
- Schools & departments

MRC Human Genetics Unit
The MRC Human Genetics Unit discovers how changes in our DNA impact our lives, combining the latest computational and experimental technologies to investigate how our genomes work to control the function of molecules, cells and tissues.

The MRC Human Genetics Unit combine the latest computational and experimental technologies to investigate how our genomes work to control the function of molecules, cells and tissues in people and populations. For more than half a century our research has been dedicated to understanding human genetic disease. Today we continue to apply our clinical and scientific expertise, harnessing the power of complex data, to improve health, and the lives of patients and their families.

Scientific aims:
- To understand the molecular basis of human genetic disease and normal development – especially that of the eye, the brain and growth
- To identify and understand the genome sequence variants involved in common disease risk and quantitative traits
- To understand how the flow of information from DNA through RNA to the organism is regulated
- To identify the forces of mutation and selection that influence human genome variation
- To investigate opportunities for novel diagnostic and therapeutic approaches
Related links
This article was published on 2022-11-08
Institute of Genetics and Cancer
Web site moved, the url: http://www.hgu.mrc.ac.uk has moved to:, https://www.ed.ac.uk/mrc-human-genetics-unit.
Please update your bookmarks.
If you have not been automatically redirected, please click the link above.
- Help & FAQ
- MRC Human Genetics Unit
- Deanery of Molecular, Genetic and Population Health Sciences
- Institute of Genetics and Cancer
- Phone +44 (0)131 332 2471, +44 (0)131 467 8456
- Email [email protected]
United Kingdom
Organisation profile
Website: www.ed.ac.uk/igmm
Director: Professor Wendy Bickmore, FRSE, FMEdSci
The Medical Research Council Human Genetics Unit is at the forefront of research into human genetics. Its role is to advance the understanding of genetic factors implicated in human disease and normal and abnormal development and physiology.
The Unit's programmes of work cover the themes of developmental genetics, common disease genetics, chromosome biology and models for human genetic diseases. The unit is one of the largest MRC research establishments supporting approximately 220 scientists, support staff, fellows, PhD students and visiting scientists.
The Unit is a partner in the MRC Institute of Genetics and Molecular Medicine (IGMM) at The University of Edinburgh - an exciting development bringing together the MRC Human Genetics Unit, the Centre for Genomics and Experimental Medicine and the Edinburgh Cancer Research Centre.
Fingerprint
- Nested Gene Biochemistry, Genetics and Molecular Biology 100%
- Protein Biochemistry, Genetics and Molecular Biology 65%
- Mutation Biochemistry, Genetics and Molecular Biology 56%
- Association Biochemistry, Genetics and Molecular Biology 53%
- Mouse Biochemistry, Genetics and Molecular Biology 50%
- Genetics Biochemistry, Genetics and Molecular Biology 40%
- Phenotype Biochemistry, Genetics and Molecular Biology 31%
- Development Biochemistry, Genetics and Molecular Biology 28%
Collaborations and top research areas from the last five years
Dive into details.
Select a country/territory to view shared publications and projects

- Deanery of Molecular, Genetic and Population Health Sciences - Personal Chair of Germline Biology
- Royal (Dick) School of Veterinary Studies - Chair position in Reproductive Biotechnologies
Person: Academic: Research Active

Stuart Aitken
- Deanery of Molecular, Genetic and Population Health Sciences - Bioinformatician

