EOG), and electromyogram (EMG). The PSG signals are segmented into 30-second units, and the continuous segments obtained are referred to as sleep stages, with each segment belonging to a specific stage category. The criteria for determining the stage category of each epoch are known as R&K (Rechtschaffen 1968 ) and AASM (Iber 2007 ), with the former established in 1968 and the latter being the most recent and commonly used. R&K divides sleep into three basic stages: wakefulness (W), rapid eye movement (REM), and non-rapid eye movement (NREM). NREM can be subdivided into S1, S2, S3, and S4. AASM merging S3 and S4 into a single stage, resulting in five sleep stages: W, N1 (S1), N2 (S2), N3 (S3-S4), and REM. Based on these standards, researchers sometimes describe sleep stages differently. We have listed the various descriptions used in the studies included in this paper in Table 1 . Different stages exhibit distinct characteristics during sleep. The N2 stage is typically marked by significant waves such as sleep spindles and K complexes (Parekh et al. 2019 ). Moreover, sleep is a continuous and dynamic process, and there exists contextual information between consecutive epochs (forming sequences) (Rechtschaffen 1968 ; Iber 2007 ). For instance, if isolated N3 occur between several consecutive N2, doctors still classify them as N2 (Wu et al. 2020 ).
Manual classification is time-intensive and laborious (Malhotra et al. 2013 ). In response to the immense demand in healthcare, numerous methods for automatically analyzing EEG for sleep staging have been proposed. These automatic sleep stage classification (ASSC) methods are developed using machine learning (ML) algorithms. Early ASSC was a combination of manual feature extraction and traditional ML. Researchers manually extract features from the time-domain, frequency-domain of signals and use traditional ML methods, such as support vector machines (SVM), for feature classification to achieve automation (Li et al. 2017 ; Sharma et al. 2017 ). However, manual feature engineering is very tedious and requires additional prior knowledge (Jia et al. 2021 ; Eldele et al. 2021 ). Moreover, due to the significant variability in EEG among different individuals (Subha et al. 2010 ), it is challenging to extract well-generalized features. Therefore, self-learning methods based on deep learning have begun to be used for sleep staging.
In recent years, deep learning (DL) has become a popular approach for automatic sleep stage classification. This may be because DL methods can automatically extract sleep features and complete classification in an end-to-end manner (Zhang et al. 2022a ), avoiding the cumbersome feature extraction and explicit classification steps. In the current context of automatic sleep stage classification based on deep learning (DL-ASSC), there are three key points worth noting. On one hand, signals form the basis of ASSC. Various studies have extensively explored multiple types of signals, which can be broadly categorized into three classes: the first category is PSG, including EEG, EOG, and EMG (Guillot et al. 2020 ; Seo et al. 2020 ; Supratak et al. 2017 ); the second category is cardiorespiratory signals, including electrocardiogram (ECG), photoplethysmography (PPG), respiratory effort, etc. (Goldammer et al. 2022 ; Kotzen et al. 2022 ; Olsen et al. 2022 ); the third category is contactless signals, mainly radar, Wi-Fi and audio signals (Zhai et al. 2022 ; Yu et al. 2021a ; Tran et al. 2023 ). On the other hand, the same signal can be represented in various forms, and different input representation into a DL model might yield different performance (Biswal et al. 2018 ). Popular data representations fall into three categories: the first category involves directly inputting raw one-dimensional (1D) signals into the network (Seo et al. 2020 ; Supratak et al. 2017 ); the second category uses transformed domain data of the signal as model input, commonly seen in two-dimensional (2D) time-frequency spectrograms [usually obtained from the original signal through continuous wavelet transform (Kuo et al. 2022 ) or short-time Fourier transform (Guillot et al. 2020 )]; the third category combines both, typically employing a dual-stream structure where different input forms are processed separately in each branch (Phan et al. 2021 ; Jia et al. 2020a ). Last but not least, the ASSC methods employing various DL models continue to emerge. Convolutional neural networks (CNNs), initially designed for the field of image processing, are commonly used by researchers for feature extraction. As a widely recognized foundational model, CNNs are widely applied in sleep stage classification, either directly using one-dimensional CNNs on raw signals or employing more common 2D CNNs on transformed domain representations of the signals. Another type of classical models takes the spotlight: recurrent neural networks (RNNs) and its two variants, long short-term memory (LSTM) and gated recurrent unit (GRU). RNNs are adept at handling time series data and can capture temporal information in sleep data. Moreover, in 2017, Google introduced the Transformer (Vaswani et al. 2017 ), which utilizes the multi-head self-attention (MHSA) mechanism and quickly became an indispensable technique in time-series data modeling. Compared with RNNs, MHSA can also effectively capture the time dependence of sleep data when applied to sleep stage classification. In practical applications, researchers often choose to customize (design) a deep neural network (DNN) to adapt to different needs and tasks. In the ASSC based on deep learning methods, the most commonly used architecture in existing research is feature extraction + sequence encoding. The feature extractor first maps the input signal to an embedding space, and then models the temporal information (context-dependent information) through the sequence encoder. CNN is a common choice for the feature extractor, and the sequence encoder is often implemented by RNN-like models or attention mechanisms.
DL-SSC research has achieved significant progress, and some studies have achieved clinically acceptable performance (Phan and Mikkelsen 2022 ). This topic has been addressed in several review articles. However, earlier publications such as those by Fiorillo et al. ( 2019 ) and Faust et al. ( 2019 ) do not encompass the developments of recent years. More comprehensive review papers have recently emerged, but they still have some limitations. For instance, the work by Alsolai et al. ( 2022 ) focuses more on feature extraction techniques and machine learning methods, with less emphasis on the latest end-to-end deep learning approaches. Sri et al. ( 2022 ) and Loh et al. ( 2020 ) reviewed the performance of different deep learning models using PSG signals but did not cover aspects such as signal representation and preprocessing. The studies by Phan and Mikkelsen ( 2022 ) and Sun et al. ( 2022 ) only considered EEG and ECG signals, excluding other types of signals. We have summarized these works in Table 2 . Therefore, this paper provides a comprehensive review of recent years’ sleep stage classification based on deep learning. We have examined all the elements required for DL-SSC, including signals, datasets, data preprocessing, data representations, deep learning models, evaluation methods, etc. Specifically, the main topices discussed in this paper include: (1) signals that can be used in DL-SSC; (2) methods to represent data, i.e., how signals can be input into DL models for further processing; (3) effective DL models and their performance.
This paper is organized as follows. Section 2 describes the sources of literature and the search process. Section 3 discusses available signals and summarizes some public datasets. Section 4 discusses PSG-based research, including preprocessing, different data representations, and DL models. Sections 5 and 6 will cover research based on cardiorespiratory signals and non-contact signals, respectively. Finally, Sect. 7 and Sect. 8 will discuss and summarize the findings.
2 Review methodology
We conduct a literature search and screening through the following process, Fig. 1 is a visual representation of this process. We searched well-known literature databases, namely Google Scholar, Web of Science, and PubMed. The relevant studies on sleep stage classification using three different types of signals were identified using the following common keywords and their combinations: (“Deep Learning” OR “Deep Machine Learning” OR “Neural Network”) AND (“Sleep Stage Classification” OR “Sleep Staging” OR “Sleep Scoring”). The keywords specific to each signal type were: (“Polysomnography” OR “Electroencephalogram” OR “Electrooculogram” OR “Electromyogram”), (“Electrocardiogram” OR “Photoplethysmography”), (“Radar” OR “Wi-Fi” OR “Microphone”). For deep neural network models, no specific keywords were set, and the publication or release year of the literature was restricted to 2016 or later. After excluding some irrelevant or duplicate studies, the literature was assessed based on the following criteria, which define the inclusion and exclusion standards of the relevant studies:
Task—only studies that performed sleep stage classification tasks were included.
Signal—studies that used one or a combination of the signals mentioned in the text for sleep staging were included. Studies using other signals, such as functional near-infrared spectroscopy (fNIRS), were excluded due to their scarcity (Huang et al. 2021 ; Arif et al. 2021 ).
Method—only studies employing deep learning-based methods were included, i.e., those using neural networks with at least two hidden layers. Traditional machine learning methods were generally not reviewed, but a few studies that used a combination of deep neural networks and machine learning classifiers for feature extraction and classification (Phan et al. 2018 ) were included.
Time—the focus was on studies conducted after 2016 (the earliest relevant study included in this paper was published in 2016).
Finally, the publicly available datasets reviewed in this paper were found through three approaches: mentioned in the articles included in this review, using the Google search engine with the keywords “Sleep stage Dataset” and corresponding signal types, and the Physionet Footnote 1 and NSRR Footnote 2 websites.

Schematic diagram of the literature selection process. It is divided into five steps: database paper search, duplicate removal, relevance screening, determination of topic compliance, and final inclusion in the review. In the diagram, n represents the number of papers, and the subscripts indicate different types of signals: 1 represents PSG, etc., 2 represents ECG, etc., and 3 represents non-contact signals. The paper search also includes additional database identifiers. This process ensures that the final included papers can summarize the main research content of recent years
3 Signals, datasets and performance metrics
3.1 signals.
The standard signal for sleep studies is PSG. In addition to this, signals containing cardiorespiratory information such as ECG, PPG, respiratory effort, etc., are commonly used. In recent years, signals like radar and Wi-Fi have also been explored due to their simplicity and comfort (Hong et al. 2019 ). Commonly used signals are listed in Table 3 .
3.1.1 PSG signals
PSG signal refer to the signals obtained from polysomnogram recordings, which are used to monitor sleep stages. It records a set of signals during sleep using multiple electrodes, including various physiological parameters such as brain activity, eye movements, and muscle activity (Kayabekir 2019 ). Electrodes on the scalp are responsible for recording electrical signals related to brain neuron activity, known as EEG. Electrodes near the eyes record electrical signals associated with eye movements, known as EOG. Electromyogram or EMG typically requires needle electrodes inserted into muscles to obtain electrical signals related to muscle activity, and during sleep monitoring, EMG is usually recorded near the chin. These three signals together are referred to as PSG. PSG serves as the standard signal for quantifying sleep stages and sleep quality (Yildirim et al. 2019 ; Tăutan et al. 2020 ).
EEG contains information necessary for ML or DL analysis in various domains such as time domain, frequency domain, and time-frequency domain. In the time domain, EEG features are mainly reflected in the changes in amplitude over time. Event-related potentials (ERPs) and statistical features can be obtained through time-domain averaging (Aboalayon et al. 2016 ). The frequency domain mainly describes the distribution characteristics of EEG power across different frequencies. The fast Fourier transform (FFT) can be used to obtain five basic frequency bands as shown in Table 4 , each with different implications (Aboalayon et al. 2016 ). EEG is a non-stationary signal generated by the superposition of electrical activities of numerous neurons (Li et al. 2022d ). It possesses variability and time-varying characteristics, meaning it has different statistical properties at different times and frequency bands, and it undergoes rapid changes within short periods (Wang et al. 2021 ; Stokes and Prerau 2020 ). Time-frequency analysis is particularly suitable for such non-stationary signals. Common methods include short-time Fourier transform (STFT), continuous wavelet transform (CWT), and Hilbert-Huang transform (HHT), among others. Time-frequency analysis can simultaneously reveal changes in signals over time and frequency (Jeon et al. 2020 ; Tyagi and Nehra 2017 ). Figure 2 shows the time waveforms and time-frequency spectrogram of N1 and N2 stages. Due to its rich information features from multiple perspectives, EEG can be used in sleep stage classification tasks in various forms. For example, Biswal et al. ( 2018 ) constructed neural networks using raw EEG or time-frequency spectra as inputs. They also compared machine learning methods with expert handcrafted features as inputs, and the results showed that deep learning methods outperformed machine learning methods. EOG and EMG signals exhibit different characteristics in different sleep stages and can provide information for identifying sleep stages. For instance, during the REM stage, eye movements are more intense, whereas during the NREM stage, eye movements are relatively stable (Iber 2007 ). The amplitude of EMG near the chin during the W stage is variable but typically higher than that in other sleep stages (Iber 2007 ). However, EOG and EMG are usually used as supplements to EEG. Combining EEG, EOG, and EMG in multimodal sleep stage classification is a popular approach (Phan et al. 2021 ; Jia et al. 2020a ). Multimodal approaches can generally improve performance, but continuous attachment of multiple electrodes might affect the natural sleep state of the subjects. Therefore, single-channel EEG is currently the most popular choice in research (Fan et al. 2021 ).

Single-channel EEG time waveform and STFT time-frequency spectrogram in N1 and N2 stages. a , b N1 and N2 stage (time waveform); c , d N1 and N2 stage (STFT time-frequency spectrogram)
3.1.2 Cardiorespiratory signals
PSG often needs to be conducted in specialized laboratories and is challenging for long-term monitoring. In contrast, cardiac and respiratory activities are easier to monitor. Many studies have also confirmed the correlation between sleep and cardiac activity (Bonnet and Arand 1997 ; Tobaldini et al. 2013 ). This has led people to explore an alternative approach to sleep monitoring apart from PSG.
Research indicates a strong connection between sleep and the activity of the autonomic nervous system (ANS) (Bonnet and Arand 1997 ; Tobaldini et al. 2013 ). When the human body is sleeping, it will be repeatedly controlled by the sympathetic and vagus nerves. When sleep changes from wakefulness to the N3 stage, the blood pressure and heart rate controlled by the ANS will also change accordingly (Shinar et al. 2006 ; Papadakis and Retortillo 2022 ). This manifests as different features in cardiac and respiratory activities corresponding to changes in sleep stages. For example, one of the features during REM is highly recognizable breathing frequency and potentially more irregular and rapid heart rate (HR). HR during NREM might be more stable, and during the W stage, there is low-frequency heart rate variability (HRV) and significant body movement (Sun et al. 2020 ). These discriminative features determine the applicability of cardiorespiratory signals in SSC. Cardiorespiratory signals encompass signals containing information about both heart and respiratory activities, primarily including ECG, PPG, and respiratory effort. ECG is a technique used to record cardiac electrical activity, which can directly reflect a person’s respiratory and circulatory systems (Sun et al. 2022 ). In SSC, raw ECG signals are not directly used; instead, derived signals are employed, such as HR (Sridhar et al. 2020 ), HRV (Fonseca et al. 2020 ), ECG-derived respiration (EDR) (Li et al. 2018 ), RR intervals (RRIs) (Goldammer et al. 2022 ), RR peak sequences (Sun et al. 2020 ), and others. An example of an ECG is shown in Fig. 3 : the instantaneous heart rate sequence derived from the ECG and the corresponding overnight sleep stage changes (Sridhar et al. 2020 ). PPG is a low-cost technique measuring changes in blood volume, commonly used to monitor heart rate, blood oxygen saturation, and other information. PPG is simple to implement and can be collected at the hand using photodetectors embedded in watches or rings (Kotzen et al. 2022 ; Radha et al. 2021 ; Walch et al. 2019 ). HR and HRV can be derived from PPG, indirectly reflecting sleep stages. A small portion of research also uses raw PPG for classification (Kotzen et al. 2022 ; Korkalainen et al. 2020 ). Figure 4 shows examples of PPG signal waveforms corresponding to the five sleep stages (Korkalainen et al. 2020 ). Similar to EEG, ECG and PPG also have their auxiliary signals. Common choices include combining signals from chest or abdominal respiratory efforts with accelerometer signals (Olsen et al. 2022 ; Sun et al. 2020 ). For instance, in Goldammer et al. ( 2022 ), the authors derived RR intervals from ECG and combined them with breath-by-breath intervals (BBIs) derived from chest respiratory efforts for W/N1/N2/N3/REM classification. In Walch et al. ( 2019 ), the authors used PPG and accelerometer signals collected from the “Apple Watch” to classify W/NREM/REM sleep stages. It’s worth noting that most studies in cardiac and respiratory signal research focus on four-stage (W/L/D/REM, L: light sleep, D: deep sleep) or three-stage (W/NREM/REM) classification.

The instantaneous heart rate time series derived from the ECG signal throughout the night, and the corresponding changes in sleep stages throughout the night (Sridhar et al. 2020 )

The waveforms of the original PPG signals corresponding to the five different sleep stages (Korkalainen et al. 2020 )
3.1.3 Contactless signals
The use of cardiorespiratory signals can effectively reduce the inconvenience caused to patients during sleep monitoring (compared to PSG). However, it still involves physical contact with the subjects. The development of non-contact sensors (such as biometric radar, Wi-Fi, microphones, etc.) has changed this situation.
In recent years, radar technology has been used for vital sign and activity monitoring (Fioranelli et al. 2019 ; Hanifi and Karsligil 2021 ; Khan et al. 2022 ). In these systems, radar sensors emit low-power radio frequency (RF) signals and extract vital signs, including heart rate, respiration rate, movement, and falls, from reflected signals. Wi-Fi technology has subsequently been developed, utilizing Wi-Fi channel state information (CSI) to monitor vital signs more cost-effectively (Soto et al. 2022 ; Khan et al. 2021 ). For example, research by Diraco et al. ( 2017 ) used ultra-wideband (UWB) radar and DL methods to monitor vital signs and falls, and Adib ( 2019 ) achieved HR measurement and emotion recognition using Wi-Fi. Previous studies have demonstrated that HR, respiration, and movement information can be extracted from RF signals reflected off the human body, which fundamentally still falls under the category of cardiorespiratory signals, and they are also related to sleep stages. Therefore, in principle, we can perform contactless SSC using technologies such as radar or Wi-Fi (Zhao et al. 2017 ). Subsequent research has proven the feasibility of wireless signals for SSC (Zhai et al. 2022 ; Zhao et al. 2017 ; Yu et al. 2021a ). Additionally, some research has achieved good results in sleep stage classification by recording nighttime breathing and snoring information through acoustic sensors (Hong et al. 2022 ; Tran et al. 2023 ). However, compared to other methods, audio signals might raise concerns about privacy.
3.2 Public datasets
Data is one of the most crucial components in DL. In recent years, the field of sleep stage classification has seen the emergence of several public databases, with the two most prominent ones being PhysioNet (Goldberger et al. 2000 ) and NSRR (Zhang et al. 2018 ). Widely used datasets such as Sleep-EDF2013 (SEFD13), Sleep-EDF2018 (SEDF18), and CAP-Sleep are all derived from the open-access PhysioNet database. The Sleep-EDF (SEDF) series is perhaps the most extensively utilized dataset. SEDF18 comprises data from each subject with 2 EEG channels, 1 EOG channel, and 1 chin EMG channel. The data is divided into two parts: SC (without medication) and ST (with medication). SC includes 153 (nighttime) recordings from 78 subjects who did not take medication. ST comprises 44 recordings from 22 subjects who took medication. The data is annotated using R&K rules, and EEG and EOG have a sampling rate of 100 Hz. Another notable database is NSRR, from which datasets like SHHS (Quan et al. 1997 ) and MESA (Chen et al. 2015 ) are derived. Table 5 summarizes some the public datasets.
Public datasets have significantly propelled the development of DL-SSC research, and their existence is highly beneficial. For instance, they can serve as common references and benchmarks, as well as be directly utilized for data augmentation or transfer learning to enhance model performance. However, existing datasets also present certain challenges. On one hand, different datasets vary in sampling rates and channels. Automated (DL) methods are often designed based on specific datasets, causing these methods to handle only particular input shapes (Guillot et al. 2021 ). A common solution is to perform operations like resampling and channel selection on different datasets to standardize the input shape (Lee et al. 2024 ). On the other hand, class imbalance issues are prevalent in sleep data. Class imbalance refers to a situation where certain categories in the dataset have significantly fewer samples than others. Due to the inherent nature of sleep, the duration of each stage in sleep recordings is not equal (Fan et al. 2020 ). We have compiled the sample distribution of several datasets in Table 6 . The results indicate that the N2 constitutes around 40% of the total samples, while N1 have substantially fewer samples. This sample imbalance might introduce biases in model training. In current research, N1 stage recognition generally performs the worst. For example, in the study by Eldele et al. ( 2021 ), the macro F1-score for the N1 class was only around 40.0, while other categories scored around 85. This class imbalance is intrinsic to sleep and cannot be eliminated. However, its impact can be mitigated through certain methods, which we will discuss in Sect. 4.1.2 .
3.3 Performance metrics
The essence of sleep staging is a multi-classification problem, commonly evaluated using performance metrics such as accuracy ( ACC ), macro F1-score ( F1 ), and Cohen’s Kappa coefficient. Accuracy refers to the ratio between the number of correctly classified samples by the model and the total number of samples. The calculation formula is as follows:
where true positive ( TP ) is the number of samples correctly predicted as positive class by the model, and true negative ( TN ) is the number of samples correctly predicted as negative class by the model. TP and TN both represent instances where the model’s prediction matches the actual class, indicating correct predictions. False positive ( FP ) is the number of negative class samples incorrectly predicted as positive class by the model, and false negative ( FN ) is the number of positive class samples incorrectly predicted as negative class by the model. FP and FN represent instances where the model’s prediction does not match the actual class, indicating incorrect predictions.
ACC is a commonly used evaluation metric in classification problems, but it may show a “pseudo-high” characteristic when dealing with imbalanced datasets (Thölke et al. 2023 ). In contrast, the F1-score takes into account both precision ( PR ) and recall ( RE ) of the model. PR is the proportion of truly positive samples among all samples predicted as positive by the model (Yacouby and Axman 2020 ). RE is the proportion of truly positive samples among all actual positive samples, as predicted by the model. In classification problems, each class has its own F1-score, known as per-class F1-score. Taking the average of F1-scores for all classes yields the more commonly used macro F1-score ( MF1 ). The calculation formula is as follows:
Cohen’s Kappa coefficient (abbreviated as Kappa ) measures the agreement between observers and is used to quantify the consistency between the model’s predicted results and the actual observed results (Hsu and Field 2003 ). The calculation formula is as follows:
where \({P_{ec}}\) is the observed agreement (the proportion of samples with consistent actual and predicted labels), and \({P_{ei}}\) is the chance agreement (the expected probability of agreement between predicted and actual labels, calculated based on the distribution of actual and predicted labels). Kappa ranges from -1 to +1, with higher values indicating better agreement.
Among these three commonly used performance metrics, accuracy corresponds to the ratio of correctly classified samples to the total number of samples, ranging from 0 (all misclassified) to 1 (perfect classification). ACC represents the overall measure of a model’s correct predictions across the entire dataset. The basic element of calculation is an individual sample, with each sample having equal weight, contributing the same to ACC . Once the concept of class is considered, there are majority and minority classes, with the majority class obviously having higher weight than the minority class. Therefore, in the face of class-imbalanced datasets, the high recognition rate and high weight of the majority class can obscure the misclassification of the minority class (Grandini et al. 2020 ). This means that high accuracy does not necessarily indicate good performance across all classes.
MF1 is the macro-average of the F1-scores of each class. MF1 evaluates the algorithm from the perspective of the classes, treating all classes as the basic elements of calculation, with equal weight in the average, thus eliminating the distinction between majority and minority classes (the effect of large and small classes is equally important) (Grandini et al. 2020 ). This means that high MF1 indicates good performance across all classes, while low MF1 indicates poor performance in at least some classes.
The Cohen’s Kappa coefficient is used to measure the consistency between the classification results of the algorithm and the ground truth (human expert classification), ranging from -1 to 1, but typically falling between 0 and 1. From formula 7 , it can be seen that the Kappa considers both correct and incorrect classifications across all classes. In the case of class imbalance, even if the classifier performs well on the majority class, misclassifications on the minority class can significantly reduce the Kappa (Ferri et al. 2009 ). To illustrate this with a simple binary classification problem, assume there are 100 samples in total for classes 0 and 1, with a ratio of 9 : 1. If a poorly performing model always predicts class 0, even if it is entirely wrong on class 1, the ACC would still be as high as 90%. Calculating the F1-score, it is found that class 0 has a score of 1.0 and class 1 has a score of 0, resulting in an MF1 of only 0.5. MF1 equally considers the majority and minority classes, fairly reflecting the poor classification performance. The Kappa value would be 0, indicating no correlation between the model’s predictions and the ground truth. Even though the overall accuracy is high, it does not indicate real classification ability. In summary, this confirms that in the face of class-imbalanced datasets, MF1 and the Kappa can provide more reliable and comprehensive evaluations than accuracy.
4 ASSC based on PSG signals
The essence of automatic sleep stage classification lies in the analysis of sleep data and the extraction of relevant information. In the process of data analysis, appropriate preprocessing and data representation methods can help the model learn and interpret these signals more effectively. This section will provide detailed explanations regarding the preprocessing of PSG signals, method of data representation, and deep learning models.
4.1 Preprocessing methods and class imbalance problems
Preprocessing plays a crucial role in the classification of sleep stages. Appropriate preprocessing methods have a positive impact on subsequent feature extraction, whether it is manual feature extraction in traditional machine learning or high-dimensional feature extraction in deep learning (Wang and Yao 2023 ). Class imbalance is a persistent problem in sleep stage classification, as shown in Table 6 . In this section, we will discuss preprocessing methods and approaches to handling class imbalance problems (CIPs).
4.1.1 Preprocessing methods
In PSG studies, most research is actually based on single-channel EEG, while a small portion uses combinations of EEG and other signals. The original EEG signal is a typical low signal-to-noise ratio signal, usually weak in amplitude and contains a lot of undesirable background noise that needs to be eliminated before actual analysis (Al-Saegh et al. 2021 ). Additionally, there is sometimes a need to enhance the original EEG to better meet the requirements. Based on these needs and reasons, the following preprocessing methods have appeared in existing studies.
Notch filtering: Used to eliminate 50 Hz or 60 Hz power line interference noise (power frequency interference) (Zhu et al. 2023 ).
Bandpass filtering: Used to remove noise and artifacts. The cutoff frequencies for filtering are inconsistent across different studies, even for the same signal from the same dataset. For example, Phyo et al. ( 2022 ) and Jadhav et al. ( 2020 ) applied bandpass filtering with cutoff frequencies of 0.5–49.9 Hz and 0.5–32 Hz for the EEG Fpz-Cz channel of the SEDF dataset, respectively.
Downsampling: Signals from different datasets have varying sampling rates. When utilizing multiple datasets, downsampling is often performed to standardize the rates. Downsampling also reduces computational complexity (Fan et al. 2021 ).
Data scaling and clipping: Scaling adjusts the signal values proportionally to facilitate subsequent processing by adjusting the amplitude range. Clipping is done to prevent large disturbances caused by outliers during model training. Guillot et al. ( 2021 ) first scaled the data to have a unit-interquartile range (IQR) and zero-median and then clipped values greater than 20 times the IQR.
Normalization: Normalization should also belong to the large category of data scaling, and is listed here separately for convenience. The most common preprocessing step, normalization plays a significant role in deep learning. It scales the data proportionally to fit within a specific range or distribution. Normalization unifies the data of different features into the same range, ensuring that each feature has an equal impact on the results during model training, thereby improving the training effectiveness. Z-score normalization (standardization) is the most commonly used method, where data is transformed into a normal distribution with a mean of 0 and a standard deviation of 1 after Z-score normalization. Olesen et al. ( 2021 ) applied Z-score normalization to each signal during preprocessing to adapt to differences in devices and baselines while evaluating the generalization ability of the model across five datasets. Additionally, it is important to note that data scaling and data normalization should not be confused, despite their similarities and occasional interchangeability. It is crucial to understand that both methods transform the values of numerical variables, endowing the transformed data points with specific useful properties. In simple terms: scaling changes the range of the data, while normalization changes the shape of the data distribution. Specifically, data scaling focuses more on adjusting the amplitude range of the data, such as between 0 to 100 or 0 to 1. Data normalization, on the other hand, is a relatively more aggressive transformation that focuses on changing the shape of the data distribution, adjusting the data to a common distribution, typically a Gaussian (normal) distribution (Ali et al. 2014 ). These two techniques are usually not used simultaneously; in practice, the choice is generally made based on the specific characteristics of the data and the needs of the model. The characteristics of the data can be examined for the presence of outliers, the numerical range of features, and their distribution. For example, when data contains a small number of outliers, scaling is often more appropriate than normalization. In particular, the median and IQR-based scaling method used by Guillot et al. ( 2021 ) (often referred to as robust scaling) is especially suitable for data with outliers because it uses the median and interquartile range to scale the data, preventing extreme values from having an impact. However, outliers can significantly affect the mean and standard deviation of the data, thus impacting the effectiveness of normalization based on the mean and standard deviation. Different models also have different requirements. For instance, distance-based algorithms (such as SVM) typically require data scaling, while algorithms that assume data is normally distributed commonly use normalization.
4.1.2 Class imbalance problems
In the preceding text, we have discussed the problem of class imbalance in sleep data (Table 6 ). Deep learning heavily relies on data, and when learning from such imbalanced data, the majority class tends to dominate, leading to a rapid decrease in its error rate (Fan et al. 2020 ). The result of training might be a model biased towards learning the majority class, performing poorly on minority classes. Moreover, when the number of samples in the minority class is very low, the model might overfit to these samples’ features, achieving high performance on the training set but poor generalization to unseen data (Spelmen and Porkodi 2018 ). The class imbalance problem in sleep cannot be eradicated but can only be suppressed through certain measures. The most common approach in existing research is data augmentation (DA), which falls within the preprocessing domain, while another category manifests during the training process.
DA is a method to expand the number of samples without significantly increasing existing data (Zhang et al. 2022a ). Typically, it generates new samples for minority classes to match the sample counts in each class, constructing a new dataset (Fan et al. 2020 ). Three methods are generally used in existing research to generate new augmented data.
Oversampling: Aims to increase the number of samples in minority classes. Different class distributions are balanced through oversampling, enhancing the recognition ability of classification algorithms for minority classes. To prevent the model from favoring the majority class excessively, Supratak et al. ( 2017 ) used “oversampling with replication” during training. By replicating minority stages from the original dataset, all stages had the same number of samples, avoiding overfitting. Mousavi et al. ( 2019 ) used “oversampling with SMOTE (synthetic minority over-sampling technique)” (Chawla et al. 2002 ). SMOTE synthesizes similar new samples by considering the similarity between existing minority samples.
Morphological transformation: A common image enhancement method in image processing is geometric transformation, including rotation, flipping, random scaling, etc. Similar transformations can be performed on physiological signals. Common operations include translation (along the time axis), horizontal or vertical flipping, etc. Noise can also be added, further introducing variability (Fan et al. 2021 ). Zhao et al. ( 2022 ) applied random morphological transformations, deciding whether to perform cyclic horizontal shifting and horizontal flipping on each EEG epoch with a 50% probability.
Generative adversarial networks (GANs): GAN itself is a deep learning model proposed by Ian Goodfellow and colleagues in 2014 (Goodfellow et al. 2014 ). The core of GAN is the competition between two neural networks (generator and discriminator), with the ultimate goal of generating realistic data. GAN are widely used in image generation in the field of images and have similar applications in physiological signals. For instance, Zhang and Liu ( 2018 ) proposed a conditional deep convolutional generative adversarial network (cDCGAN) based on GAN to augment EEG training data in the brain-computer interface field. This network can automatically generate artificial EEG signals, effectively improving the classification accuracy of the model in scenarios with limited training data. In Kuo’s study (Kuo et al. 2022 ), another variant of GAN, self-attention GAN (SAGAN) (Zhang et al. 2019 ), was used. SAGAN is a variant of GAN for image generation tasks. The authors applied continuous wavelet transform to the original EEG signal and used SAGAN to augment the obtained spectrograms. A detailed introduction to the GAN model can be found in Sect. 4.3 .
Another category of methods does not belong to preprocessing but is manifested during model training. Firstly, there is class weight adjustment, usually performed in the loss function. The basic idea is to introduce class weights into the loss function, giving more weight to minority classes, thus focusing more on the classification performance of minority classes during training. Commonly used methods include weighted cross-entropy loss function (Zhao et al. 2022 ) and focal loss function (Lin et al. 2017 ; Neng et al. 2021 ). Secondly, there are ensemble learning strategies, which balance the model’s attention to different classes by combining predictions from multiple base models, thus improving performance. Neng et al. ( 2021 ) trained 13 basic CNN models, selected the top 3 best-performing ones to form an ensemble model, achieving an average accuracy of 93.78%. Research focusing on addressing data imbalance problems is summarized in Table 7 .
We reviewed current methods for mitigating class imbalance in sleep stage classification. Various methods can achieve performance improvements, but their applicability needs further discussion. For DA, the key is to introduce additional information while minimizing changes to the physiological or medical significance of the signals, thereby increasing data diversity (Rommel et al. 2022 ). Oversampling typically involves replicating existing samples or synthesizing similar samples based on existing ones. GAN, through adversarial training, can implicitly learn the mapping from latent space to the overall distribution of sleep data, generating samples that better fit the original data distribution and are more diverse (Fan et al. 2020 ). However, in morphological transformation methods, the essence is to obtain new samples by flipping, translating, etc., the original samples. For weak signals like EEG, simple waveform fluctuations can lead to different medical interpretations. Morphological transformations may not bring about sample diversity and could introduce erroneously annotated new samples, severely disrupting model learning. These were demonstrated by Fan et al. ( 2020 ). They compared EEG data augmentation methods such as repeating the minority classes (DAR), morphological change (DAMC), and GAN (DAGAN) on the MASS dataset. The results showed that DAMC performed the worst among all methods, only improving accuracy by 0.9%, while DAGAN improved performance by 3.8%. However, DAGAN introduced additional model training and resource costs. In Fan et al.’s experiments, GAN required 71.69 h of training and 19.63 min to generate synthetic signals, whereas morphological transformations only needed 201 min.
Class weight adjustment is typically done in the loss function, introducing minimal additional computation but usually bringing in new hyperparameters. For instance, the weighted cross-entropy loss function is calculated as follows:
where \(\textit{y}_i^k\) is the actual label value of the \(\textit{i}\) th sample, \(\hat{y}_i^k\) is the predicted probability for class \(\textit{k}\) of the \(\textit{i}\) -th sample, \(\textit{M}\) is the total number of samples, and \(\textit{K}\) is the total number of classes. \(\textit{w}_k\) represents the weight of class \(\textit{k}\) , which is typically provided as a hyperparameter by the researchers. Other functions, such as focal loss, introduce two hyperparameters: the modulation factor and class weight. Ensemble learning requires training multiple base models for combined predictions, which adds extra overhead.
In summary, each existing method has its pros and cons. In terms of low cost and ease of application, oversampling and morphological transformation have more advantages. Although weighted loss functions also have low costs, they come with hyperparameter issues. GAN have performance advantages. When researchers can accommodate the additional overhead brought by GAN or pursue higher performance, GAN are worth trying. Additionally, Fan et al. ( 2020 ) and Rommel et al. ( 2022 ) conducted in-depth comparisons and analyses of different data augmentation methods, and interested readers can refer to their works.
4.2 Data representation
When using DL methods to process sleep data, a crucial issue is transforming sleep signals into suitable representations for subsequent learning by DL models. The choice of representation largely depends on the model’s requirements and the nature of the signal (Altaheri et al. 2023 ). Appropriate input forms enable the model to effectively learn and interpret sleep information. Signals can be directly represented as raw values, in which case they are in the time domain. Through signal processing methods such as wavelet transforms, Fourier transforms, etc., transformed domain representations of the signal can be obtained. Moreover, a combination of these two approaches is also commonly used. Figure 5 displays the representation methods and their proportions used in the reviewed articles. Figure 6 a categorizes the representation methods applicable to PSG data, while Fig. 6 b provides examples of these different representations.

The proportional representation of each data representation in this review paper (*Spatial-frequency images)

Classification of PSG data representation methods, and examples of each representation method. a Classification of representations; b Raw multi-channel signal, [TP \(\times\) C]; c STFT gets [T \(\times\) F] time-frequency spectrogram; d STFT gets [T \(\times\) F \(\times\) C] time-frequency spectrogram; e FFT gets [F \(\times\) C] spatial-frequency spectrum. TP Time Point (sampling point), C Channel (electrode), T Time window (time segment), F Frequency
4.2.1 Raw signal
In the time domain, the raw signal represents the main information as variations in signal amplitude over time. When signals come from multiple channels (electrodes), they can be represented as a 2D matrix of [TP (time point) \(\times\) C (channel)] (for a single-channel, it would be [TP \(\times\) 1]). This can be visualized as shown in Fig. 6 b. Traditionally, specific manually designed time-domain features are extracted from raw signals for input, such as power spectral density features (Xu and Plataniotis 2016 ). However, DL methods can automatically learn complex features from extensive data. This allows researchers to bypass the manual feature extraction step, directly inputting 1D raw signals with limited or no preprocessing into neural networks. In recent years, this straightforward and effective approach has become increasingly mainstream (as indicated in Fig. 5 ). Existing studies have directly input raw signals into various DL models, achieving good performance. This includes classic CNN architectures like ResNet (He et al. 2016 ; Seo et al. 2020 ) and U-Net (Ronneberger et al. 2015 ; Perslev et al. 2019 ), as well as CNN models proposed by researchers (Tsinalis et al. 2016 ; Goshtasbi et al. 2022 ; Sors et al. 2018 ). It also encompasses models such as RNNs (long short-term memory or gated recurrent unit) and Transformer (Phyo et al. 2022 ; Olesen et al. 2021 ; Lee et al. 2024 ; Pradeepkumar et al. 2022 ). In these works, minimal or no preprocessing has been applied. For example, Seo et al. ( 2020 ) utilized an improved ResNet to extract representative features from raw single-channel EEG epochs and explicitly emphasized that their method was an “end-to-end model trained in one step”, requiring no additional data preprocessing methods.
4.2.2 Transform domain
Transformed domain data are typically obtained from the raw signals through methods such as short-time Fourier transform, continuous wavelet transform, Hilbert-Huang transform, fast Fourier transform, and others. STFT, CWT and HHT fall under time-frequency analysis methods, providing time-frequency spectrograms that encompass both time and frequency information. The spectrogram can be regarded as a specific type of image, offering better understanding of the signal’s time-frequency features and patterns of change. As depicted in Fig. 6 c and d, spectrograms can be represented as [T (time window) \(\times\) F (frequency)] or in the case of multiple channels, [T \(\times\) F \(\times\) C]. Different time-frequency analysis methods have variances between them. For instance, STFT utilizes a fixed-length window for signal analysis, thus can be considered a static method concerning time and frequency resolution. In contrast, CWT employs multiple resolution windows, providing dynamic features (Herff et al. 2020 ; Elsayed et al. 2016 ). To our knowledge, there is a lack of comprehensive research comparing the performance of different time-frequency analysis methods. For EEG, the energy across different sleep stages not only varies in frequency but also in spatial distribution (Jia et al. 2020a ). This spatial information can be introduced through the “spatial-frequency spectrum”, typically implemented using FFT (Cai et al. 2021 ), as shown in Fig. 6 e.
Phan et al. ( 2019 ) transformed 30-second epochs of EEG, EOG, and EMG signals into power spectra using STFT (window size of 2 s, 50% overlap, Hamming window, and 256-point FFT). This resulted in a multi-channel image of [T \(\times\) F \(\times\) C], where C = 3. The authors input these spectrogram data into a bidirectional hierarchical RNN model with attention mechanisms for sleep stage classification. The spatial-frequency spectrum introduced EEG electrode spatial information to enhance classification accuracy. Jia et al. ( 2020a ) first conducted frequency domain feature analysis on power spectral density using FFT for five EEG frequency bands (delta, theta, alpha, beta, gamma) closely related to sleep. They placed the frequency domain features of different electrodes in the same frequency band on a 16 \(\times\) 16 2D map, resulting in five 2D maps representing different frequency bands. Each 2D map was treated as a channel of the image, producing a 5-channel image for each sample representing the spatial distribution of frequency domain features from different frequency bands.
In addition to using a single type of input form, some studies simultaneously use both. In these studies, it is often considered that individual time-domain, frequency-domain, or spatial-domain features alone are insufficient to completely differentiate sleep stages. Their combination offers complementarity, supplementing classification information (Jia et al. 2020a ; Cai et al. 2021 ; Phan et al. 2021 ; Fang et al. 2023 ). Researchers usually construct a multi-branch network to process different forms of data separately. Features from multiple branches are fused using specific strategies to achieve better classification results. For example, Jia et al. ( 2020a ) established a multi-branch model, simultaneously inputting spatial-frequency spectrum from 20 EEG channels and raw signals (EEG, EOG, EMG) into the model.
4.3 Deep learning models
In automatic sleep stage classification, DL has become the mainstream method in recent years compared to traditional ML techniques. Figure 7 a and b provide a comparative overview of the workflows between the two methods. DL methods automate the feature extraction and classification steps present in ML, enabling an end-to-end approach. In this section, different deep learning models used in relevant studies will be introduced. DL models can be categorized into two subclasses based on their functionality: discriminative models and generative models, as well as hybrid models formed by combinations of these, as depicted in Fig. 8 .

a General workflow of machine learning; b General workflow of deep learning

Classification of deep learning models
4.3.1 Discriminative models
Discriminative models refer to DL architectures that can learn different features from input signals through nonlinear transformations and classify them into predefined categories using probability predictions (Altaheri et al. 2023 ). Discriminative models are commonly utilized in supervised learning tasks and serve both feature extraction and classification purposes. In the context of sleep stage classification, the two major types of discriminative models widely used are CNN and RNN.
4.3.1.1 CNN
CNN, one of the most common DL models, is primarily used for tasks such as image classification in computer vision, and in recent years, it has been applied to biological signal classification tasks like ECG and EEG (Yang et al. 2015 ; Morabito et al. 2016 ). CNN is composed of a series of neural network layers arranged in a specific order, typically including five layers: input layer, convolutional layer, pooling layer, fully connected layer, and output layer (Yang et al. 2015 ; Morabito et al. 2016 ), as illustrated in Fig. 9 . Starting from the input layer, the initial few layers learn low-level features, while later layers learn high-level features (Altaheri et al. 2023 ). The convolutional layer is the core building block of a CNN, where feature extraction from the input data is achieved through convolutional kernels. For example, in a 2D convolution, if the input data is a 224 \(\times\) 224 matrix and the convolutional kernel is a 3 \(\times\) 3 matrix (which can be adjusted), the values within this matrix are referred to as weight parameters. The convolutional kernel is applied to a specific region of the input data, computing the dot product between the data in that region and the kernel. The result of this dot product is provided to the output array. After this computation, the kernel moves by one unit length, known as the “stride,” and the process is repeated. This procedure continues until the convolutional kernel has scanned the entire input matrix. The dot product results from a series of scans constitute the final output, known as the feature map, representing the features extracted by the convolution. Note that the kernel remains unchanged during its sliding process, meaning all regions of the input share the same set of weight parameters, which is referred to as “weight sharing” and is one of the critical reasons for CNN’s success. Pooling layer performs a similar but distinct operation by scanning the input with a pooling kernel. For instance, in the commonly used max pooling, if the pooling kernel size is 3 \(\times\) 3, the result of each pooling operation is the maximum value from a 3 \(\times\) 3 region of the input matrix. The essence of pooling is downsampling, aimed at reducing network complexity or computational load. Typically, a series of consecutive convolution-pooling operations are used to extract data features. The feature maps obtained from convolution and pooling are usually flattened and then fed into one or more fully connected layers. As shown in Fig. 9 , in the fully connected layers, each node in the input feature map is fully connected to each node in the output feature map, whereas convolutional layers have partial connections. The fully connected layers often use the softmax function to classify the input appropriately, generating probability values between 0 and 1. CNN is one of the most important models in sleep stage classification, with 76% of the studies reviewed in this paper utilizing CNN, as shown in Fig. 10 . The CNN variants used in existing research include both standard CNN architectures, as well as various modified versions of CNN. For example, the residual CNN (He et al. 2016 ), inception-CNN (Szegedy et al. 2015 ), dense-convolutional (DenseNet) (Huang et al. 2017 ), 3D-CNN (Ji et al. 2023 ), and multi-branch CNN (used in ensemble learning) (Kuo et al. 2021 ), among others, are listed in Table 8 , and their structures are shown in Fig. 11 .