Craig Anderson
- Deanery of Molecular, Genetic and Population Health Sciences - Research Fellow
- MRC Human Genetics Unit - Research Fellow
Person: Academic: Research Active (Research Assistant)
Research output
- 3339 Article
- 88 Meeting abstract
- 88 Review article
- 51 Literature review
- 48 Conference contribution
- 40 Abstract
- 35 Comment/debate
- 27 Editorial
- 12 Chapter (peer-reviewed)
- 4 Other chapter contribution
- 3 Conference article
- 3 Doctoral Thesis
- 2 Other report
- 2 Book/Film/Article review
- 2 Working paper
- 1 Commissioned report
- 1 Entry for encyclopedia/dictionary
- 1 Special issue
- 1 Digital or Visual Products
- 1 Performance
- 1 Web publication/site
- 1 Other contribution
Research output per year
Effects of Monovalent and Divalent Cations on the Rheology of Entangled DNA
Research output : Contribution to journal › Article › peer-review
- Rheology 100%
- Hydrogel 16%
DNA lesion bypass and the stochastic dynamics of transcription coupled repair
- Transcription 100%
- Dynamics 100%
- DNA Base 25%
- RNA Polymerase II 25%
- In Vivo 25%
Vertebrate centromeres in mitosis are functionally bipartite structures stabilized by cohesin
- Centromere 100%
- Cohesin 100%
- Condensin 10%
- Chromosome Segregation 10%
- Electron 10%
- 30 Participation in workshop, seminar, course
- 28 Invited talk
- 18 Oral presentation
- 12 Public Engagement – Festival/Exhibition
- 9 Public Engagement – Public lecture/debate/seminar
- 8 Participation in conference
- 6 Public Engagement – Schools engagement
- 5 Membership of peer review panel or committee
- 4 Professional Development and Training
- 4 Contribution to the work of national or international committees and working groups
- 4 Editorial activity
- 4 Types of External academic engagement - Research and Teaching at External Organisation
- 3 External Examiner or Assessor
- 3 Types of Public engagement and outreach - Media article or participation
- 2 Hosting an academic visitor
- 1 Examination
- 1 Membership of board
- 1 Membership of public/government advisory/policy group or panel
- 1 Types of Public engagement and outreach - Festival/Exhibition
Activities per year
Edinburgh Chromatin, Epigenetics and Transcriptional Regulation Network
Douglas Vernimmen (Organiser), Sari Pennings (Organiser), Nezha Benabdallah (Speaker), Yatendra Kumar (Speaker), Jeroen Witteveldt (Speaker) & Stefan Bresson (Speaker)
Activity : Participating in or organising an event types › Participation in workshop, seminar, course
Eukaryotic mRNA processing
Javier Caceres (Advisor)
Activity : Participating in or organising an event types › Participation in conference
Douglas Vernimmen (Organiser), Sari Pennings (Organiser), Hannah Long (Speaker), Rafal Czapiewski (Speaker) & Michael O'Dwyer (Speaker)
CMVM Student experience award
Navarro, Pau (Recipient), 10 Nov 2022
Prize : Prize (including medals and awards)
Elected as a member of the Latin American Academy of Sciences (ACAL)
Caceres, Javier (Recipient), 2021
Prize : Election to learned society
Elected as Fellow of the Royal Society
Jackson, Andrew (Recipient), 2020
- 173 Finished
Projects per year
Energy poverty: How does the home influence the risk, recovery, and intergenerational recurrence of childhood respiratory infections?
Swann, O. , Whyte, M., Jackson, A. , Henderson, N. , Gilbert, N. & Walmsley, S.
1/06/23 → 31/05/24
Project : Research
Cis-Regulatory mechanisms in genetic eye disease (Transition Programme)
1/04/23 → 31/03/28
Mechanisms for Microcephaly, cancer and Autoinflammation
Jackson, A.
Code and datasets for structural analyses in Natan et al, "Assembly in the translation milieu imposes evolutionary constraints on homomeric proteins"
Marsh, J. (Creator), Edinburgh DataShare, 10 Oct 2017
DOI : 10.7488/ds/2227
eHistology Atlas: Plates 02-15 Early Gestation (E5.5 – E8.5)
Graham, E. (Creator), Moss, J. (Creator), Burton, N. (Creator), Roochun, Y. (Creator), Armit, C. (Creator), Richardson, L. (Creator) & Baldock, R. (Creator), Edinburgh DataShare, 21 Jan 2016
https://datashare.is.ed.ac.uk/handle/10283/943
eHistology Atlas: Plates 30-41 Late Gestation (E13.5 – E17.5)
Graham, E. (Creator), Moss, J. (Creator), Burton, N. (Creator), Roochun, Y. (Creator), Armit, C. (Creator), Richardson, L. (Creator) & Baldock, R. (Creator), Edinburgh DataShare, 22 Dec 2015
https://datashare.is.ed.ac.uk/handle/10283/945
Press/Media
Precision medicine funding announcement.
David Hunt & Rebecca Devon
1 item of Media coverage
Press/Media : Press Release
Science behind DecodeME points to better treatments for Covid-19
Veronique Vitart
1 Media contribution
Press/Media : Public Engagement Activities
Evidence for 28 genetic disorders discovered by combining healthcare and research data
Press/Media : Research