Basic principles of CNN

The proportional representation of each DL model in this review paper

a The upper part is a single residual connection block, and the lower part is a cascade of multiple residual blocks; b Replacing conv-layer with attention module; c Inception structure proposed in GoogLeNet (Szegedy et al. 2015 ; d Using an ensemble learning approach, the outputs of three basic CNNs are fed into a neural network with a hidden fully connected layer for further learning; e A novel CNN variant: DenseNet (Huang et al. 2017 ) is a convolutional neural network architecture that directly connects each layer to all subsequent layers to enhance feature reuse, facilitate gradient flow, and reduce the number of parameters
Zhou et al. ( 2021 ) proposed a lightweight CNN model that utilized the inception structure (as shown in Fig. 11 c) to increase network width while reducing the number of parameters. This model took EEG’s STFT spectrogram as input. In a multimodal deep neural network model proposed in Zhao et al. ( 2021a ), which included two parallel 13-layer 1D-CNNs, residual connections (as shown in Fig. 11 a) were used to address potential gradient vanishing problems. EEG and ECG features were extracted separately in their respective convolutional branches and were later merged through simple concatenation for input into the classification module. Jia et al. ( 2020a ) proposed a CNN model using EEG, EOG, and EMG. The model had multiple convolutional branches, each extracting different features from raw signals, and features from images generated by FFT from EEG. Features from different data representations were concatenated and input into the classification module. Kanwal et al. ( 2019 ) combined EEG and EOG to create RGB images, which were then transformed into high bit depth FFT features using 2D-FFT and classified using DenseNet (as shown in Fig. 11 e). Conversely, in Liu et al. ( 2023b ), an end-to-end deep learning model for automatic sleep staging based on DenseNet was designed and built. This model took raw EEG as input and employed two convolutional branches to extract features at different frequency levels. Significant waveform features were extracted using DenseNet modules and enhanced with coordinate attention mechanisms, achieving an overall accuracy of 90% on SEDF. Kuo et al. ( 2021 ) designed a CNN model that utilized CWT time-frequency spectrograms as input and combined Inception and residual connections. They also trained other classic CNN models and selected the top 3 models with the highest accuracy as base CNNs. These outputs were further learned using a fully connected network with a hidden layer, implementing ensemble learning (as shown in Fig. 11 d). In Fang et al. ( 2023 ), authors used an ensemble strategy based on Boosting to combine multiple weak classifiers. Additionally, various CNN variants have been introduced in other studies, such as architectures incorporating different attention modules, as seen in Liu et al. ( 2023a ) and Liu et al. ( 2022b ) (as shown in Fig. 11 b).
4.3.1.2 RNN
In many real-world scenarios, the input elements exhibit a certain degree of contextual dependency (temporal dependency) rather than being independent of each other. For instance, the variation of stock prices over time and sleep stage signals both reflect this dependency. To capture such relationships, models need to possess a memory capability, enabling them to make predictive outputs based on both current elements and features of previously input elements. This requirement has led to the widespread use of RNN in sleep stage classification tasks. A typical RNN architecture is illustrated in Fig. 12 a, which includes an input layer, an output layer, and a hidden layer. Define \(\textit{x}_t\) as the input at time \(\textit{t}\) , \(\textit{o}_t\) as the output, \(\textit{s}_t\) as the memory, \(\textit{U}\) , \(\textit{V}\) , and \(\textit{W}\) as the weight parameter. As shown on the right side of Fig. 12 a, when unfolded along the time axis, the RNN repetitively uses the same unit structure at different time steps, incorporating the memory from the previous time step into the hidden layer during each iteration. \(\textit{U}\) , \(\textit{V}\) , and \(\textit{W}\) are shared across all time steps, enabling all previous inputs to influence future outputs through this recurrence. RNN possess memory capabilities, making them suitable for the demands of sleep stage classification tasks. However, the memory capacity of RNN is limited: it is generally assumed that inputs closer to the current time have a greater impact, while earlier inputs have a lesser impact, restricting RNN to short-term memory. Additionally, RNN face challenges such as high training costs (due to the inability to perform parallel computations in their recurrent structure) and the problem of vanishing gradients (Yifan et al. 2020 ). To address these issues, two widely used variants of RNN were proposed: LSTM and GRU. The basic unit composition of LSTM is depicted in Fig. 12 b. Unlike RNN, which have a single hidden state s representing short-term memory, LSTM introduce \(\textit{h}\) as the hidden state (short-term memory). Moreover, LSTM add a cell state \(\textit{c}\) capable of storing long-term memory. The basic unit is controlled by three gates: the input gate, the forget gate, and the output gate. These “gates” are implemented using the sigmoid function, which outputs a probability value between 0 and 1, indicating the amount of information allowed to pass through. Among the three gates in LSTM, the forget gate determines how much of the previous cell state \(\textit{c}_{t-1}\) is retained in the current cell state \(\textit{c}_t\) , based on the current input \(\textit{x}_t\) and the previous output \(\textit{h}_{t-1}\) . After forgetting the irrelevant information, new memories need to be supplemented based on the current input. The input gate determines how much of \(\textit{x}_t\) updates the cell state \(\textit{c}_t\) based on \(\textit{x}_t\) , \(\textit{h}_{t-1}\) , and the output of the forget gate. The output gate controls how much of the cell state ct is output based on \(\textit{x}_t\) and \(\textit{h}_{t-1}\) . By introducing the cell state \(\textit{c}\) and gate structures, LSTM can maintain longer memories and overcome issues such as vanishing gradients. However, LSTM are still essentially recurrent structures and thus cannot perform parallel computations (Yifan et al. 2020 ). GRU, another common variant of RNN, simplifies the architecture by having only two gate structures, reducing the number of parameters and increasing computational efficiency, though it still lacks the capability for parallel computation (Chung et al. 2014 ).

a Typical basic structure of RNN; b Basic unit of LSTM
Phan et al. ( 2018 ) designed a bidirectional RNN with an attention mechanism to learn features from single-channel EEG signal’s STFT transformation. The authors first divided the EEG epoch into multiple small frames. Using STFT, they transformed these into continuous frame-by-frame feature vectors, which were then input into the model shown in Fig. 13 for training. The training objective was to enable the model to encode the information of the input sequence into high-level feature vectors. Note that this is not an end-to-end process; the RNN was used as a feature extractor, while the classification was performed by a linear SVM classifier. The final classification is done through SVM. As an improvement, they later proposed a bidirectional hierarchical LSTM model combined with attention. The model takes STFT transformations of signals (EEG, EOG, EMG) as input. Based on attention, bidirectional LSTM encodes epochs into attention feature vectors, which are further modeled by bidirectional GRU (Phan et al. 2019 ). Inspired by their work, Guillot et al. ( 2020 ) enhanced a model based on GRU and positional embedding, reducing the number of parameters. In the study by Xu et al. ( 2020 ), four LSTM models were constructed, each with different input signal lengths (1, 2, 3, and 4 epochs). It was found that each model exhibited varying sensitivity to different sleep stages. The authors combined models with distinct stage sensitivities, resulting in improved classification accuracy.

Attention-based bidirectional RNN (Phan et al. 2018 )
4.3.1.3 Hybrid
There exists rich temporal contextual information between consecutive sleep stages, which should not be ignored whether in expert manual staging or computer-assisted staging. For instance, if one or more sleep spindles or K-complexes are observed in the second half of the preceding epoch or the first half of the current epoch, the current epoch is classified as N2 stage. Moreover, sleep exhibits continuous stage transition patterns like N1-N2-N1-N2, N2-N2-N3-N2 (Iber 2007 ; Tsinalis et al. 2016 ). Both intra-epoch features and inter-epoch dependencies within the epoch sequence should be considered simultaneously (Seo et al. 2020 ). This is a challenge that individual CNN or RNN models cannot effectively address. Hence, the most common type of model in sleep stage classification is actually the hybrid of CNN and RNN (CRNN), which is designed to simultaneously handle feature extraction and model long-term dependencies. As shown in Fig. 14 , hybrid models can be generalized into two main components: feature extractor (FE) and sequence encoder (SE). CNN is commonly used as FE, responsible for extracting epoch features and encoding invariant information over time; RNN is typically used as SE, focusing on representing relationships between epochs and encoding temporal relationships within the epoch sequence (Supratak et al. 2017 ; Phyo et al. 2022 ; Phan and Mikkelsen 2022 ).

A hybrid model consisting of Feature Extractor (FE) and Sequence Encoder (SE). \(x_1\) - \(x_L\) constitute an epoch sequence, FE extracts features at the intra-epoch level, and SE captures contextual information at the inter-epoch level. L \(\ge\) 1 (integer)
Such hybrid structure is implemented in DeepSleepNet, proposed by Supratak et al. ( 2017 ). The model extracts invariant features from raw single-channel EEG using a dual-branch CNN with different kernel sizes and encodes temporal information into the model with bidirectional LSTM featuring residual connections. DeepSleepNet achieved an accuracy of 82.0% on SEDF. In subsequent improvements, the authors significantly reduced the parameter count of the CRNN structure (approximately 6% of DeepSleepNet) and improved the performance to 85.4% (Supratak and Guo 2020 ). Seo et al. ( 2020 ) utilized the epoch sequence of raw single-channel EEG as input, employed an improved ResNet-50 network to extract representative features at the sub-epoch level, and captured intra- and inter-epoch temporal context from the obtained feature sequence with bidirectional LSTM. Performance comparisons were made with input sequences of different lengths (L) ranging from 1 to 10, with the model achieving the best accuracy of 83.9% on SEDF and 86.7% on SHHS datasets when L=10. Neng et al. divided sleep data into three levels: frame, epoch, and sequence, where frame is a finer division of epoch, and sequence represents epoch sequences (Neng et al. 2021 ). Based on this, they designed models with frame-level CNN, epoch-level CRNN, and sequence-level RNN, essentially aiming at modeling long-term dependencies. The input sequence length of the model was 25 epochs, and it achieved an accuracy of 84.29% on SEDF.
CRNN is the most widely used approach, but RNN suffers from long training times and challenges in parallel training. Hence, researchers have explored attention mechanisms and Transformer architectures based on self-attention (Vaswani et al. 2017 ), which have shown excellent performance in sequential tasks. The self-attention mechanism excels at capturing the inherent relationships and dependencies within input sequences. As depicted in Fig. 15 , the basic structure of self-attention involves computing the relationship between each position in the input sequence and every other position, yielding a weight distribution. By performing a weighted summation of the input sequence based on this distribution, an output sequence encapsulating internal dependencies is produced (Guo et al. 2022 ). The core of the Transformer is the self-attention mechanism, which is divided into two main parts: the encoder and the decoder. In existing research, the encoder part is typically used. The Transformer encoder comprises several key components: positional encoding, multi-head self-attention, feed-forward neural network, layer normalization, and residual connections, as illustrated in Fig. 16 a. The first operation of the encoder is to encode the position of the input sequence. MHSA can model the relationships within the input time series, but it cannot perceive the local positional information of the input sequence (Foumani et al. 2024 ). Therefore, positional information is first added to the input using fixed positional encoding based on sine and cosine functions of different frequencies (Vaswani et al. 2017 ):
where \(\textit{t}\) represents the input sequence data, \(\textit{p}\) represents the matrix calculated by the positional encoding function PE, \(\textit{p}os\) is the position index in the input sequence, d is the dimension of the input embeddings, and \(\textit{i}\) is the index of the dimension in the positional encoding vector. Next, MHSA modeling is performed. MHSA is an extension of self-attention that divides the input sequence into H sub-sequences, utilizing H parallel self-attention heads to capture different interactive information in various projection spaces (each head has different parameters). These H heads can capture different features and relationships of the input elements, and their fusion results in a richer global representation. As shown in Fig. 16 b, taking the \(\textit{h}\) -th head and sub-sequence input \(\textit{x}\) as an example, three linear projections are first obtained for \(\textit{x}\) , resulting in three copies of \(\textit{x}\) (query ( \(\textit{q}\) ), key ( \(\textit{k}\) ), and value ( \(\textit{v}\) ) matrices). This can be represented as:
where \(\textit{x}_q^h\) , \(\textit{x}_k^h\) , and \(\textit{x}_v^h\) represent the \(\textit{q}\) , \(\textit{k}\) , and \(\textit{v}\) copies, respectively, and \(\textit{W}_q^h\) , \(\textit{W}_k^h\) , and \(\textit{W}_v^h\) represent the learnable projection matrices. The self-attention output of the \(\textit{h}\) -th head is:
where \(\textit{d}_k\) is the dimension of the h -th head. Assuming there are H heads, each head’s output can be represented as \(O^i\) ( \(1 \le i \le H\) ). Concatenating the outputs of all heads and applying another linear projection \(W_o\) yields the final output of MHSA. This can be represented as:
After the multi-head self-attention mechanism, each encoder layer also includes a feed-forward neural network. This network typically consists of two fully connected layers and a nonlinear activation function, such as ReLU. It operates on the inputs at each position to generate new representations for each element. Layer normalization follows the multi-head self-attention and feed-forward neural network, helping to stabilize the training process and accelerate convergence. It normalizes the inputs of each layer so that the output has a mean of 0 and a standard deviation of 1. Residual connections, which appear alongside layer normalization, add the input of a sub-layer directly to its output. This connection helps to address the problem of vanishing gradients in deep networks and speeds up the training process. These components together form a standard Transformer encoder layer, and the encoder typically stacks multiple such layers. Each layer produces higher-level abstract representations, with the output of one layer serving as the input to the next, thereby extracting deeper features step by step. Compared to the recursive computations of RNN, the self-attention mechanism can parallelize the entire sequence, making it easily accelerated by GPU, similar to CNN (Guo et al. 2022 ). Furthermore, the self-attention mechanism can effortlessly obtain global information. These factors contribute to its widespread application in sequence data tasks, including sleep stage classification problems.