- Edinburgh DataShare
- College of Medicine & Veterinary Medicine
- Edinburgh Medical School
- Molecular, Genetic and Population Health Sciences
- Institute of Genetics and Cancer
Medical Research Council Human Genetics Unit
Titles Date Accessioned
Full Text Search:
mrc-human-genetics-unit
Sub-communities within this community
E-mouse atlas, evolutionary genetics and genomics research group, collections in this community, biomedical genomics, quantitative trait locus (qtl) identification, regulatory genomics in evolution and disease, research data from the boulter-kendall research group, scottish diabetes research network type 1 bioresource, recent submissions.
Leveraging molecular-QTL co-association to predict novel disease-associated genetic loci using a graph convolutional neural network
Decodeme questionnaire , croatian cohorts data dictionary .

Prevalence of Y chromosome haplogroups by area of birth in UK Biobank
Additional data pertaining to publication: meta-gwas reveals novel genetic variants associated with urinary excretion of uromodulin , emriboseq ribo context analysis , gwas summary statistics pertaining to the publication "genetic regulation of post-translational modification of two distinct proteins" , superseded - gwas summary statistics pertaining to the publication "same role but different actors: genetic regulation of post-translational modification of two distinct proteins" , supplementary tables for david w clark's phd thesis "inbreeding depression on human complex traits" , traveller genes data dictionary , pathogenic loss-of-function, gain-of-function and dominant-negative mutations have profoundly different effects on protein structure: implications for variant effect prediction , whole slide images of armadillo liver from m.leprae dataset , viking ii data dictionary .

Wnt Pathway Analysis mapped gene-expression data

Wnt Pathway Analysis mapped gene-expression point-cloud data
Interface size drives cotranslational assembly of protein complexes , supporting data for: "the properties of human disease mutations at protein interfaces" , extended supplementary tables for thesis: "omics measures of ageing and disease susceptibility" by erin macdonald-dunlop .

Mutational bias in spermatogonia impacts the anatomy of regulatory sites in the human genome
Data pertaining to publication: meta-gwas reveals novel genetic variants associated with urinary excretion of uromodulin .
- Accessibility
- DataShare Privacy Notice
- Service level definition
- Freedom of information
- Share full article
Advertisement
Supported by
Special Issue
For Whom the Cell Mutates: The Origins of Genetic Quirks
By Sean B. Carroll
- March 14, 2011
There was very little that was safe or conventional in Ernest Hemingway’s life. The great writer hurled himself into danger in three wars, managed to survive two plane crashes on the same big-game safari in remote Africa, and precipitated many domestic dramas with a variety of love affairs in the course of his four marriages. “Moderation” appears to have been one word lacking from his otherwise superb vocabulary, even when it came to cats.
The image of the macho big-game hunter and marlin fisherman is hard to reconcile with that of a pet-hoarder, but Hemingway surrounded himself with felines. The author loved the animals and took in so many strays that at one point his house in Cuba, Finca Vigia (Lookout Farm), had 57 cats. Every animal had a name, including such unmasculine monikers as Princessa, Furhouse, and Littless Kitty. Hemingway loved their company when writing, especially when he was alone for lengths of time on the island. He incorporated many favorite pets into his short stories and novels. His loyal, longtime companion Boise merited 35 pages in “Islands in the Stream,” including this autobiographical passage:
“That night, when he had sat in the big chair reading with Boise at his side in the chair, he had thought that he did not know what he would do if Boise should be killed. He thought, from his actions and desperation, that the cat felt the same way about the man.”
But of all the cats in Hemingway’s life, the most famous are those that have taken up residence at his former home in Key West, Fla. In late 1931, Hemingway and his second wife, Pauline, moved into a two-story house on Whitfield Street. It was in Key West that Hemingway established a routine of writing in the morning, and then spending the hot afternoons fishing from the bridges, docks or a boat, or relaxing with friends.
It was an extremely productive lifestyle. Over a 12-year span in Key West, he worked on or completed “A Farewell to Arms,” “Green Hills of Africa,” “Death in the Afternoon,” “For Whom the Bell Tolls,” “To Have or Have Not,” and the short stories “The Snows of Kilimanjaro” and “The Short Happy Life of Francis Macomber.”
The house in Key West, which the author owned until his death 50 years ago this summer, is now a museum and a permanent home to about 50 cats. But of course, such an extraordinary man would not be associated with just ordinary cats; about half of the animals bear extra toes, typically on their forepaws. Most cats are, like humans, pentadactyls. That is, they have five digits on their forepaws. The so-called Hemingway cats have six digits, with the extra digit the homolog of the human thumb, which gives the paw a mittenlike appearance. When found in felines, the condition, formally known as preaxial polydactyly, is now commonly referred to as a “Hemingway cat.”
The origins of the cats on the Hemingway grounds are shrouded in legend, and it remains difficult to sort out facts from tall tales in many matters concerning the famous writer. One version of the cat story offered today is that Hemingway was given a six-toed tomcat, Snowball, by a ship’s captain in the mid-1930s, and that all of the six-toed residents are descended from this founding father.