The basic structure of the self-attention mechanism
a Transformer encoder: It is composed of N standard encoder layers stacked together. The encoder layer consists of positional encoding, multi-head self-attention, feed-forward neural network, layer normalization, and residual connections; b The self-attention calculation process of the \(\textit{h}\) -th head
Attention and Transformer encoders (as shown in Fig. 16 a) are often combined with CNNs to form hybrid models, where they also play the role of SE. For example, in the CNN-Attention model constructed by Zhu et al. ( 2020 ), CNN is used to encode epoch features, and self-attention is employed to learn temporal dependencies. AttnSleep, proposed by Eldele, uses CNN for feature extraction and employs a Transformer-encoder module combined with causal convolutions for encoding temporal context (Eldele et al. 2021 ). A CNN-Transformer model for real-time sleep stage classification on energy-constrained wireless devices was proposed in Yao and Liu ( 2023 ). The model, applied to single-channel input data of size (3000, 1) (signal length 30 s, sampling rate 100 Hz), extracts features of size (19, 128) through 4 consecutive convolutional layers. The Transformer-encoder is then used to learn temporal information from these features. The downsized model was tested on an Arduino development board, achieving an accuracy of 80% on the SEDF dataset. Lee et al. ( 2024 ) and Pradeepkumar et al. ( 2022 ) also introduced their CNN-Transformer approaches. Additionally, Phan et al. ( 2022b ) proposed a model called SleepTransformer, which entirely eliminates the need for convolutional and recurrent operations. SleepTransformer no longer relies on CNN for epoch feature extraction but instead relies entirely on Transformer’s encoder to serve as FE and SE.
4.3.2 Generative models
In sleep stage classification, one popular generative DL model is GAN. It is important to note that the task reviewed in this paper is a classification task. GAN itself is used for data generation, and although it has a discriminator that performs binary classification, its sole purpose is to distinguish between real data and data synthesized by the generator, ultimately aiding the generator in producing realistic data. In the current context, GAN is typically used in the data augmentation phase to mitigate issues such as insufficient EEG training data or class imbalance, as described in Sect. 4.1.2 . The data augmented by GAN still requires a classification model to achieve classification. Several studies have compared the effects of GAN with traditional data augmentation methods (such as SMOTE, morphological transformations, etc.) (Fan et al. 2020 ; Yu et al. 2023 ). The results of these studies indicate that sleep data augmentation based on GAN significantly improves classification performance. Fan et al. ( 2020 ) compared five data augmentation methods: repeating minority class samples, signal morphological transformations, signal segmentation and recombination, dataset-to-dataset transfer, and GAN. The results showed that GAN increased accuracy by 3.79% and 4.51% on MASS and SEDF, respectively, achieving the most remarkable performance improvement. Cheng et al. ( 2023a ) designed a new GAN model (SleepEGAN), using the model from Supratak and Guo ( 2020 ) as the generator and discriminator of GAN, combined with a CRNN classifier to perform the classification task. After SleepEGAN augmentation on the SHHS dataset, the number of samples in the N1 stage increased from 10,304 to 46,272, and the overall classification accuracy improved to 88.0% (the second-best method achieved 84.2%). In Cheng’s study, original signals were augmented, while in Kuo et al. ( 2022 ), self-attention GAN was used to augment spectrogram images, and ResNet was employed for classification. On their private dataset, the combination of spectrogram, self-attention GAN and ResNet achieved an accuracy of 95.70%, whereas the direct classification approach was only 87.50%. Moreover, Yu et al. ( 2023 ); Zhou et al. ( 2022 ); Ling et al. ( 2022 ), and other studies also utilized GAN for data augmentation. In Yu et al. ( 2023 ), the generator and discriminator of the GAN model were both based on Transformer-encoder. Figure 8 displays the proportion of deep learning methods included in the reviewed studies, and Tables 9 , 10 and 11 summarizes key information extracted from the papers. In these tables, we have compiled information on various types of input data, datasets, preprocessing methods, deep learning models, and their reported performance in recent papers.
5 ASSC based on cardiorespiratory signals
Currently, PSG remains the “gold standard” signal in sleep research. However, the time-consuming and labor-intensive nature of PSG data collection can disrupt a subject’s natural sleep patterns. Due to these limitations, sleep monitoring based on PSG struggles to transition from sleep labs to everyday life. Recent studies have demonstrated the correlation between sleep and respiratory or circulatory systems (Sun et al. 2020 ). In contrast, signals reflecting such activities, such as ECG, PPG, etc., offer unique advantages in terms of signal acquisition, cost, and subject comfort. For example, PPG can be collected using smartwatches. Hence, researchers have started exploring how to perform sleep stage classification using signals from the heart and lungs.
In studies based on heart and lung signals, various preprocessing methods and input formats are employed. However, unlike PSG, most studies do not directly use raw ECG or PPG signals but instead use derived time series (derived signals) such as HR, HRV, RRIs, etc. (Goldammer et al. 2022 ; Sun et al. 2020 ; Sridhar et al. 2020 ; Fonseca et al. 2020 ). These studies typically involve four steps: signal collection, extraction of derived time series, preprocessing, and neural network classification. Firstly, most studies still use public datasets, with only a few using their own data. For instance, in Fonseca et al. ( 2020 ), data from 11 sleep labs in five European countries and the United States were used for training, while data from another lab in the Netherlands served as a reserved validation set. The study involved 389 subjects, which is relatively small compared to some public datasets. The second step involves extracting derived time series. This often involves different algorithms aimed at extracting the required derived signals from the raw signal. Commonly derived signals include HR, HRV, RRIs, EDR, RR peak sequences, etc. Goldammer et al. ( 2022 ) used ECG and chest respiratory effort data from SHHS. RRIs were extracted from the raw ECG using a filter band algorithm, while breath-to-breath intervals were extracted from chest respiratory effort data using another algorithm. These algorithms can be found in Afonso et al. ( 1999 ) and Baillet et al. ( 2011 ). Sridhar et al. ( 2020 ) used ECG data provided by SHHS, MESA, etc. To extract heart rate information, they first normalized the raw ECG and then detected R-waves using the Pan-Tompkins algorithm, a common algorithm for automatic R-wave detection (Pan and Tompkins 1985 ). The time differences between consecutive R-waves form the interbeat interval (IBI) time series. Taking the reciprocal of IBI yields the required heart rate information (Sridhar et al. 2020 ). Sun et al. ( 2020 ) also used the Pan-Tompkins algorithm for ECG R-peak detection. However, after obtaining the time points of R-peaks, they converted the ECG into a binary sequence (1 at R-peaks, 0 elsewhere). The third step is preprocessing. In fact, this step is not consistent across studies; different studies preprocess either the raw signal or both the raw and derived signals. Common preprocessing methods include interpolation resampling, normalization, and outlier removal. In Goldammer et al. ( 2022 ), both RRIs and BBIs were linearly interpolated, resampled at a frequency of 4 Hz, and z-score normalized. The first and last five minutes of each signal were considered outliers (poor signal quality) and were truncated. Sridhar et al. ( 2020 ) simultaneously processed the raw ECG and derived signals. The raw ECG was normalized before extracting HR, and after obtaining HR, each night was independently z-score normalized and linearly interpolated and resampled to a sampling rate of 2Hz. Padding with zeros was performed to fix the size at 10 h. Sun et al. ( 2020 ) also identified potential non-physiological artifact segments based on voltage values. The final step involves using neural networks for classification. In heart and lung signals, CRNN remains popular. For example, Sun et al. ( 2020 ) constructed multiple neural networks, each comprising CNN and LSTM components. The former learned features related to each epoch, while the latter learned temporal patterns between consecutive epochs.
Apart from using derived time series as input, some studies have chosen raw signals or images as input. In Kotzen et al. ( 2022 ) and Korkalainen et al. ( 2020 ), preprocessed PPG was directly input into neural network models for classification. Olsen et al. ( 2022 ) used both PPG and accelerometer data, with PPG coming from clinical collection and wearable devices. All accelerometer and PPG data were resampled to 32 Hz, and outlier removal was performed after cropping the data. After STFT, time-frequency representations of both data types were obtained. The authors used a CNN model similar to U-Net to receive these time-frequency data as input, achieving an accuracy of 69.0% on the reserved validation set. Key information extracted from the heart and lung related research is summarized in Table 12 .
6 ASSC based on contactless signals
In recent years, monitoring physiological signals through non-contact methods has emerged as a promising field in e-health. These methods aim to provide a viable alternative to contact-based signal acquisition. Contact-based methods, such as those involving EEG, EOG, and ECG mentioned in Sect. 4 and Sect. 5 , require direct skin contact via sensors or electrodes. These methods are often impractical for subjects with severe burns, skin diseases, sensitive skin (as in elderly patients or infants), and they typically necessitate the involvement of healthcare personnel, as the correct placement of electrodes can be challenging for laypersons. Non-contact methods, which eliminate physical contact during data collection, include technologies like radar, Wi-Fi, and microphones. These signals can be seamlessly integrated into the environment, having minimal impact on the subject, and enabling remote and unobtrusive data collection (Nocera et al. 2021 ). This characteristic is particularly advantageous for long-term tasks such as sleep monitoring. Consequently, many researchers have recently begun exploring the combination of non-contact signals and deep learning techniques in this domain. Table 13 presents a summary of recent studies in this area. Figure 17 shows a flow chart of contactless sleep stage classification using radar or Wi-Fi. Signal acquisition is usually implemented by a pair of transmitters and receivers. After preprocessing, features such as motion and breathing are extracted and fed into the DL model for classification.
Flowchart of sleep stage classification using radar or Wi-Fi. From top to bottom: signal acquisition, feature extraction and deep learning model classification. The transmitter transmits wireless signals, interacts with human activities in the middle, and the receiver receives signals containing physiological information. After preprocessing, features such as movement, breathing, and heartbeat are extracted and finally sent to the DL model for classification
Radar and Wi-Fi both fall under the category of radio frequency (RF) signals and are currently widely applied in remote vital signs monitoring and activity recognition. RF-based non-contact transmission can capture reflections caused by physiological activities such as thoracic respiration and heartbeats. These reflection signals are often complex due to the presence of large-scale body movements, resulting in a non-linear combination of vital sign information and other motion data (Chen et al. 2021 ). Since the vital sign information is subtle but persistent, powerful tools like deep learning are required to extract and map this data to sleep stages for classification. Radar is an excellent non-contact sensor that can directly measure relevant information about a target, such as distance, speed, and angle, through the emission, reflection, and reception of electromagnetic waves. In Table 13 , we review eight papers that classify sleep stages using radar. These studies exhibit distinct characteristics, which we detail below:
6.1.1 Radar equipment
Among the eight reviewed studies, various types of radar equipment were used (two studies did not specify the type). These included impulse-radio ultra-wideband (IR-UWB) radar (Park et al. 2024 ; Kwon et al. 2021 ; Toften et al. 2020 ), continuous wave (CW) Doppler radar (Chung et al. 2018 ; Favia 2021 ), and microwave radar (Wang and Matsushita 2023 ). CW Doppler radar appeared twice, IR-UWB three times, and microwave radar once. Although these numbers lack statistical significance, another review on radar in healthcare reported similar findings, showing that UWB and CW radars have usage rates of 26% and 29% respectively in healthcare applications (Nocera et al. 2021 ). This suggests that these radar types may be more suited for sleep monitoring tasks, though fair comparative experiments are needed to confirm this. Notably, Zhai et al. ( 2022 ) compared radar working frequencies, collecting nighttime sleep radar signals at 6 GHz and 60 GHz, respectively, for W/L/D/REM classification. They found that the lower frequency 6 GHz signals achieved an accuracy of 79.2%, whereas the 60 GHz signals achieved only 75.2%.
6.1.2 Datasets
There are no publicly available datasets in the existing research. Among the eight studies, only the data collected by Zhao et al. ( 2017 ) is available upon simple request.
6.1.3 Preprocessing
The preprocessing methods show no consistency, with techniques including downsampling to reduce computational complexity, normalization to constrain data distribution, and high-pass or band-pass filtering to remove noise. Unique to this review is the “moving average method” used to remove clutter (Park et al. 2024 ), appearing exclusively in this context.
6.1.4 Data representation
Regarding the use of radar signals, Wang and Matsushita ( 2023 ), Kwon et al. ( 2021 ), Toften et al. ( 2020 ), and Chung et al. ( 2018 ) all chose to input hand-crafted features into their models. These features included motion characteristics, respiratory features, and heart rate features, likely due to the weaker nature of radar signals compared to direct signals like EEG and ECG. Additionally, Park et al. ( 2024 ) and Zhao et al. ( 2017 ) used spectral forms of the signals. Zhai et al. ( 2022 ) and Favia ( 2021 ) used raw one-dimensional signals, preprocessed with filtering and normalization, as model inputs. Favia also compared raw signal inputs with STFT spectral inputs, finding that models using raw data outperformed those using spectral inputs. They noted that it would be simplistic to conclude that raw data is inherently better suited for the task, suggesting that multiple factors, such as non-optimal windowing or FFT points in STFT, or the model’s suitability for the task, could be influencing the results.
6.1.5 Deep learning models
Similar to Sect. 4 , hybrid models like CNN-RNN (Toften et al. 2020 ) and CNN-Transformer (Park et al. 2024 ) dominate the landscape for radar signals, appearing five times, whereas RNNs alone appear only once. Additionally, multilayer perceptron (MLP) models, which are rarely used alone in PSG studies, appear twice in this context (Wang and Matsushita 2023 ; Chung et al. 2018 ). Although we reviewed the models and their performance, it is important to note that these are not fair comparisons, highlighting the potential value of a comparative study in this field.
6.1.6 Number of sleep stage categories
Almost all studies in Table 13 performed classification into the four sleep stages W/L/D/REM (or just W/Sleep), likely because non-contact signals struggle to distinguish between N1 and N2 stages. In fact, even when using PSG signals, N1 and N2 stages are often confused in existing research (Supratak and Guo 2020 ).
In recent years, Wi-Fi signals have been utilized for tasks such as activity recognition, respiratory detection, and sleep monitoring. Compared to radar equipment, Wi-Fi is undoubtedly a cheaper and more embedded technology within real-life environments. As a mature technology already prevalent in households, Wi-Fi has been explored for sleep monitoring. As early as 2014, Liu et al. ( 2014 ) proposed Wi-sleep, which uses off-the-shelf Wi-Fi devices to continuously collect fine-grained channel state information (CSI) during sleep. Wi-sleep extracts rhythm patterns associated with breathing and sudden changes due to body movements from the CSI data. Their tests showed that Wi-sleep can track human breathing and posture changes during sleep. In recent years, researchers have begun exploring the use of Wi-Fi signals to identify sleep stages. Although related studies are few (as shown in Table 13 ), we believe this technology holds great potential because it is inexpensive, requires no specialized equipment, and is entirely unobtrusive. Table 13 includes three Wi-Fi related studies: Yu et al. (2021b), Liu et al. ( 2022a ), and Maheshwari and Tiwari ( 2019 ).
6.2.1 Datasets
All studies used private datasets, making direct performance comparison challenging.
6.2.2 Signal types
The authors of the three studies chose to use amplitude and phase information of fine-grained CSI for subsequent operations. Another channel information type in Wi-Fi sensing is received signal strength (RSS), which provides coarse-grained channel data and can be used for indoor localization, object tracking, and monitoring heart and respiratory rates (Liu et al. 2022a ). However, RSS is more susceptible to obstruction and electromagnetic environment changes, which might explain the common choice of CSI.
6.2.3 Preprocessing
Due to the influence of surrounding environments and hardware noise, raw CSI data is often very noisy (Maheshwari and Tiwari 2019 ). Furthermore, Wi-Fi devices receive signals from different subcarriers with various spatial paths, each interacting differently with human body parts (Yu et al. 2021b). This introduces high-dimensional data issues in Wi-Fi sensing. To improve the signal-to-noise ratio and extract the main information from each path (dimensionality reduction), Maheshwari and Tiwari ( 2019 ) used principal component analysis (PCA), while Yu et al. (2021b) combined maximum ratio combining (MRC) with PCA to integrate signals from all subcarriers.
6.2.4 Data representation and deep learning models
Liu et al. ( 2022a ) designed a CNN-based model for W/L/D/REM sleep stage classification, using one-dimensional amplitude and phase signals as input, achieving 95.925% accuracy on private data. Maheshwari and Tiwari ( 2019 ) and Yu et al. (2021b) used manually extracted features related to respiratory rate and movement. Maheshwari and Tiwari ( 2019 ) implemented a simple Bi-LSTM model for sleep motion classification to compute sleep stage information, while Yu et al. (2021b) used a hybrid CNN and Bi-LSTM model, incorporating conditional random fields for transition constraints between sleep stages, achieving 81.8% classification accuracy, close to results obtained with PSG signals.
6.3 Sound (Microphones)
During sleep, although the human body is unconscious, different physiological events spontaneously generate different audio signals, such as snoring and respiratory obstructions. Indeed, recent studies have explored detecting snoring (Xie et al. 2021a ) and sleep apnea (Wang et al. 2022 ) events using sleep sound signals recorded by microphones. Nighttime sounds are easy to obtain and have a mapping relationship with sleep stages. For example, respiratory frequency decreases and becomes more regular during NREM stages, while it increases and varies more during REM stages. Additionally, unconscious body movements during the night produce friction sounds with bedding, capturing movement characteristics that can further supplement sleep stage classification (Dafna et al. 2018 ). Despite the rich sleep-related information contained in sound signals, they also include a significant amount of redundant information (Zhang et al. 2017 ). Therefore, extracting these features and mapping them to sleep stages has become a focus of research in recent years, with deep learning methods gaining significant attention. Table 13 lists five studies included in this review.
6.3.1 Microphone equipment
Various types of microphones appeared in the reviewed papers. Early studies, such as Zhang et al. ( 2017 ) and Dafna et al. ( 2018 ), used a recording pen microphone and a professional microphone, respectively. More recent studies by Hong et al. ( 2022 ) and Tran et al. ( 2023 ) used more common and cost-effective smartphone microphones, exploring how existing devices can facilitate sleep research outside laboratory or hospital settings. Han et al. ( 2024 ) used in-ear microphones embedded in sleep earplugs.
6.3.2 Datasets
All studies used private datasets, but Hong et al. ( 2022 ) and Tran et al. ( 2023 ), considering the limited data volume, also utilized a large public dataset, PSG Audio (Korompili et al. 2021 ), which contains synchronized recordings of PSG signals and audio.
6.3.3 Preprocessing
Sound signals usually have high sampling frequencies, so downsampling was applied in Han et al. ( 2024 ), Zhang et al. ( 2017 ), and Hong et al. ( 2022 ). Sound signals in real environments are typically noisy, including background noise and noise from recording devices. To suppress noise and outliers, Hampel filtering (Han et al. 2024 ), adaptive noise reduction (Tran et al 2023 ), and Wiener filter-based adaptive noise reduction (Dafna et al. 2018 ) were applied. Additionally, Tran and Hong achieved data augmentation through pitch shifting.
6.3.4 Data representation and deep learning models
Recognizing sleep stages through sound still involves capturing cardiopulmonary activities and body movement information. Therefore, Dafna et al. ( 2018 ) extracted 67 features in five groups, including respiratory and body movement features, and used an artificial neural network (ANN) to classify W/NREM/REM and W/Sleep with accuracies of 86.9% and 91.7%, respectively. Han et al. ( 2024 ) extracted body activity features, snoring and sleep talking features, and physiological features such as heart and respiratory rates, using a CNN-RNN hybrid model with attention mechanisms for W/L/D/REM classification, achieving an MF1 score of 69.51. In other studies, Zhang et al. ( 2017 ), Hong et al. ( 2022 ), and Tran et al. ( 2023 ) used spectral representations of audio signals. Zhang et al. extracted STFT spectra and Mel-frequency cepstral coefficients (MFCC), using CNNs for classification. Hong and Tran used the Mel spectrogram, the most common audio analysis tool, and implemented hybrid models of CNNs, RNNs, and multi-head attention for classification, achieving accuracies of 70.3% and 59.4%, respectively, in four-class classification. Although the performance was lower, their research showed that combining deep learning with smartphones could achieve sleep stage classification in uncontrolled home environments. Among the five studies, hybrid models were predominant.
In summary, we have reviewed studies on automatic sleep stage classification using non-contact signals and deep learning, focusing on radar, Wi-Fi, and microphone audio signals. This field also includes other forms of research, such as sleep stage monitoring through near-infrared cameras (Carter et al. 2023 ) or home surveillance cameras (Choe et al. 2019 ). We have organized relevant information in Table 13 for readers to explore further.
7 Discussions and challenges
7.1 discussions.
This section discusses and summarizes research on deep learning for sleep stage classification, focusing on three main aspects, available signals, data representations, and deep learning models, as well as their performance.
7.1.1 Signals (sleep physiological data)
Deep learning is a data-driven approach that relies on large amounts of data and uses deep neural networks to address real-world problems. The first crucial step in solving sleep stage classification problems is collecting signals (data) containing information about sleep physiological activities. However, in current research, this step is often overlooked due to the availability of public datasets. Researchers typically improve models or algorithms using existing data. The existing data includes not only traditional PSG signals but also “new signals” such as cardiac and non-contact signals.
Among the signal types reviewed in this paper, PSG, as the “gold standard,” dominates in terms of both the number of related studies and performance, as shown in Tables 9 , 10 , 11 , 12 , and 13 . In PSG systems, single-channel EEG is currently the most popular modality. On the one hand, single-channel EEG alone can achieve good performance(Supratak et al. 2017 ; Phyo et al. 2022 ); on the other hand, it simplifies signal acquisition. However, there are still issues, as EEG is collected through electrodes distributed at different positions on the head, resulting in variations in the information and quality of signals obtained from different electrodes, which can lead to different model performance. Supratak et al. ( 2017 ) tested two EEG channels, Fpz-Cz (frontal) and Pz-Oz (occipital), from the Sleep-EDF-2013 dataset, achieving an overall accuracy of 82.0% on the Fpz-Cz channel, but only 79.8% on the Pz-Oz channel. Additionally, model performance can be improved by increasing the number of EEG channels or by supplementing with EOG and EMG signals (Cui et al. 2018 ; Jia et al. 2021 ; Olesen et al. 2021 ). However, when using EEG, EOG, or EMG simultaneously to form multimodal inputs, extra attention must be paid to the differences and fusion between modalities. To fairly compare these scenarios, we refer to the study by Zhu et al. ( 2023 ), in which the authors compared single-channel EEG (Fpz-Cz channel), EEG+EOG, and EEG+EOG+EMG on the Sleep-EDF-2018 dataset. Table 14 shows their results, with model performance increasing as the number of channels increases, especially with a significant improvement brought by the addition of EOG. In the study by Fan et al. ( 2021 ), the authors performed sleep stage classification using only a single-channel EOG, but the accuracy was only around 76%.
Cardiac and non-contact signals essentially fall into the same category of data, as the information contained in non-contact signals also pertains to cardiac activity. The main advantage of these signals over PSG lies in the comfortable and convenient signal acquisition methods. For example, PPG signals can be collected using a simple photoplethysmographic sensor integrated into smartwatches, while non-contact signals like Wi-Fi are ubiquitous in daily life. Although the association between cardiac signals and sleep conditions has long been recognized, these signals have only recently been utilized for sleep stage classification, thanks to advancements in deep learning techniques. Compared to EEG, research on these types of signals is still limited. In these studies, derived time series such as HR (Sridhar et al. 2020 ), HRV (Fonseca et al. 2020 ), and EDR (Li et al. 2018 ) extracted from raw signals are commonly used for classification. Therefore, they involve an additional step of “derived time series extraction” compared to PSG. Moreover, studies based on cardiac or non-contact signals mostly perform well in the W/L/D/REM four-stage classification but struggle with the more detailed AASM 5-stage classification. This limitation may stem from the inherent characteristics of these signals, which contain less sleep information compared to EEG.
7.1.2 Data representation
Automated sleep stage classification essentially involves extracting sleep physiological information from physiological signals using deep learning tools. Physiological signals, whether PSG or cardiac signals, can be represented in various forms, including raw one-dimensional signals, spectrogram images, derived time series, or combinations thereof. For PSG systems, inputting raw signal values directly into deep learning models is a popular choice, as demonstrated in Fig. 5 . This straightforward approach has proven to be effective, driving the widespread adoption of this method. Additionally, spectral representations obtained through signal analysis methods such as STFT, CWT, and HHT are commonly used. Some researchers have noticed the benefits of combining these two representations. In cardiac and non-contact signals, most studies use derived signals extracted from raw data as inputs. Some also explore using raw signals or transformed domain data (Korkalainen et al. 2020 ; Olsen et al. 2022 ). PSG-related research is more abundant, so using raw signals is a popular choice. However, for cardiac and non-contact signals, due to the limited number of studies and lack of uniform methods, it is challenging to determine the most popular data representation method. It is essential to note that in terms of data representation, raw signals seem to be more straightforward and avoid information loss during the transformation process.
It is actually difficult to draw this conclusion. Because different studies vary greatly in terms of data preprocessing, data volume, validation methods, and network structures, simply comparing the reported performance in various studies is insufficient to support this conclusion. We believe this could be a direction for future work, that is, conducting a fair and comprehensive comparison including models, signal types, input forms, etc. Additionally, in Phan and Mikkelsen ( 2022 ) and Phan et al. ( 2021 ), the authors argue that different input forms should not be compared and should be seen as different mappings of underlying data distributions.
7.1.3 Deep learning models
In the studies we reviewed, about 35% adopted a fully CNN-based deep learning structure, while approximately 41% proposed combining CNN with other deep learning models, such as recurrent (RNN, LSTM, etc.), Transformer, and generative (GAN) models. Research involving CNN accounts for around 76% of the total studies. The widespread use of CNN can be justified by the following points. Firstly, the CNN structure can extract deep discriminative features and spatial patterns from sleep signals, thus CNN is used for direct classification or as a feature extractor. Secondly, CNN resources are abundant and have achieved success in many fields, such as image and video processing, with numerous accessible CNN-related resources (open code). Therefore, researchers have more opportunities to learn and use CNN, and can even transfer CNNs from other fields to the current subject, as Seo et al. ( 2020 ) used the well-known ResNet (He et al. 2016 ) from the image domain. Thirdly, various representations of sleep physiological signals, including raw one-dimensional signals, two-dimensional spectral representations obtained from various transformations, and extracted feature sequences, can all be accepted and processed by different forms of CNNs. Some studies have demonstrated that CNNs outperform other deep learning methods. In Stuburić et al. ( 2020 ), the authors tested the performance of CNN and LSTM networks on a combination of heartbeat, respiration, and motion signals (one-dimensional time series data). The CNN consisted of three convolutional layers and two fully connected layers, while the LSTM had only one LSTM hidden layer and two fully connected layers. The authors conducted five-class (W/N1/N2/N3/REM) and four-class (W/L/D/REM) tests, with the CNN and LSTM achieving overall accuracies of 40% and 32% in the five-class test, and 55% and 51% in the four-class test, respectively, proving that CNN outperformed LSTM. Despite the simplicity of the LSTM used, the authors claimed that its computational cost was still much higher than that of the CNN, a significant drawback of RNN-type models. Parekh et al. ( 2021 ) tested various well-known visual CNN models on the Sleep-EDF-2018 dataset, including AlexNet (Krizhevsky et al. 2012 ), VGG (Simonyan and Zisserman 2014 ), ResNet, DenseNet, SqueezeNet (Iandola et al. 2016 ), and MobileNet (Howard et al. 2017 ), with input being grayscale images visualizing single-channel EEG waveforms. All models were pre-trained on the large image vision dataset ImageNet (Deng et al. 2009 ). The experimental results showed that almost every model achieved around 95% accuracy, which is impressive. Another study (Phan et al. 2022a ) compared the CRNN hybrid model (DeepSleepNet (Supratak et al. 2017 )), pure RNN model (SeqSleepNet (Phan et al. 2019 )), pure Transformer model (SleepTransformer (Phan et al. 2022b )), FCNN-RNN model (fully convolutional neural network hybrid RNN), and a time-frequency combined input model (XSleepNet (Phan et al. 2021 ), essentially a combination of FCNN-RNN and SeqSleepNet, receiving raw input and time-frequency transformed input to leverage their complementarity). The experiments were conducted on a pediatric sleep dataset, and the results showed that the time-frequency combined input model performed best (ACC = 88.9), while the pure Transformer model performed the worst (ACC = 86.9), possibly due to the limited data. In another study, Yeckle and Manian ( 2023 ) compared the performance of LeNet (LeCun et al. 1998 ), ResNet, MLP, LSTM, and CNN-LSTM hybrid models under the same conditions using single-channel one-dimensional EEG signals on the Sleep-EDF dataset. The results showed that LeNet performed best, achieving an accuracy of 85%. This may be due to the small amount of data used, only 20 subjects’ data. Overall, there is a relative lack of sufficient comparison of CNN structures in existing studies, such as the impact of different numbers of convolutional layers and fully connected layers, different activation functions, and different pooling methods on sleep staging.
A very small portion of studies entirely use RNN (including LSTM and GRU variants), which is much fewer than expected, given that RNN has always shown good performance in learning time series features. One explanation for this phenomenon is that RNN-type models consume a lot of training time and memory, especially for longer sequences. Although there is no fully relevant comparison in existing studies, Eldele et al. ( 2021 ) recorded the training times of their proposed model AttnSleep and DeepSleepNet (Supratak et al. 2017 ). Both models use similar multi-scale convolution to extract single-channel EEG features, but the former uses multi-head attention to model temporal dependencies, while the latter uses two-layer bidirectional LSTM. On Sleep-EDF-2018, AttnSleep required only 1.7 h of training time, while DeepSleepNet required 7.2 h, nearly four times the difference. Furthermore, although the number is small, almost all RNN-based studies included in this paper used LSTM, except for one study that used GRU (Guillot et al. 2020 ). There is currently a lack of sufficient comparison between the two, and it is recommended to test both to determine which method performs better. Another reason for the rare occurrence of RNN is the emergence of more promising alternatives, namely Transformer models based on multi-head attention. In recent studies, such as Maiti et al. ( 2023 ), Zhu et al. ( 2023 ), Yubo et al. ( 2022 ), Pradeepkumar et al. ( 2022 ), Eldele et al. ( 2021 ), Phan et al. ( 2022b ), and Dai et al. ( 2023 ), it has appeared frequently. Siddhad et al. ( 2024 ) compared the effectiveness of Transformer, CNN, and LSTM in EEG classification. Their test results on a private age and gender EEG dataset showed that Transformer outperformed the other two methods. In the binary classification problem of gender, the Transformer achieved 94.53% accuracy, while the other two only had around 86%; in the six-class age task, the Transformer still had 87.79% accuracy, while the other two had only around 67%. However, in the current subject (ASSC), there is a lack of fair comparison between Transformer and RNN-type models.
Hybrid models, which are the most numerous type, usually combine CNN with other model structures. These models have shown strong spatial feature extraction and temporal feature modeling capabilities in many studies, with the former typically achieved by CNN and the latter by RNN or Transformer, especially the CNN-Transformer hybrid, which is gradually becoming a trend. Additionally, a small number of studies in the review used representative generative models: GANs, aimed at alleviating issues of insufficient training data or imbalanced sample classes through GANs.
In recent years, deep learning-based automatic sleep stage classification has achieved significant progress. The overall accuracy on PSG signals typically exceeds 80%. Although it’s difficult to deem this result entirely satisfactory, it seems that deep learning methods have reached a plateau in terms of performance. Given this situation, it may be challenging to achieve better performance simply by designing new model architectures. Instead, we should focus on the practical application of these models. In real-world applications, sleep data originate from various institutions, devices, demographic characteristics, and collection conditions, leading to substantial differences in data distribution. For instance, individuals of different ages, genders, races, or those with different medical conditions exhibit variations in sleep structure. Moreover, different PSG equipment and acquisition settings may result in differences in resolution, channel numbers, and signal-to-noise ratios, further increasing data heterogeneity. This diversity makes it difficult for existing DL models to generalize their performance beyond their local testing environments.
A promising solution emerging in current research to address this issue is transfer learning and domain adaptation. The core idea is to transfer pre-trained models (usually trained on large source domains) across different data domains, enabling them to adapt to the target data domain. This includes supervised, semi-supervised, and unsupervised methods. Supervised domain adaptation typically involves fine-tuning pre-trained models using annotated samples available from the target domain (Phan et al. 2020 ). The target domain could be a small clinical sleep dataset or the sleep records of an individual. A representative work in this area is the study by Phan et al. ( 2020 ), who extensively explored the transferability of features learned by sleep stage classification networks. However, it is evident that supervised domain adaptation requires a sufficient number of labeled samples from the target domain to be effective, which is not always feasible. Therefore, semi-supervised or unsupervised domain adaptation is employed. Banluesombatkul et al. ( 2020 ) proposed a transfer learning framework based on model-agnostic meta-learning (MAML), a typical semi-supervised framework that can quickly adjust models to new domains with only a few labeled samples. In the works of Yoo et al. ( 2021 ), Zhao et al. ( 2021b ), and Nasiri and Clifford ( 2020 ), adversarial training-based frameworks for unsupervised domain adaptation are introduced. These methods achieve domain adaptation by matching the feature distributions of the source and target domains through domain classifiers and specifically designed models. While these studies seem promising, they still rely on specially designed networks rather than a universal framework.
7.1.4 The computational cost of models
In current research, three basic models-CNN, RNN, and Transformer-are widely used, but their computational costs and performance vary, necessitating reasonable selection based on requirements. Stuburić et al. ( 2020 ) tested a three-layer CNN model and a one-layer LSTM model on one-dimensional cardiopulmonary data, achieving 40% and 32% accuracy in the W/N1/N2/N3/REM five-class classification and 55% and 51% in the W/L/D/REM four-class classification, respectively, with CNN outperforming LSTM. Despite the simplicity of the LSTM used, the authors claimed its computational cost was much higher than that of the CNN. Eldele et al. ( 2021 ) proposed the CNN-Transformer hybrid model AttnSleep with 0.51 M parameters; Supratak et al. ( 2017 ) proposed the CNN-LSTM model DeepSleepNet with 21 M parameters. These are two very similar models with significant differences only in the LSTM and Transformer parts. In addition to the parameter count, on Sleep-EDF-2018, AttnSleep required only 1.7 h of training, whereas DeepSleepNet needed 7.2 h-nearly a fourfold difference-with AttnSleep achieving better performance. These studies in fact demonstrate that RNN models neither provide a performance advantage nor are resource-efficient.
Liu et al. ( 2023a ) proposed the CNN model MicroSleepNet, which can run on smartphones. MicroSleepNet has only 48.2 K parameters but outperforms the 21 M parameter DeepSleepNet (82.8% vs. 82.0%). Compared to the SleepTransformer model built entirely on the Transformer architecture, there is a performance gap (79.5% vs. 81.4%), but the high parameter count of 3.7 M limits SleepTransformer’s deployment and real-time inference on mobile devices (Phan et al. 2022b ). Pradeepkumar et al. ( 2022 ) and Yao and Liu ( 2023 ) optimized the CNN-Transformer hybrid models for light weight, but their parameter counts still reached 320 K and 300 K, with performance lower than MicroSleepNet (79.3% accuracy for Pardeepkumar et al. and 77.5% for Yao et al.). Zhou et al. ( 2023 ) also proposed a fully CNN-based lightweight model with only about 42 K parameters, outperforming other models that include LSTM or multi-head attention mechanisms.
The above indicates that RNNs result in higher parameter counts and computational resource consumption without performance advantages. Without considering computational cost and pursuing high performance, CNN and Transformer models are suitable. In scenarios requiring low parameter and computational cost, introducing Transformer structures does not significantly improve CNN model performance, and simple CNN structures can achieve competitive results in sleep stage classification tasks.
7.1.5 Other learning methods (self-supervised, semi-supervised learning)
Above, we provided a detailed discussion of sleep stage classification based on deep learning methods. Our investigation covered popular practices in signal processing, data representation, and modeling in the context of sleep stage classification. Additionally, during the survey, we identified some relatively niche research methods, mainly focusing on self-supervised or semi-supervised learning approaches. Unsupervised, self-supervised and semi-supervised learning is considered in contrast to supervised learning methods. In supervised learning, which is a primary paradigm in machine learning, the algorithm or model’s objective is to learn the mapping relationship between inputs and outputs from labeled training data (Qi and Luo 2020 ). However, in the real world, a significant portion of data is unlabeled, especially in medical fields. This limitation results in the waste of a large amount of unlabeled data, and emerging unsupervised, self-supervised, or semi-supervised methods aim to address this issue. Unsupervised learning, as the name suggests, involves training algorithms or models entirely on unlabeled data, allowing the model to autonomously learn the intrinsic structure of the data. Self-supervised learning is a subset of unsupervised learning, where the model learns from data by creating a “pretext task” to generate useful representations for subsequent tasks. Self-supervised learning doesn’t require data labels; instead, it uses some form of information inherent in the data as “pseudo-labels” for training. Pretext tasks need to be designed based on the application; for example, in computer vision, a pretext task might involve predicting the color of a certain part of an image (Misra and Maaten 2020 ). If the network can successfully accomplish this task, it indicates that it has learned general features in the data (Yun et al. 2022 ). Self-supervised learning is often categorized into three types: generative-based methods, contrastive-based methods, and adversarial-based methods (Zhang et al. 2023 ). Among these, contrastive-based methods, commonly known as contrastive learning, is most frequently used in sleep stage classification and is one of the widely adopted strategies in self-supervised learning. Contrastive learning aims to learn data representations by contrasting positive and negative samples (Chen et al. 2020 ). Most methods use two data augmentation techniques to generate different views of input samples x and y , denoted as \(\textit{x}_1\) , \(\textit{y}_1\) , and \(\textit{x}_2\) , \(\textit{y}_2\) . The model’s learning objective is to maximize the similarity between views from the same sample ( \(\textit{x}_1\) - \(\textit{x}_2\) , \(\textit{y}_1\) - \(\textit{y}_2\) ) and minimize the similarity between views from different samples ( \(\textit{x}_1\) - \(\textit{y}_2\) , \(\textit{x}_2\) - \(\textit{y}_1\) ) (Jaiswal et al. 2020 ). Through this contrastive training, the model’s representation learning capability is enhanced, making it better suited for subsequent tasks. Semi-supervised learning is a “middle-ground” approach that utilizes both labeled and unlabeled data, bridging the gap between supervised and unsupervised (self-supervised) learning (Chen et al. 2020 ). Semi-supervised learning typically handles unlabeled data with unsupervised or self-supervised methods, while labeled data is learned using traditional supervised methods (Li et al. 2022c ). Although novel learning methods such as semi-supervised, self-supervised, and even unsupervised methods are widely used in fields like computer vision, their entry into the field of sleep analysis has been much slower, with relatively limited research at present.
Jiang et al. ( 2021 ) designed a contrastive learning-based backbone network for EEG, employing seven data transformation methods, including adding Gaussian noise and flipping, with the pretext task of matching transformation pairs from the same sample. Through contrastive training, a robust EEG feature extractor was obtained, and a classifier head (a fully connected output layer) was added to the backbone for subsequent sleep stage classification tasks (backbone network parameter freezing). Li et al. ( 2022c ) designed a semi-supervised learning model for pediatric sleep staging. For unlabeled data, they used contrastive learning based on data augmentation for self-supervised learning, with the pretext task of predicting the data augmentation method used. For labeled data, the authors employed a supervised contrastive learning strategy (Khosla et al. 2020 ), incorporating label information into contrastive learning. This supervised contrastive learning strategy proposed by Khosla et al. ( 2020 ) aligns well with sleep stage classification problems, as evidenced in Lee et al. ( 2024 ) and Huang et al. ( 2023 ). In fact, existing labeled sleep data is relatively abundant Lee et al. ( 2024 ), and mining information from sleep data itself is challenging, making the performance of self-supervised algorithms less satisfactory. In such circumstances, not utilizing existing data is likely a waste. In Table 15 , we extracted and summarized the information from these studies based on self-supervised or semi-supervised learning. The table extracts information from recent papers on self-supervised, semi-supervised, or supervised learning based on methods such as contrastive learning. In these papers, there are primarily two methods for creating pairs of samples required for contrastive learning. The first method involves creating sample pairs based on data augmentation, while the second method utilizes contrastive predictive coding (CPC) (Oord et al. 2018 ). Data augmentation methods are as described above, and the core of CPC lies in the model predicting future samples based on existing samples for learning (Brüsch et al. 2023 ). Additionally, we have compiled pretext tasks set in various studies and provided brief descriptions of their main content.
7.2 Challenges
In existing research, most work can be summarized as following the pattern of “proposing a model-applying public data-performance improvement.” This approach is undoubtedly meaningful; however, as of now, the problem of automatic sleep stage classification seems to be largely addressed (Phan and Mikkelsen 2022 ). Although various new models continue to push the performance metrics, it is challenging to ascertain whether such improvements have practical significance. Researchers, besides focusing on designing or building new models to achieve performance enhancement, should also address other challenges and explore innovative opportunities. In our investigation, we have identified three main areas for potential improvement: sleep data, deep learning models, and future scalable research.
The use of large and diverse datasets is lacking in relevant research: Existing studies often focus on several commonly used datasets, such as Sleep-EDF-2018, SHHS, and MASS. However, in fact, large database websites such as PhysioNet and NSRR have already openly released many large datasets. These data can be accessed with simple applications (some without), covering various populations including the elderly, children, and individuals with cardiovascular and pulmonary diseases. However, existing research tends to concentrate on classic benchmark datasets like Sleep-EDF-2018. Although this facilitates the comparison of algorithm or model performance, it fails to validate the generalization of models on heterogeneous data. We believe this is worth exploring; an excellent deep learning model should not only perform well on Sleep-EDF but should also be applicable to other datasets with minimal adjustments. This can be achieved through methods such as unsupervised domain adaptation (Yoo et al. 2021 ) and knowledge distillation (Duan et al. 2023 ), which can compensate for differences in data distributions.
Class imbalance in sleep data: Sleep data suffer from severe class imbalance issues, particularly in the N1 stage. Existing research often struggles with accurately identifying the N1 stage. For instance, in Jia et al. ( 2021 ), the F1-score for the N1 stage was only 56.2 (using the SEDF dataset), while other stages scored 87.2 or higher. Class imbalance is an inherent issue in sleep data. In fact, this paper statistically analyzes research focusing on class imbalance, with major mitigation methods including oversampling, morphological transformation, GAN synthetic samples, and adjusting loss functions and ensemble learning. We believe GAN synthetic samples are a more promising direction. This is because physiological signals are highly sensitive, and slight variations can lead to different medical interpretations. Oversampling or morphological transformation may have difficulty controlling whether the generated new samples are reasonable. However, GANs, through adversarial training between the generator and discriminator, have the potential to guide the generator to produce distributions extremely similar to real samples. Meanwhile, issues exist with loss functions and ensemble learning as well. The former typically brings about hyperparameter selection problems, while the latter entails high training costs.
The impact of noise and denoising processes on sleep stage classification systems: Noise is a pervasive issue during the acquisition of signals, whether they come from EEG, radar, Wi-Fi, or other sources, potentially leading to inaccuracies in sleep stage classification. In our review, we found that many studies incorporate denoising steps during data preprocessing. However, as far as we know, only Zhu et al. ( 2022 ) have investigated the impact of removing internal artifacts (noise) from EEG on deep learning-based sleep stage classification systems. They developed a novel method for removing internal EOG or EMG artifacts from sleep EEG and fed the denoised and original signals into deep neural networks in both time-domain one-dimensional and transformed domain (STFT) forms for classification. Their comparative results showed that when using the original time-domain signals, the presence of artifacts improved the accuracy of the W stage but reduced the accuracy for N1, N2, N3, and REM stages. Conversely, when using the signals in their time-frequency domain form, the artifacts had minimal impact. They concluded that appropriate artifact removal from EEG signals is advisable. Similar studies on other types of signals are currently lacking, and future research could benefit from exploring these findings to apply them to other signals.
Potential issues with noisy labels: The development of ASSC aims to bypass the labor-intensive process of manually annotating sleep data. However, until this field reaches full maturity, researchers must rely on expert manual annotations to train deep learning models. This reliance introduces a potential problem: the accuracy of these dataset annotations is uncertain. The uncertainty stems from various factors, including the quality of the data and the expertise of the annotators. For example, in the open-source dataset ISRUC released by Khalighi et al. ( 2016 ), the annotation labels were provided by two experts. They reported a Cohen’s Kappa coefficient of 0.9 in the healthy population (subgroup-3) and a lower value of 0.82 in the sleep disorder population (subgroup-2). This indicates that even expert annotations can result in misclassifications or disagreements, raising concerns about label reliability, commonly referred to as the issue of noisy labels. When labels are noisy, deep neural networks with a large number of parameters can overfit these erroneous labels. Zhang et al. ( 2021 ) conducted experiments on datasets with noisy labels and demonstrated that deep neural networks could easily fit training data with any proportion of corrupted labels, leading to poor generalization on test data. Moreover, they showed that popular regularization methods do not mitigate the impact of noisy labels, making them more detrimental than other types of noise, such as input data noise. Several methods have been proposed to train models robust to noisy labels, but most focus on image classification (Karimi et al. 2020 ). Unlike images, time series data, such as EEG and ECG, present additional challenges. These data types are harder to interpret and may have more ambiguities. Our survey found relatively little focus on addressing noisy labels in the context of ASSC. Fiorillo et al. ( 2023b ) analyzed discrepancies among multiple annotators’ labels within the SSC. They used annotations from multiple annotators to train two lightweight models on three multi-annotator datasets. During training, they incorporated label smoothing and a soft-consensus distribution to calibrate the classification framework. Their approach, where models learn to align with the consensus among multiple annotators, suggests robustness to label noise even with annotator disagreements. In other domains, such as emotion recognition, Li et al. ( 2022a ) addressed the issue of noisy labels in EEG data. They employed capsule networks combined with a joint optimization strategy for classification in the presence of noisy labels. Similarly, in the ECG domain, Liu et al. ( 2021 ) used a CNN model with a specially designed data cleaning method and a new loss function to effectively suppress the impact of noisy labels on arrhythmia classification. Furthermore, Song et al. ( 2022 ) provided a comprehensive review of methods for handling noisy labels in deep learning, including non-deep learning, machine learning, and deep learning approaches. Although their survey focuses on the image domain, these methods could potentially be adapted for ASSC. For instance, in the work of Vázquez et al. ( 2022 ), a state-of-the-art self-learning label correction method (Han et al. 2019 ) used for image classification was adapted for ECG tasks.
Impact of diseases on sleep stage classification: Current research typically uses sleep data from healthy individuals to validate algorithm performance. However, sleep disorders (Boostani et al. 2017 ; Malafeev et al. 2018b) and other neurological diseases (Patanaik et al. 2018b; Stephansen et al. 2018 ) can alter sleep structures, making accurate sleep stage identification in such patients highly challenging. Timplalexis et al. ( 2019 ) examined the differences in sleep stage classification using machine learning methods across healthy individuals, untreated sleep disorder patients, and medicated sleep disorder patients: EEG patterns in healthy individuals are easier to distinguish, while sleep disorders and medication seem to distort EEG, reducing classification accuracy by approximately 3%. They attempted to apply algorithms trained on healthy data to sleep disorder patients, resulting in a significant drop in accuracy. Korkalainen et al. ( 2019 ) observed that with increasing severity of obstructive sleep apnea, the classification accuracy based on single-channel EEG decreased. This confirms that diseases can cause potential changes in sleep structures and signal patterns. However, current studies lack exploration of the underlying reasons: How exactly do diseases like obstructive sleep apnea affect sleep stages? Can models circumvent or correct these impacts? Additionally, other studies have explored predicting the occurrence of sleep disorders [e.g., sleep apnea (Wang et al. 2023 )] using deep learning during sleep. Cheng et al. ( 2023b ) developed a multitask model capable of predicting both sleep stages and sleep disorders, but there was no interaction between the two tasks. We believe that increasing interaction and feedback between multitask branches might help the model more accurately identify sleep stages in diseased populations.
Deep learning models
Interpretability of deep learning models: One of the primary obstacles to the clinical application of automatic sleep staging based on deep learning is the black box suspicion. Deep learning algorithms are often perceived as “black boxes,” making it challenging to understand why they make specific decisions. To address this issue, using models with stronger interpretability is a good approach, such as the self-attention mechanism model Transformer. In the study proposed by Phan et al. ( 2022b ), the SleepTransformer achieved high interpretability. They input a sequence of continuous sleep epochs into the model, first extracting sleep-related features within each epoch. Subsequently, they visualize attention scores between epochs, representing the influence of different adjacent epochs (i.e., context) in the input sequence on the identification of the target epoch. This approach closely mimics the process of manual classification by human experts. Additionally, feature visualization techniques, such as t-distributed Stochastic Neighbor Embedding (t-SNE) (Van der Maaten and Hinton 2008 ) and Gradient-weighted Class Activation Mapping (Grad-CAM) (Selvaraju et al. 2017 ), are also options to enhance model interpretability by observing the model features.
Performance challenges of non-invasive, non-contact methods: Non-invasive and non-contact methods have the advantage of being comfortable and non-intrusive, unlike PSG systems, which are invasive and uncomfortable. However, their association with sleep stages is relatively weak, making it challenging to explore and resulting in poor performance in existing research. Additionally, signals like Wi-Fi or radar face challenges in multi-person environments. We envision that this can be addressed by designing more effective models or algorithms and extracting more efficient and richer features.
Future scalable research
Acceptability of results from new methods to experts: The acceptability of sleep stage classification results obtained through contactless signals by doctors or experts remains an open question. As a novel approach that has emerged in recent years alongside advancements in wireless communication and electronic technologies, the reliability and acceptance of these methods are not well-established. In existing research, PSG remains the universally recognized gold standard for sleep stage classification. Experts can trust PSG results and base their diagnoses on them. However, when it comes to contactless signals, such as those obtained through radar or Wi-Fi, the acceptance and reliability of these methods by medical professionals are unknown and pose significant challenges. Future efforts may involve large-scale data collection and expert surveys to address this issue.
Extending from sleep stages to other diseases: The accurate classification of sleep stages aims to assist in diagnosing and preventing other diseases, such as sleep disorders and neurological diseases. When sleep stage classification is linked to the prediction and diagnosis of specific diseases, ASSC may become more practically significant. In fact, some datasets are designed to explore the relationship between sleep and certain diseases. For example, the SHHS dataset aims to investigate the potential relationship between sleep-disordered breathing and cardiovascular diseases. Xie et al. demonstrated that using overnight polysomnography and machine learning methods to predict ischemic stroke is feasible (Xie et al. 2018 , 2021b ). They extracted sleep stages, EEG-related features, and relevant clinical feature information from the data of SHHS participants who had experienced a stroke, and successfully predicted stroke in 17 out of 20 patients using their proposed prediction model. Although this is excellent work, their predictions rely on manually annotated sleep stage information by experts. Future research might combine automatic sleep stage classification models with prediction models to create an end-to-end integrated model, achieving fully automated monitoring, and potentially expanding to other diseases. This would be a significant advancement.
8 Conclusions
This paper studies and reviews deep learning methods for automatic sleep stage classification. Unlike traditional approaches, deep learning methods can automatically learn advanced and latent complex features from sleep data, eliminating the need for additional feature extraction steps. The paper comprehensively analyzes the signals, datasets, data representation methods, preprocessing techniques, deep learning models, and performance evaluations in sleep stage classification. We provide an overview of traditional PSG studies, and our survey reveals researchers’ focus on extracting different features from PSG data using various new models or methods. Most of these studies are based on large publicly available PSG datasets, and some of them have shown promising performance. Additionally, we discuss research involving less invasive and non-contact signals, namely cardiorespiratory signals and contactless signals. Compared to PSG, cardiorespiratory and contactless signals offer the advantages of convenient and comfortable signal acquisition, although their performance currently lags behind. Our review indicates that by combining deep learning with different types of signals, ASSC can be flexibly implemented without being confined to specialized PSG equipment, which is crucial for bringing sleep stage classification out of the laboratory. We believe that future research should focus on three key areas: firstly, the accuracy of cardiorespiratory and contactless signal classification; secondly, the robustness of the models in various real-world environments (e.g., home settings); and thirdly, the generalization capability of the models when faced with new data. These are not the only research directions that need attention, but they play a significant role in the practical application of ASSC.
Data Availability
No datasets were generated or analysed during the current study.
https://physionet.org/ .
https://sleepdata.org/ .
Aboalayon KAI, Faezipour M, Almuhammadi WS et al (2016) Sleep stage classification using EEG signal analysis: a comprehensive survey and new investigation. Entropy 18(9):272. https://doi.org/10.3390/e18090272
Article Google Scholar
Adib F (2019) Seeing with radio wi-fi-like equipment can see people through walls, measure their heart rates, and gauge emotions. IEEE Spectr 56(6):34–39. https://doi.org/10.1109/MSPEC.2019.8727144
Afonso VX, Tompkins WJ, Nguyen TQ et al (1999) Ecg beat detection using filter banks. IEEE Trans Biomed Eng 46(2):192–202. https://doi.org/10.1109/10.740882
Ali PJM, Faraj RH, Koya E et al (2014) Data normalization and standardization: a technical report. Mach Learn Tech Rep 1(1):1–6
Google Scholar
Al-Saegh A, Dawwd SA, Abdul-Jabbar JM (2021) Deep learning for motor imagery EEG-based classification: a review. Biomed Signal Process Control 63:102172. https://doi.org/10.1016/j.bspc.2020.102172
Alsolai H, Qureshi S, Iqbal SMZ et al (2022) A systematic review of literature on automated sleep scoring. IEEE Access 10:79419–79443
Altaheri H, Muhammad G, Alsulaiman M et al (2023) Deep learning techniques for classification of electroencephalogram (EEG) motor imagery (MI) signals: a review. Neural Comput Appl 35(20):14681–14722. https://doi.org/10.1007/s00521-021-06352-5
Arif S, Khan MJ, Naseer N et al (2021) Vector phase analysis approach for sleep stage classification: a functional near-infrared spectroscopy-based passive brain-computer interface. Front Hum Neurosci 15:658444
Baek J, Lee C, Yu H et al (2022) Automatic sleep scoring using intrinsic mode based on interpretable deep neural networks. IEEE Access 10:36895–36906. https://doi.org/10.1109/ACCESS.2022.3163250
Baglioni C, Battagliese G, Feige B et al (2011) Insomnia as a predictor of depression: a meta-analytic evaluation of longitudinal epidemiological studies. J Affect Disord 135(1–3):10–19. https://doi.org/10.1016/j.jad.2011.01.011
Baillet S, Friston K, Oostenveld R (2011) Academic software applications for electromagnetic brain mapping using MEG and EEG. Comput Intell Neurosci 2011:12–12. https://doi.org/10.1155/2011/972050
Banluesombatkul N, Ouppaphan P, Leelaarporn P et al (2020) Metasleeplearner: a pilot study on fast adaptation of bio-signals-based sleep stage classifier to new individual subject using meta-learning. IEEE J Biomed Health Inform 25(6):1949–1963
Banville H, Chehab O, Hyvärinen A et al (2021) Uncovering the structure of clinical EEG signals with self-supervised learning. J Neural Eng 18(4):046020. https://doi.org/10.1088/1741-2552/abca18
Biswal S, Sun H, Goparaju B et al (2018) Expert-level sleep scoring with deep neural networks. J Am Med Inform Assoc 25(12):1643–1650. https://doi.org/10.1093/jamia/ocy131
Biswal S, Kulas J, Sun H, et al (2017) Sleepnet: automated sleep staging system via deep learning. arXiv preprint arXiv:1707.08262 https://doi.org/10.48550/arXiv.1707.08262
Bonnet M, Arand D (1997) Heart rate variability: sleep stage, time of night, and arousal influences. Electroencephalogr Clin Neurophysiol 102(5):390–396. https://doi.org/10.1016/S0921-884X(96)96070-1
Boostani R, Karimzadeh F, Nami M (2017) A comparative review on sleep stage classification methods in patients and healthy individuals. Comput Methods Programs Biomed 140:77–91
Brüsch T, Schmidt MN, Alstrøm TS (2023) Multi-view self-supervised learning for multivariate variable-channel time series. In: 2023 IEEE 33rd International Workshop on Machine Learning for Signal Processing (MLSP), IEEE, pp 1–6, https://doi.org/10.1109/MLSP55844.2023.10285993
Cai X, Jia Z, Jiao Z (2021) Two-stream squeeze-and-excitation network for multi-modal sleep staging. In: 2021 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), IEEE, pp 1262–1265, https://doi.org/10.1109/BIBM52615.2021.9669375
Carter J, Jorge J, Venugopal B, et al (2023) Deep learning-enabled sleep staging from vital signs and activity measured using a near-infrared video camera. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp 5940–5949
Casal R, Di Persia LE, Schlotthauer G (2021) Classifying sleep-wake stages through recurrent neural networks using pulse oximetry signals. Biomed Signal Process Control 63:102195. https://doi.org/10.1016/j.bspc.2020.102195
Chawla NV, Bowyer KW, Hall LO et al (2002) Smote: synthetic minority over-sampling technique. J Artif Intell Res 16:321–357. https://doi.org/10.1613/jair.953
Chen X, Wang R, Zee P et al (2015) Racial/ethnic differences in sleep disturbances: the multi-ethnic study of atherosclerosis (mesa). Sleep 38(6):877–888. https://doi.org/10.5665/sleep.4732
Cheng YH, Lech M, Wilkinson RH (2023) Simultaneous sleep stage and sleep disorder detection from multimodal sensors using deep learning. Sensors 23(7):3468
Cheng X, Huang K, Zou Y, et al (2023a) Sleepegan: A gan-enhanced ensemble deep learning model for imbalanced classification of sleep stages. arXiv preprint arXiv:2307.05362 https://doi.org/10.48550/arXiv.2307.05362
Chen T, Kornblith S, Norouzi M, et al (2020) A simple framework for contrastive learning of visual representations. In: International conference on machine learning, PMLR, pp 1597–1607
Chen Z, Zheng T, Cai C, et al (2021) Movi-fi: Motion-robust vital signs waveform recovery via deep interpreted rf sensing. In: Proceedings of the 27th Annual International Conference on Mobile Computing and Networking, pp 392–405, https://doi.org/10.1145/3447993.3483251
Choe J, Schwichtenberg AJ, Delp EJ (2019) Classification of sleep videos using deep learning. In: 2019 IEEE Conference on Multimedia Information Processing and Retrieval (MIPR), IEEE, pp 115–120
Chung KY, Song K, Cho SH et al (2018) Noncontact sleep study based on an ensemble of deep neural network and random forests. IEEE Sens J 18(17):7315–7324
Chung J, Gulcehre C, Cho K, et al (2014) Empirical evaluation of gated recurrent neural networks on sequence modeling. arXiv preprint arXiv:1412.3555 https://doi.org/10.48550/arXiv.1412.3555
Clarke G, Harvey AG (2012) The complex role of sleep in adolescent depression. Child Adolesc Psychiatr Clin 21(2):385–400. https://doi.org/10.1016/j.chc.2012.01.006
Cui Z, Zheng X, Shao X et al (2018) Automatic sleep stage classification based on convolutional neural network and fine-grained segments. Complexity. https://doi.org/10.1155/2018/9248410
Dafna E, Tarasiuk A, Zigel Y (2018) Sleep staging using nocturnal sound analysis. Sci Rep 8(1):13474
Dai Y, Li X, Liang S et al (2023) Multichannelsleepnet: a transformer-based model for automatic sleep stage classification with psg. IEEE J Biomed Health Inform. https://doi.org/10.1109/JBHI.2023.3284160
Deng J, Dong W, Socher R, et al (2009) Imagenet: A large-scale hierarchical image database. In: 2009 IEEE conference on computer vision and pattern recognition, Ieee, pp 248–255
Devuyst S, Dutoit T, Kerkhofs M (2005) The dreams databases and assessment algorithm. Zenodo: Geneva, Switzerland Zenodo https://zenodo.org/records/2650142#.ZG1w6XZBw2w
Diraco G, Leone A, Siciliano P (2017) Detecting falls and vital signs via radar sensing. In: 2017 IEEE SENSORS, IEEE, pp 1–3, https://doi.org/10.1109/ICSENS.2017.8234405
Duan L, Zhang Y, Huang Z, et al (2023) Dual-teacher feature distillation: A transfer learning method for insomniac psg staging. IEEE Journal of Biomedical and Health Informatics
Efe E, Ozsen S (2023) Cosleepnet: automated sleep staging using a hybrid CNN-LSTM network on imbalanced EEG-EOG datasets. Biomed Signal Process Control 80:104299
Eldele E, Chen Z, Liu C et al (2021) An attention-based deep learning approach for sleep stage classification with single-channel EEG. IEEE Trans Neural Syst Rehabil Eng 29:809–818. https://doi.org/10.1109/TNSRE.2021.3076234
Elsayed M, Badawy A, Mahmuddin M, et al (2016) Fpga implementation of dwt eeg data compression for wireless body sensor networks. In: 2016 IEEE Conference on Wireless Sensors (ICWiSE), IEEE, pp 21–25, https://doi.org/10.1109/ICWISE.2016.8187756
Fan J, Sun C, Chen C et al (2020) Eeg data augmentation: towards class imbalance problem in sleep staging tasks. J Neural Eng 17(5):056017. https://doi.org/10.1088/1741-2552/abb5be
Fan J, Sun C, Long M et al (2021) Eognet: a novel deep learning model for sleep stage classification based on single-channel EOG signal. Front Neurosci 15:573194. https://doi.org/10.3389/fnins.2021.573194
Fang Y, Xia Y, Chen P et al (2023) A dual-stream deep neural network integrated with adaptive boosting for sleep staging. Biomed Signal Process Control 79:104150. https://doi.org/10.1016/j.bspc.2022.104150
Faust O, Razaghi H, Barika R et al (2019) A review of automated sleep stage scoring based on physiological signals for the new millennia. Comput Methods Programs Biomed 176:81–91. https://doi.org/10.1016/j.cmpb.2019.04.032
Favia A (2021) Deep learning for sleep state detection using cw doppler radar technology. Master’s thesis, Aalto University
Ferri C, Hernández-Orallo J, Modroiu R (2009) An experimental comparison of performance measures for classification. Pattern Recogn Lett 30(1):27–38
Fioranelli F, Le Kernec J, Shah SA (2019) Radar for health care: recognizing human activities and monitoring vital signs. IEEE Potentials 38(4):16–23. https://doi.org/10.1109/MPOT.2019.2906977
Fiorillo L, Puiatti A, Papandrea M et al (2019) Automated sleep scoring: a review of the latest approaches. Sleep Med Rev 48:101204. https://doi.org/10.1016/j.smrv.2019.07.007
Fiorillo L, Favaro P, Faraci FD (2021) Deepsleepnet-lite: a simplified automatic sleep stage scoring model with uncertainty estimates. IEEE Trans Neural Syst Rehabil Eng 29:2076–2085. https://doi.org/10.1109/TNSRE.2021.3117970
Fiorillo L, Monachino G, van der Meer J et al (2023) U-sleep’s resilience to aasm guidelines. NPJ Digit Med 6(1):33
Fiorillo L, Pedroncelli D, Agostini V et al (2023) Multi-scored sleep databases: how to exploit the multiple-labels in automated sleep scoring. Sleep 46(5):zsad028
Fonseca P, van Gilst MM, Radha M et al (2020) Automatic sleep staging using heart rate variability, body movements, and recurrent neural networks in a sleep disordered population. Sleep 43(9):zass048. https://doi.org/10.1093/sleep/zsaa048
Foumani NM, Tan CW, Webb GI et al (2024) Improving position encoding of transformers for multivariate time series classification. Data Min Knowl Disc 38(1):22–48
Article MathSciNet Google Scholar
Goldammer M, Zaunseder S, Brandt MD et al (2022) Investigation of automated sleep staging from cardiorespiratory signals regarding clinical applicability and robustness. Biomed Signal Process Control 71:103047. https://doi.org/10.1016/j.bspc.2021.103047
Goldberger AL, Amaral LA, Glass L et al (2000) Physiobank, physiotoolkit, and physionet: components of a new research resource for complex physiologic signals. Circulation 101(23):e215–e220. https://doi.org/10.1161/01.CIR.101.23.e215
Goodfellow I, Pouget-Abadie J, Mirza M, et al (2014) Generative adversarial nets. Adv Neural Inf Process Syst 27
Goshtasbi N, Boostani R, Sanei S (2022) Sleepfcn: a fully convolutional deep learning framework for sleep stage classification using single-channel electroencephalograms. IEEE Trans Neural Syst Rehabil Eng 30:2088–2096. https://doi.org/10.1109/TNSRE.2022.3192988
Grandini M, Bagli E, Visani G (2020) Metrics for multi-class classification: an overview. arXiv preprint arXiv:2008.05756
Guillot A, Sauvet F, During EH et al (2020) Dreem open datasets: multi-scored sleep datasets to compare human and automated sleep staging. IEEE Trans Neural Syst Rehabil Eng 28(9):1955–1965. https://doi.org/10.1109/TNSRE.2020.3011181
Guillot A, Sauvet F, During EH et al (2021) Robustsleepnet: transfer learning for automated sleep staging at scale. IEEE Trans Neural Syst Rehabil Eng 29:1441–1451. https://doi.org/10.1109/TNSRE.2021.3098968
Guo MH, Xu TX, Liu JJ et al (2022) Attention mechanisms in computer vision: a survey. Computational Visual Media 8(3):331–368. https://doi.org/10.1007/s41095-022-0271-y
Han F, Yang P, Feng Y et al (2024) Earsleep: In-ear acoustic-based physical and physiological activity recognition for sleep stage detection. Proceedings of the ACM on Interactive, Mobile, Wearable and Ubiquitous Technologies 8(2):1–31
Hanifi K, Karsligil ME (2021) Elderly fall detection with vital signs monitoring using cw doppler radar. IEEE Sens J 21(15):16969–16978. https://doi.org/10.1109/JSEN.2021.3079835
Han J, Luo P, Wang X (2019) Deep self-learning from noisy labels. In: Proceedings of the IEEE/CVF international conference on computer vision, pp 5138–5147
Herff C, Krusienski DJ, Kubben P (2020) The potential of stereotactic-EEG for brain-computer interfaces: current progress and future directions. Front Neurosci 14:123. https://doi.org/10.3389/fnins.2020.00123
He K, Zhang X, Ren S, et al (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp 770–778
Hong H, Zhang L, Zhao H et al (2019) Microwave sensing and sleep: noncontact sleep-monitoring technology with microwave biomedical radar. IEEE Microwave Mag 20(8):18–29. https://doi.org/10.1109/MMM.2019.2915469
Hong J, Tran HH, Jung J et al (2022) End-to-end sleep staging using nocturnal sounds from microphone chips for mobile devices. Nat Sci Sleep. https://doi.org/10.2147/NSS.S361270
Howard AG, Zhu M, Chen B, et al (2017) Mobilenets: Efficient convolutional neural networks for mobile vision applications. arXiv preprint arXiv:1704.04861
Hsu LM, Field R (2003) Interrater agreement measures: comments on Kappan, Cohen’s kappa, Scott’s π , and Aickin’s α . Underst Stat 2(3):205–219. https://doi.org/10.1207/S15328031US0203_03
Huang J, Ren L, Zhou X et al (2022) An improved neural network based on senet for sleep stage classification. IEEE J Biomed Health Inform 26(10):4948–4956. https://doi.org/10.1109/JBHI.2022.3157262
Huang X, Schmelter F, Irshad MT et al (2023) Optimizing sleep staging on multimodal time series: Leveraging borderline synthetic minority oversampling technique and supervised convolutional contrastive learning. Comput Biol Med 166:107501. https://doi.org/10.1016/j.compbiomed.2023.107501
Huang M, Jiao X, Jiang J, et al (2021) An overview on sleep research based on functional near infrared spectroscopy. Sheng wu yi xue Gong Cheng xue za zhi= Journal of Biomedical Engineering= Shengwu Yixue Gongchengxue Zazhi 38(6):1211–1218
Huang G, Liu Z, Van Der Maaten L, et al (2017) Densely connected convolutional networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp 4700–4708
Iandola FN, Han S, Moskewicz MW, et al (2016) Squeezenet: Alexnet-level accuracy with 50x fewer parameters and \(<\) 0.5 mb model size. arXiv preprint arXiv:1602.07360
Iber C (2007) The aasm manual for the scoring of sleep and associated events: rules, terminology, and technical specification. (No Title)
Jadhav P, Rajguru G, Datta D et al (2020) Automatic sleep stage classification using time-frequency images of cwt and transfer learning using convolution neural network. Biocybern Biomed Eng 40(1):494–504. https://doi.org/10.1016/j.bbe.2020.01.010
Jahrami HA, Alhaj OA, Humood AM et al (2022) Sleep disturbances during the covid-19 pandemic: a systematic review, meta-analysis, and meta-regression. Sleep Med Rev 62:101591. https://doi.org/10.1016/j.smrv.2022.101591
Jaiswal A, Babu AR, Zadeh MZ et al (2020) A survey on contrastive self-supervised learning. Technologies 9(1):2. https://doi.org/10.3390/technologies9010002
Jeon H, Jung Y, Lee S et al (2020) Area-efficient short-time fourier transform processor for time-frequency analysis of non-stationary signals. Appl Sci 10(20):7208. https://doi.org/10.3390/app10207208
Ji X, Li Y, Wen P (2023) 3dsleepnet: a multi-channel bio-signal based sleep stages classification method using deep learning. IEEE Trans Neural Syst Rehabil Eng. https://doi.org/10.1109/TNSRE.2023.3309542
Jia Z, Cai X, Zheng G et al (2020) Sleepprintnet: a multivariate multimodal neural network based on physiological time-series for automatic sleep staging. IEEE Trans Artif Intell 1(3):248–257. https://doi.org/10.1109/TAI.2021.3060350
Jia Z, Lin Y, Wang J, et al (2020b) Graphsleepnet: adaptive spatial-temporal graph convolutional networks for sleep stage classification. In: IJCAI, pp 1324–1330
Jia Z, Lin Y, Wang J, et al (2021) Salientsleepnet: Multimodal salient wave detection network for sleep staging. arXiv preprint arXiv:2105.13864 https://doi.org/10.48550/arXiv.2105.13864
Jiang X, Zhao J, Du B, et al (2021) Self-supervised contrastive learning for eeg-based sleep staging. In: 2021 International Joint Conference on Neural Networks (IJCNN), IEEE, pp 1–8, https://doi.org/10.1109/IJCNN52387.2021.9533305
Kanwal S, Uzair M, Ullah H, et al (2019) An image based prediction model for sleep stage identification. In: 2019 IEEE International Conference on Image Processing (ICIP), IEEE, pp 1366–1370, https://doi.org/10.1109/ICIP.2019.8803026
Karimi D, Dou H, Warfield SK et al (2020) Deep learning with noisy labels: exploring techniques and remedies in medical image analysis. Med Image Anal 65:101759
Kayabekir M (2019) Sleep physiology and polysomnogram, physiopathology and symptomatology in sleep medicine. In: Updates in Sleep Neurology and Obstructive Sleep Apnea. IntechOpen
Khalighi S, Sousa T, Santos JM et al (2016) Isruc-sleep: a comprehensive public dataset for sleep researchers. Comput Methods Programs Biomed 124:180–192. https://doi.org/10.1016/j.cmpb.2015.10.013
Khan MI, Jan MA, Muhammad Y et al (2021) Tracking vital signs of a patient using channel state information and machine learning for a smart healthcare system. Neural Comput Appl. https://doi.org/10.1007/s00521-020-05631-x
Khan F, Azou S, Youssef R et al (2022) IR-UWB radar-based robust heart rate detection using a deep learning technique intended for vehicular applications. Electronics 11(16):2505. https://doi.org/10.3390/electronics11162505
Khosla P, Teterwak P, Wang C et al (2020) Supervised contrastive learning. Adv Neural Inf Process Syst 33:18661–18673
Korkalainen H, Aakko J, Nikkonen S et al (2019) Accurate deep learning-based sleep staging in a clinical population with suspected obstructive sleep apnea. IEEE J Biomed Health Inform 24(7):2073–2081
Korkalainen H, Aakko J, Duce B et al (2020) Deep learning enables sleep staging from photoplethysmogram for patients with suspected sleep apnea. Sleep 43(11):zsaa098. https://doi.org/10.1093/sleep/zsaa098
Korompili G, Amfilochiou A, Kokkalas L et al (2021) Psg-audio, a scored polysomnography dataset with simultaneous audio recordings for sleep apnea studies. Scientific Data 8(1):197
Kotzen K, Charlton PH, Salabi S et al (2022) Sleepppg-net: a deep learning algorithm for robust sleep staging from continuous photoplethysmography. IEEE J Biomed Health Inform 27(2):924–932. https://doi.org/10.1109/JBHI.2022.3225363
Krizhevsky A, Sutskever I, Hinton GE (2012) Imagenet classification with deep convolutional neural networks. Adv Neural Inf Process Syst 25
Kuo CE, Chen GT, Liao PY (2021) An EEG spectrogram-based automatic sleep stage scoring method via data augmentation, ensemble convolution neural network, and expert knowledge. Biomed Signal Process Control 70:102981. https://doi.org/10.1016/j.bspc.2021.102981
Kuo CE, Lu TH, Chen GT et al (2022) Towards precision sleep medicine: self-attention gan as an innovative data augmentation technique for developing personalized automatic sleep scoring classification. Comput Biol Med 148:105828. https://doi.org/10.1016/j.compbiomed.2022.105828
Kwon HB, Choi SH, Lee D et al (2021) Attention-based lSTM for non-contact sleep stage classification using IR-UWB radar. IEEE J Biomed Health Inform 25(10):3844–3853. https://doi.org/10.1109/JBHI.2021.3072644
LeCun Y, Bottou L, Bengio Y et al (1998) Gradient-based learning applied to document recognition. Proc IEEE 86(11):2278–2324
Lee S, Yu Y, Back S et al (2024) Sleepyco: automatic sleep scoring with feature pyramid and contrastive learning. Expert Syst Appl 240:122551. https://doi.org/10.1016/j.eswa.2023.122551
Li X, Cui L, Tao S et al (2017) Hyclasss: a hybrid classifier for automatic sleep stage scoring. IEEE J Biomed Health Inform 22(2):375–385. https://doi.org/10.1109/JBHI.2017.2668993
Li Q, Li Q, Liu C et al (2018) Deep learning in the cross-time frequency domain for sleep staging from a single-lead electrocardiogram. Physiol Meas 39(12):124005. https://doi.org/10.1088/1361-6579/aaf339
Li C, Hou Y, Song R et al (2022) Multi-channel EEG-based emotion recognition in the presence of noisy labels. Sci China Inf Sci 65(4):140405
Li C, Qi Y, Ding X et al (2022) A deep learning method approach for sleep stage classification with EEG spectrogram. Int J Environ Res Public Health 19(10):6322. https://doi.org/10.3390/ijerph19106322
Li Y, Luo S, Zhang H et al (2022) Mtclss: multi-task contrastive learning for semi-supervised pediatric sleep staging. IEEE J Biomed Health Inform. https://doi.org/10.1109/JBHI.2022.3213171
Li T, Gong Y, Lv Y et al (2023) Gac-sleepnet: a dual-structured sleep staging method based on graph structure and Euclidean structure. Comput Biol Med 165:107477
Lin TY, Goyal P, Girshick R, et al (2017) Focal loss for dense object detection. In: Proceedings of the IEEE international conference on computer vision, pp 2980–2988
Ling H, Luyuan Y, Xinxin L et al (2022) Staging study of single-channel sleep EEG signals based on data augmentation. Front Public Health 10:1038742. https://doi.org/10.3389/fpubh.2022.1038742
Li Z, Sun S, Wang Y, et al (2022d) Time-frequency analysis of non-stationary signal based on sliding mode singular spectrum analysis and wigner-ville distribution. In: 2022 3rd International Conference on Information Science and Education (ICISE-IE), IEEE, pp 218–222, https://doi.org/10.1109/ICISE-IE58127.2022.00051
Liu Z, Luo S, Lu Y et al (2022) Extracting multi-scale and salient features by MSE based u-structure and CBAM for sleep staging. IEEE Trans Neural Syst Rehabil Eng 31:31–38. https://doi.org/10.1109/TNSRE.2022.3216111
Liu G, Wei G, Sun S et al (2023) Micro sleepnet: efficient deep learning model for mobile terminal real-time sleep staging. Front Neurosci. https://doi.org/10.3389/fnins.2023.1218072
Liu Z, Qin M, Lu Y et al (2023) Densleepnet: densenet based model for sleep staging with two-frequency feature fusion and coordinate attention. Biomed Eng Lett. https://doi.org/10.1007/s13534-023-00301-y
Liu X, Cao J, Tang S, et al (2014) Wi-sleep: Contactless sleep monitoring via wifi signals. In: 2014 IEEE Real-Time Systems Symposium, IEEE, pp 346–355
Liu M, Lin Z, Xiao P, et al (2022a) Human biometric signals monitoring based on wifi channel state information using deep learning. arXiv preprint arXiv:2203.03980 https://doi.org/10.48550/arXiv.2203.03980
Liu X, Wang H, Li Z (2021) An approach for deep learning in ecg classification tasks in the presence of noisy labels. In: 2021 43rd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), IEEE, pp 369–372
Loh HW, Ooi CP, Vicnesh J et al (2020) Automated detection of sleep stages using deep learning techniques: a systematic review of the last decade (2010–2020). Appl Sci 10(24):8963
Maheshwari S, Tiwari AK (2019) Ai-enabled wi-fi network to estimate human sleep quality based on intensity of movements. In: 2019 IEEE International Conference on Advanced Networks and Telecommunications Systems (ANTS), IEEE, pp 1–6
Maiti S, Sharma SK, Bapi RS (2023) Enhancing healthcare with eog: a novel approach to sleep stage classification. arXiv preprint arXiv:2310.03757 https://doi.org/10.48550/arXiv.2310.03757
Malafeev A, Laptev D, Bauer S et al (2018) Automatic human sleep stage scoring using deep neural networks. Front Neurosci 12:781
Malhotra A, Younes M, Kuna ST et al (2013) Performance of an automated polysomnography scoring system versus computer-assisted manual scoring. Sleep 36(4):573–582. https://doi.org/10.5665/sleep.2548
Malik J, Lo YL, Ht Wu (2018) Sleep-wake classification via quantifying heart rate variability by convolutional neural network. Physiol Meas 39(8):085004. https://doi.org/10.1088/1361-6579/aad5a9
Misra I, Maaten Lvd (2020) Self-supervised learning of pretext-invariant representations. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, pp 6707–6717
Morabito FC, Campolo M, Ieracitano C, et al (2016) Deep convolutional neural networks for classification of mild cognitive impaired and alzheimer’s disease patients from scalp eeg recordings. In: 2016 IEEE 2nd International Forum on Research and Technologies for Society and Industry Leveraging a better tomorrow (RTSI), IEEE, pp 1–6, https://doi.org/10.1109/RTSI.2016.7740576
Mousavi S, Afghah F, Acharya UR (2019) Sleepeegnet: automated sleep stage scoring with sequence to sequence deep learning approach. PLoS ONE 14(5):e0216456. https://doi.org/10.1371/journal.pone.0216456
Nasiri S, Clifford GD (2020) Attentive adversarial network for large-scale sleep staging. In: Machine Learning for Healthcare Conference, PMLR, pp 457–478
Neng W, Lu J, Xu L (2021) Ccrrsleepnet: a hybrid relational inductive biases network for automatic sleep stage classification on raw single-channel eeg. Brain Sci 11(4):456. https://doi.org/10.3390/brainsci11040456
Nocera A, Senigagliesi L, Raimondi M et al (2021) Machine learning in radar-based physiological signals sensing: a scoping review of the models, datasets and metrics. Mach Learn 19:1
Olesen AN, Jørgen Jennum P, Mignot E et al (2021) Automatic sleep stage classification with deep residual networks in a mixed-cohort setting. Sleep 44(1):zsaa161. https://doi.org/10.1093/sleep/zsaa161
Olsen M, Zeitzer JM, Richardson RN et al (2022) A flexible deep learning architecture for temporal sleep stage classification using accelerometry and photoplethysmography. IEEE Trans Biomed Eng 70(1):228–237. https://doi.org/10.1109/TBME.2022.3187945
Oord Avd, Li Y, Vinyals O (2018) Representation learning with contrastive predictive coding. arXiv preprint arXiv:1807.03748 https://doi.org/10.48550/arXiv.1807.03748
O’reilly C, Gosselin N, Carrier J et al (2014) Montreal archive of sleep studies: an open-access resource for instrument benchmarking and exploratory research. J Sleep Res 23(6):628–635. https://doi.org/10.1111/jsr.12169
Pan J, Tompkins WJ (1985) A real-time QRS detection algorithm. IEEE Trans Biomed Eng 3:230–236. https://doi.org/10.1109/TBME.1985.325532
Papadakis Z, Retortillo SG (2022) Acute partial sleep deprivation and high-intensity exercise effects on cardiovascular autonomic regulation and lipemia network. In: International Journal of Exercise Science: Conference Proceedings, p 12
Parekh A, Mullins AE, Kam K et al (2019) Slow-wave activity surrounding stage n2 k-complexes and daytime function measured by psychomotor vigilance test in obstructive sleep apnea. Sleep 42(3):zsy256. https://doi.org/10.1093/sleep/zsy256
Parekh N, Dave B, Shah R et al (2021) Automatic sleep stage scoring on raw single-channel eeg: A comparative analysis of cnn architectures. 2021 Fourth International Conference on Electrical. Computer and Communication Technologies (ICECCT), IEEE, pp 1–8
Park J, Yang S, Chung G, et al (2024) Ultra-wideband radar-based sleep stage classification in smartphone using an end-to-end deep learning. IEEE Access
Patanaik A, Ong JL, Gooley JJ et al (2018) An end-to-end framework for real-time automatic sleep stage classification. Sleep 41(5):zsy041
Perslev M, Darkner S, Kempfner L et al (2021) U-sleep: resilient high-frequency sleep staging. NPJ Digi Med 4(1):72. https://doi.org/10.1038/s41746-021-00440-5
Perslev M, Jensen M, Darkner S, et al (2019) U-time: a fully convolutional network for time series segmentation applied to sleep staging. Adv Neural Inf Process Syst 32
Phan H, Mikkelsen K (2022) Automatic sleep staging of EEG signals: recent development, challenges, and future directions. Physiol Measurement 43(4):04TR01. https://doi.org/10.1088/1361-6579/ac6049
Phan H, Andreotti F, Cooray N et al (2019) Seqsleepnet: end-to-end hierarchical recurrent neural network for sequence-to-sequence automatic sleep staging. IEEE Trans Neural Syst Rehabil Eng 27(3):400–410. https://doi.org/10.1109/TNSRE.2019.2896659
Phan H, Chén OY, Koch P et al (2020) Towards more accurate automatic sleep staging via deep transfer learning. IEEE Trans Biomed Eng 68(6):1787–1798
Phan H, Chén OY, Tran MC et al (2021) Xsleepnet: multi-view sequential model for automatic sleep staging. IEEE Trans Pattern Anal Mach Intell 44(9):5903–5915. https://doi.org/10.1109/TPAMI.2021.3070057
Phan H, Mertins A, Baumert M (2022) Pediatric automatic sleep staging: a comparative study of state-of-the-art deep learning methods. IEEE Trans Biomed Eng 69(12):3612–3622
Phan H, Mikkelsen K, Chén OY et al (2022) Sleeptransformer: automatic sleep staging with interpretability and uncertainty quantification. IEEE Trans Biomed Eng 69(8):2456–2467. https://doi.org/10.1109/TBME.2022.3147187
Phan H, Andreotti F, Cooray N, et al (2018) Automatic sleep stage classification using single-channel eeg: learning sequential features with attention-based recurrent neural networks. In: 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), IEEE, pp 1452–1455, https://doi.org/10.1109/EMBC.2018.8512480
Phyo J, Ko W, Jeon E et al (2022) Transsleep: transitioning-aware attention-based deep neural network for sleep staging. IEEE Trans Cybern. https://doi.org/10.1109/TCYB.2022.3198997
Pradeepkumar J, Anandakumar M, Kugathasan V, et al (2022) Towards interpretable sleep stage classification using cross-modal transformers. arXiv preprint arXiv:2208.06991 https://doi.org/10.48550/arXiv.2208.06991
Qi GJ, Luo J (2020) Small data challenges in big data era: a survey of recent progress on unsupervised and semi-supervised methods. IEEE Trans Pattern Anal Mach Intell 44(4):2168–2187. https://doi.org/10.1109/TPAMI.2020.3031898
Quan SF, Howard BV, Iber C et al (1997) The sleep heart health study: design, rationale, and methods. Sleep 20(12):1077–1085. https://doi.org/10.1093/sleep/20.12.1077
Radha M, Fonseca P, Moreau A et al (2021) A deep transfer learning approach for wearable sleep stage classification with photoplethysmography. NPJ Digi Med 4(1):135. https://doi.org/10.1038/s41746-021-00510-8
Rechtschaffen A (1968) A manual of standardized terminology, techniques and scoring system for sleep stage of human subject. (No Title)
Rommel C, Paillard J, Moreau T et al (2022) Data augmentation for learning predictive models on EEG: a systematic comparison. J Neural Eng 19(6):066020
Ronneberger O, Fischer P, Brox T (2015) U-net: convolutional networks for biomedical image segmentation. In: Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, October 5-9, 2015, Proceedings, Part III 18, Springer, pp 234–241, https://doi.org/10.1007/978-3-319-24574-4_28
Selvaraju RR, Cogswell M, Das A, et al (2017) Grad-cam: Visual explanations from deep networks via gradient-based localization. In: Proceedings of the IEEE international conference on computer vision, pp 618–626
Seo H, Back S, Lee S et al (2020) Intra-and inter-epoch temporal context network (iitnet) using sub-epoch features for automatic sleep scoring on raw single-channel eeg. Biomed Signal Process Control 61:102037. https://doi.org/10.1016/j.bspc.2020.102037
Sharma R, Pachori RB, Upadhyay A (2017) Automatic sleep stages classification based on iterative filtering of electroencephalogram signals. Neural Comput Appl 28:2959–2978. https://doi.org/10.1007/s00521-017-2919-6
Shen Q, Xin J, Liu X, et al (2023) Lgsleepnet: an automatic sleep staging model based on local and global representation learning. IEEE Transactions on Instrumentation and Measurement
Shinar Z, Akselrod S, Dagan Y et al (2006) Autonomic changes during wake-sleep transition: a heart rate variability based approach. Auton Neurosci 130(1–2):17–27. https://doi.org/10.1016/j.autneu.2006.04.006
Siddhad G, Gupta A, Dogra DP et al (2024) Efficacy of transformer networks for classification of EEG data. Biomed Signal Process Control 87:105488
Siegel JM (2009) Sleep viewed as a state of adaptive inactivity. Nat Rev Neurosci 10(10):747–753. https://doi.org/10.1038/nrn2697
Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556
Song H, Kim M, Park D, et al (2022) Learning from noisy labels with deep neural networks: a survey. IEEE transactions on neural networks and learning systems
Sors A, Bonnet S, Mirek S et al (2018) A convolutional neural network for sleep stage scoring from raw single-channel EEG. Biomed Signal Process Control 42:107–114. https://doi.org/10.1016/j.bspc.2017.12.001
Soto JC, Galdino I, Caballero E et al (2022) A survey on vital signs monitoring based on wi-fi CSI data. Comput Commun 195:99–110. https://doi.org/10.1016/j.comcom.2022.08.004
Spelmen VS, Porkodi R (2018) A review on handling imbalanced data. In: 2018 international conference on current trends towards converging technologies (ICCTCT), IEEE, pp 1–11, https://doi.org/10.1109/ICCTCT.2018.8551020
Sri TR, Madala J, Duddukuru SL, et al (2022) A systematic review on deep learning models for sleep stage classification. In: 2022 6th International Conference on Trends in Electronics and Informatics (ICOEI), IEEE, pp 1505–1511
Sridhar N, Shoeb A, Stephens P et al (2020) Deep learning for automated sleep staging using instantaneous heart rate. NPJ Digi Med 3(1):106. https://doi.org/10.1038/s41746-020-0291-x
Stephansen JB, Olesen AN, Olsen M et al (2018) Neural network analysis of sleep stages enables efficient diagnosis of narcolepsy. Nat Commun 9(1):5229
Stokes PA, Prerau MJ (2020) Estimation of time-varying spectral peaks and decomposition of EEG spectrograms. IEEE Access 8:218257–218278. https://doi.org/10.1109/ACCESS.2020.3042737
Stuburić K, Gaiduk M, Seepold R (2020) A deep learning approach to detect sleep stages. Procedia Computer Sci 176:2764–2772
Subha DP, Joseph PK, Acharya UR et al (2010) EEG signal analysis: a survey. J Med Syst 34:195–212
Sun H, Ganglberger W, Panneerselvam E et al (2020) Sleep staging from electrocardiography and respiration with deep learning. Sleep 43(7):zsz306. https://doi.org/10.1093/sleep/zsz306
Sun C, Hong S, Wang J et al (2022) A systematic review of deep learning methods for modeling electrocardiograms during sleep. Physiol Meas. https://doi.org/10.1088/1361-6579/ac826e
Supratak A, Dong H, Wu C et al (2017) Deepsleepnet: a model for automatic sleep stage scoring based on raw single-channel EEG. IEEE Trans Neural Syst Rehabil Eng 25(11):1998–2008. https://doi.org/10.1109/TNSRE.2017.2721116
Supratak A, Guo Y (2020) Tinysleepnet: an efficient deep learning model for sleep stage scoring based on raw single-channel eeg. In: 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), IEEE, pp 641–644, https://doi.org/10.1109/EMBC44109.2020.9176741
Szegedy C, Liu W, Jia Y, et al (2015) Going deeper with convolutions. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp 1–9
Tăutan AM, Rossi AC, De Francisco R, et al (2020) Automatic sleep stage detection: a study on the influence of various psg input signals. In: 2020 42nd Annual International Conference of the Ieee Engineering in Medicine & Biology Society (EMBC), IEEE, pp 5330–5334, https://doi.org/10.1109/EMBC44109.2020.9175628
Thölke P, Mantilla-Ramos YJ, Abdelhedi H et al (2023) Class imbalance should not throw you off balance: choosing the right classifiers and performance metrics for brain decoding with imbalanced data. Neuroimage 277:120253. https://doi.org/10.1016/j.neuroimage.2023.120253
Timplalexis C, Diamantaras K, Chouvarda I (2019) Classification of sleep stages for healthy subjects and patients with minor sleep disorders. In: 2019 IEEE 19th International Conference on Bioinformatics and Bioengineering (BIBE), IEEE, pp 344–351
Tobaldini E, Nobili L, Strada S et al (2013) Heart rate variability in normal and pathological sleep. Front Physiol 4:294. https://doi.org/10.3389/fphys.2013.00294
Toften S, Pallesen S, Hrozanova M et al (2020) Validation of sleep stage classification using non-contact radar technology and machine learning (somnofy®). Sleep Med 75:54–61
Tran HH, Hong JK, Jang H et al (2023) Prediction of sleep stages via deep learning using smartphone audio recordings in home environments: model development and validation. J Med Internet Res 25:e46216. https://doi.org/10.2196/46216
Tsinalis O, Matthews PM, Guo Y, et al (2016) Automatic sleep stage scoring with single-channel eeg using convolutional neural networks. arXiv preprint arXiv:1610.01683 https://doi.org/10.48550/arXiv.1610.01683
Tyagi A, Nehra V (2017) Time frequency analysis of non-stationary motor imagery eeg signals. In: 2017 International Conference on Computing and Communication Technologies for Smart Nation (IC3TSN), IEEE, pp 44–50, https://doi.org/10.1109/IC3TSN.2017.8284448
Van der Maaten L, Hinton G (2008) Visualizing data using t-sne. J Mach Learn Res 9(11):2579–2605
Van Someren EJ (2021) Brain mechanisms of insomnia: new perspectives on causes and consequences. Physiol Rev 101(3):995–1046. https://doi.org/10.1152/physrev.00046.2019
Vaswani A, Shazeer N, Parmar N, et al (2017) Attention is all you need. Adv Neural Inf Process Syst 30
Vázquez CG, Breuss A, Gnarra O et al (2022) Label noise and self-learning label correction in cardiac abnormalities classification. Physiol Meas 43(9):094001
Vilamala A, Madsen KH, Hansen LK (2017) Deep convolutional neural networks for interpretable analysis of eeg sleep stage scoring. In: 2017 IEEE 27th international workshop on machine learning for signal processing (MLSP), IEEE, pp 1–6
Walch O, Huang Y, Forger D et al (2019) Sleep stage prediction with raw acceleration and photoplethysmography heart rate data derived from a consumer wearable device. Sleep 42(12):zsz180. https://doi.org/10.1093/sleep/zsz180
Wang X, Matsushita D (2023) Non-contact determination of sleep/wake state in residential environments by neural network learning of microwave radar and electroencephalogram-electrooculogram measurements. Build Environ 233:110095
Wang Y, Yao Y (2023) Application of artificial intelligence methods in carotid artery segmentation: a review. IEEE Access. https://doi.org/10.1109/ACCESS.2023.3243162
Wang Q, Wei HL, Wang L et al (2021) A novel time-varying modeling and signal processing approach for epileptic seizure detection and classification. Neural Comput Appl 33:5525–5541. https://doi.org/10.1007/s00521-020-05330-7
Wang B, Tang X, Ai H et al (2022) Obstructive sleep apnea detection based on sleep sounds via deep learning. Nat Sci Sleep 31:2033–2045
Wang E, Koprinska I, Jeffries B (2023) Sleep apnea prediction using deep learning. IEEE Journal of Biomedical and Health Informatics
Wulff K, Gatti S, Wettstein JG et al (2010) Sleep and circadian rhythm disruption in psychiatric and neurodegenerative disease. Nat Rev Neurosci 11(8):589–599. https://doi.org/10.1038/nrn2868
Wu Y, Lo Y, Yang Y (2020) Stcn: A lightweight sleep staging model with multiple channels. In: 2020 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), IEEE, pp 1180–1183, https://doi.org/10.1109/BIBM49941.2020.9313371
Xie J, Aubert X, Long X et al (2021) Audio-based snore detection using deep neural networks. Comput Methods Programs Biomed 200:105917
Xie J, Wang Z, Yu Z et al (2021) Ischemic stroke prediction by exploring sleep related features. Appl Sci 11(5):2083
Xie J, Wang Z, Yu Z et al (2018) Enabling efficient stroke prediction by exploring sleep related features. 2018 IEEE SmartWorld, Ubiquitous Intelligence & Computing, Advanced & Trusted Computing, Scalable Computing & Communications. Cloud & Big Data Computing, Internet of People and Smart City Innovation (SmartWorld/SCALCOM/UIC/ATC/CBDCom/IOP/SCI), IEEE, pp 452–461
Xu Z, Yang X, Sun J et al (2020) Sleep stage classification using time-frequency spectra from consecutive multi-time points. Front Neurosci 14:14. https://doi.org/10.3389/fnins.2020.00014
Xu H, Plataniotis KN (2016) Affective states classification using eeg and semi-supervised deep learning approaches. In: 2016 IEEE 18th International Workshop on Multimedia Signal Processing (MMSP), IEEE, pp 1–6, https://doi.org/10.1109/MMSP.2016.7813351
Yacouby R, Axman D (2020) Probabilistic extension of precision, recall, and f1 score for more thorough evaluation of classification models. In: Proceedings of the first workshop on evaluation and comparison of NLP systems, pp 79–91, https://doi.org/10.18653/v1/2020.eval4nlp-1.9
Yang C, Li B, Li Y et al (2023) Lwsleepnet: a lightweight attention-based deep learning model for sleep staging with singlechannel EEG. Digital Health 9:20552076231188210. https://doi.org/10.1177/20552076231188206
Yang H, Sakhavi S, Ang KK, et al (2015) On the use of convolutional neural networks and augmented csp features for multi-class motor imagery of eeg signals classification. In: 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), IEEE, pp 2620–2623, https://doi.org/10.1109/EMBC.2015.7318929
Yao Z, Liu X (2023) A cnn-transformer deep learning model for real-time sleep stage classification in an energy-constrained wireless device. In: 2023 11th International IEEE/EMBS Conference on Neural Engineering (NER), IEEE, pp 1–4, https://doi.org/10.1109/NER52421.2023.10123825
Ye J, Xiao Q, Wang J et al (2021) Cosleep: a multi-view representation learning framework for self-supervised learning of sleep stage classification. IEEE Signal Process Lett 29:189–193. https://doi.org/10.1109/LSP.2021.3130826
Yeckle J, Manian V (2023) Automated sleep stage classification in home environments: an evaluation of seven deep neural network architectures. Sensors 23(21):8942
Yifan Z, Fengchen Q, Fei X (2020) Gs-rnn: a novel rnn optimization method based on vanishing gradient mitigation for hrrp sequence estimation and recognition. In: 2020 IEEE 3rd International Conference on Electronics Technology (ICET), IEEE, pp 840–844, https://doi.org/10.1109/ICET49382.2020.9119513
Yildirim O, Baloglu UB, Acharya UR (2019) A deep learning model for automated sleep stages classification using PSG signals. Int J Environ Res Public Health 16(4):599. https://doi.org/10.3390/ijerph16040599
Yoo C, Lee HW, Kang JW (2021) Transferring structured knowledge in unsupervised domain adaptation of a sleep staging network. IEEE J Biomed Health Inform 26(3):1273–1284
Young T, Palta M, Dempsey J et al (2009) Burden of sleep apnea: rationale, design, and major findings of the Wisconsin sleep cohort study. WMJ: Off Publ State Med Soc Wisconsin 108(5):246
Yu B, Wang Y, Niu K et al (2021) Wifi-sleep: sleep stage monitoring using commodity wi-fi devices. IEEE Internet Things J 8(18):13900–13913. https://doi.org/10.1109/JIOT.2021.3068798
Yubo Z, Yingying L, Bing Z et al (2022) Mmasleepnet: a multimodal attention network based on electrophysiological signals for automatic sleep staging. Front Neurosci 16:973761. https://doi.org/10.3389/fnins.2022.973761
Yun S, Lee H, Kim J, et al (2022) Patch-level representation learning for self-supervised vision transformers. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp 8354–8363
Yu L, Tang P, Jiang Z, et al (2023) Denoise enhanced neural network with efficient data generation for automatic sleep stage classification of class imbalance. In: 2023 International Joint Conference on Neural Networks (IJCNN), IEEE, pp 1–8, https://doi.org/10.1109/IJCNN54540.2023.10191282
Zhai Q, Tang T, Lu X et al (2022) Machine learning-enabled noncontact sleep structure prediction. Adv Intell Syst 4(5):2100227. https://doi.org/10.1002/aisy.202100227
Zhang GQ, Cui L, Mueller R et al (2018) The national sleep research resource: towards a sleep data commons. J Am Med Inform Assoc 25(10):1351–1358. https://doi.org/10.1093/jamia/ocy064
Zhang J, Yao R, Ge W et al (2020) Orthogonal convolutional neural networks for automatic sleep stage classification based on single-channel EEG. Comput Methods Programs Biomed 183:105089. https://doi.org/10.1016/j.cmpb.2019.105089
Zhang C, Bengio S, Hardt M et al (2021) Understanding deep learning (still) requires rethinking generalization. Commun ACM 64(3):107–115
Zhang R, Tian D, Xu D et al (2022) A survey of wound image analysis using deep learning: classification, detection, and segmentation. IEEE Access 10:79502–79515. https://doi.org/10.1109/ACCESS.2022.3194529
Zhang Y, Ren R, Yang L et al (2022) Sleep in alzheimer’s disease: a systematic review and meta-analysis of polysomnographic findings. Transl Psychiatry 12(1):136. https://doi.org/10.1038/s41398-022-01897-y
Zhang Y, Chen Y, Hu L, et al (2017) An effective deep learning approach for unobtrusive sleep stage detection using microphone sensor. In: 2017 IEEE 29th International Conference on Tools with Artificial Intelligence (ICTAI), IEEE, pp 37–44
Zhang H, Goodfellow I, Metaxas D, et al (2019) Self-attention generative adversarial networks. In: International Conference on Machine Learning, PMLR, pp 7354–7363
Zhang Q, Liu Y (2018) Improving brain computer interface performance by data augmentation with conditional deep convolutional generative adversarial networks. arXiv preprint arXiv:1806.07108 https://doi.org/10.48550/arXiv.1806.07108
Zhang K, Wen Q, Zhang C, et al (2023) Self-supervised learning for time series analysis: taxonomy, progress, and prospects. arXiv preprint arXiv:2306.10125 https://doi.org/10.48550/arXiv.2306.10125
Zhao R, Xia Y, Wang Q (2021) Dual-modal and multi-scale deep neural networks for sleep staging using EEG and ECG signals. Biomed Signal Process Control 66:102455. https://doi.org/10.1016/j.bspc.2021.102455
Zhao R, Xia Y, Zhang Y (2021) Unsupervised sleep staging system based on domain adaptation. Biomed Signal Process Control 69:102937
Zhao C, Li J, Guo Y (2022) Sleepcontextnet: a temporal context network for automatic sleep staging based single-channel eeg. Comput Methods Programs Biomed 220:106806. https://doi.org/10.1016/j.cmpb.2022.106806
Zhao M, Yue S, Katabi D, et al (2017) Learning sleep stages from radio signals: a conditional adversarial architecture. In: International Conference on Machine Learning, PMLR, pp 4100–4109
Zhou D, Xu Q, Wang J et al (2022) Alleviating class imbalance problem in automatic sleep stage classification. IEEE Trans Instrum Meas 71:1–12. https://doi.org/10.1109/TIM.2022.3191710
Zhou H, Liu A, Cui H et al (2023) Sleepnet-lite: a novel lightweight convolutional neural network for single-channel EEG-based sleep staging. IEEE Sensors Lett 7(2):1–4
Zhou D, Xu Q, Wang J, et al (2021) Lightsleepnet: a lightweight deep model for rapid sleep stage classification with spectrograms. In: 2021 43rd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), IEEE, pp 43–46, https://doi.org/10.1109/EMBC46164.2021.9629878
Zhu T, Luo W, Yu F (2020) Convolution-and attention-based neural network for automated sleep stage classification. Int J Environ Res Public Health 17(11):4152. https://doi.org/10.3390/ijerph17114152
Zhu H, Wu Y, Shen N et al (2022) The masking impact of intra-artifacts in EEG on deep learning-based sleep staging systems: a comparative study. IEEE Trans Neural Syst Rehabil Eng 30:1452–1463
Zhu H, Zhou W, Fu C et al (2023) Masksleepnet: a cross-modality adaptation neural network for heterogeneous signals processing in sleep staging. IEEE J Biomed Health Inform. https://doi.org/10.1109/JBHI.2023.3253728
Download references
Author information
Authors and affiliations.
Research Institute for Medical and Biological Engineering, Ningbo University, Ningbo, China
Peng Liu, Wei Qian & Hua Zhang
Health Science Center, Ningbo University, Ningbo, China
Department of Radiology, The Affiliated People’s Hospital of Ningbo University, Ningbo, China
Qi Hong & Qiang Li
Department of Electrical and Computer Engineering, Stevens Institute of Technology, Hoboken, USA
You can also search for this author in PubMed Google Scholar
Contributions
PL conducted literature survey and wrote the main manuscript text. WQ guided literature survey. HZ guided literature survey. YZ guided the literature survey. G.X. guided the literature survey. QH guided the literature survey. QL guided the literature survey. YY guided the literature survey and the writing of the main manuscript text. All authors reviewed the manuscript
Corresponding authors
Correspondence to Qiang Li or Yudong Yao .
Ethics declarations
Competing interests.
The authors declare no competing interests.
Additional information
Publisher's note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/ .
Reprints and permissions
About this article
Liu, P., Qian, W., Zhang, H. et al. Automatic sleep stage classification using deep learning: signals, data representation, and neural networks. Artif Intell Rev 57 , 301 (2024). https://doi.org/10.1007/s10462-024-10926-9
Download citation
Accepted : 22 August 2024
Published : 23 September 2024
DOI : https://doi.org/10.1007/s10462-024-10926-9
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Sleep stage classification
- Deep learning
- Polysomnography
- Contactless
- Cardiorespiratory
- Find a journal
- Publish with us
- Track your research

IMAGES
VIDEO
COMMENTS
UTF8 can take upto 4 bytes to represent a symbol. Question 13. UTF32 takes exactly 4 bytes to represent a symbol. Question 14. Unicode value of a symbol is called code point. True/False Questions Question 1. A computer can work with Decimal number system. False. Question 2. A computer can work with Binary number system. True. Question 3
Data Representation Common Exam Questions. Get a hint. Explain what overflow is and give an example of a situation which might cause overflow to occur. Overflow occurs when a number is too large it store in the available number of bits. An example of this occurring is when multiplying two numbers together. 1 / 25.
Data representation in computer MCQ.Questions and answers with PDF for all Computer Related Entrance & Competitive Exams Preparation. Helpful for Class 11, GATE, IBPS, SBI (Bank PO & Clerk), SSC, Railway etc.
25 Multiple choice questions. Term. Explain what overflow is and give an example of a situation which might cause overflow to occur. An analogue signal is an electrical signal that represents analogue data that varies in a continuous manner. Digital signals with voltage changes that are in discrete steps.
Data representation exercises. Exercises not as directly relevant to this year's class are marked with ⚠️. DATAREP-1. Sizes and alignments. QUESTION DATAREP-1A. True or false: For any non-array type X, the size of X (sizeof(X)) is greater than or equal to the alignment of type X (alignof(X)). Show solution. QUESTION DATAREP-1B.
1.3.4 STORAGE AND COMPRESSION | Lossy and Lossless, how files are compressed. Here we cover all points from the Cambridge Computer Science Topic 1, including questions and key terminology on Data representation including Binary, Hex, BCD, Sound, Images, storage and data compression.
computers are optimized to work with a particular xed size chunk of data, the word size is the smallest size group of bytes that a computer handle. All operations are conducted on a word-size chunks of bits. The number of values a particular group of bits can represent grows exponentially as the number of bits grows.
We also cover the basics of digital circuits and logic gates, and explain how they are used to represent and process data in computer systems. Our guide includes real-world examples and case studies to help you master data representation principles and prepare for your computer science exams. Check out the links below:
The problem of data representation is the problem of representing all the concepts we might want to use in programming—integers, fractions, real numbers, sets, pictures, texts, buildings, animal species, relationships—using the limited medium of addresses and bytes. Powers of ten and powers of two.
Test your knowledge of data representation in computer science with this quiz. Explore the fundamentals of binary digits, bits, transistors, and their role in representing and processing information in computers. Challenge yourself with our data representation quiz and flashcards, featuring questions to help you master computer science concepts.
This is a hexadecimal (base-16) number indicating the value of the address of the object. A line contains one to sixteen bytes of memory starting at this address. The contents of memory starting at the given address, such as 3d 00 00 00. Memory is printed as a sequence of bytes, which are 8-bit numbers between 0 and 255.
Quiz yourself with questions and answers for Data Representation Quiz, so you can be ready for test day. ... the number found by adding all the data in a set and dividing by the amount of numbers you added. For example 4 + 5 + 1 + 2 = 12 12 ÷ 4 = 3. The _____ of that data set is 3. Choose matching term.
We can represent numbers using only the digits 0s and 1s with the binary number system. Instead of counting the number of 1s, 5s, 10s, and 25s in coins, or 1s, 10s, 100s, and 1000s in abstract amounts, count the number of 1s, 2s, 4s, 8s, etc. For example, 1101 in binary is 1 * 8 + 1 * 4 + 0 * 2 + 1 * 1 = 13 in decimal.
b) Show how the computer would add the two floating-point numbers in part a by changing one of the numbers so they are both expressed using the same power of 2. c) Show how the computer would represent the sum in part b using the given floating-point representation. What decimal value for the sum is the computer actually storing? Explain.
Numbers - Data Representation - Computer Science Field Guide. In this section, we will look at how computers represent numbers. To begin with, we'll revise how the base 10 number system that we use every day works, and then look at binary, which is base 2. After that, we'll look at some other charactertistics of numbers that computers must deal ...
DATA REPRESENTATION Introduction In Digital Computer, data and instructions are stored in computer memory using binary code (or machine code) r epresented by Binary digIT's 1 and 0 called BIT's. The data may contain digits, alphabets or special character, which are converted to bits, understandable by the computer.
6. Histogram. A histogram is the graphical representation of data. It is similar to the appearance of a bar graph but there is a lot of difference between histogram and bar graph because a bar graph helps to measure the frequency of categorical data.
Questions and model answers on Number Systems for the CIE IGCSE Computer Science syllabus, written by the Computer Science experts at Save My Exams.
Data Representation Techniques. Classification of Computers. Computer scans are classified broadly based on their speed and computing power. 1. Microcomputers or PCs (Personal Computers): It is a single-user computer system with a medium-power microprocessor. It is referred to as a computer with a microprocessor as its central processing unit.
Computer Organization multiple choice questions and answers set contain 5 mcqs on number system and data representation in computer science. Each quiz objective question has 4 options as possible answers. Choose your option and check it with the given correct answer.
Data Representation. A network is a collection of different devices connected and capable of communicating. For example, a company's local network connects employees' computers and devices like printers and scanners. Employees will be able to share information using the network and also use the common printer/ scanner via the network.
Data. Data can be anything like a number, a name, notes in a musical composition, or the color in a photograph. Data representation can be referred to as the form in which we stored the data, processed it and transmitted it. In order to store the data in digital format, we can use any device like computers, smartphones, and iPads.
In clinical practice, sleep stage classification (SSC) is a crucial step for physicians in sleep assessment and sleep disorder diagnosis. However, traditional sleep stage classification relies on manual work by sleep experts, which is time-consuming and labor-intensive. Faced with this obstacle, computer-aided diagnosis (CAD) has the potential to become an intelligent assistant tool for sleep ...