Another account is that Hemingway had no housepets at the time and that the six-toed cats are descended from strays that came onto the property from time to time and took up residence after the writer was gone. Cats have long been present in Key West for controlling the rodent populations, and six-toed cats were popular with ship captains and sailors for the same purpose, as well as being considered good luck on voyages.
While the origins of the Hemingway House cats remains murky, the cause of their polydactyly is no longer a mystery. Researchers have recently pinpointed the precise mutation in the cats’ DNA responsible for the formation of the extra digit. The story of the origin of Hemingway’s cats is one of finding deep genetic connections among very different animals — from fruit flies to chickens, mice, cats, and yes, even humans.
In the late 1970s, one of the most challenging puzzles in all of biology was that of embryonic development — how a complex creature formed from a single fertilized egg cell. Two researchers, Christiane Nüsslein-Volhard and Eric Wieschaus, led a bold undertaking to identify all of the genes that were responsible for the process in fruit flies, then and now one of the main workhorses of basic genetic research. They identified scores of genes that played roles in building the fruit fly body and its body parts, work that led to their sharing of the 1995 Nobel Prize in Physiology or Medicine with the late Edward B. Lewis, another pioneer fruit fly geneticist.
One of the reasons for that honor was the discovery that, contrary to all biologists’ expectations, similar sets of genes to those involved in building fruit flies were also involved in building the bodies of such different animals as mice, frogs and other vertebrates, including humans. Indeed, by the 1990s, one common strategy for discovering genes involved in building vertebrate bodies, organs or body parts was to look for the counterparts of fruit fly genes in those vertebrates.
That was the approach taken by one team led by Prof. Cliff Tabin at Harvard Medical School that was eager to find the genes responsible for the formation and patterning of vertebrate limbs. Decades of research on the chicken wing had shown that the formation of the pattern of digits across the entire structure depended on some signal produced by cells in the most posterior part of the developing embryo’s wing bud. Professor Tabin’s team sought to identify that signal by isolating the chicken’s counterparts of certain fruit fly genes.
They isolated a chicken homolog of a fruit fly gene called “hedgehog.” The name had been given by Dr. Nüsslein-Volhard and Dr. Wieschaus because mutations in the fly gene caused the fruit fly larva to be covered with fine hairs, like a hedgehog.
Professor Tabin’s team was stunned and delighted to find that the chicken gene, dubbed “Sonic hedgehog” after the video game character, was turned on in the posterior of the limb bud, right where the digit-patterning activity was also located. They then found that the Sonic hedgehog protein was indeed the long-sought digit-patterning signal. For instance, they demonstrated that turning Sonic hedgehog on in the anterior part of the limb bud had the same effect as transplanting posterior tissue to the anterior part of the limb — it caused the formation of extra digits.
The induction of polydactyly by Sonic hedgehog in laboratory experiments raised the possibility that inherited cases of polydactyly might be caused by mutations in the Sonic hedgehog gene. Polydactyly is well known in mice and in humans, as well as cats, and sure enough, cats, mice and humans all have Sonic hedgehog genes. But inspection of the Sonic hedgehog genes of polydactyl individuals did not reveal any mutations that would cause the Sonic hedgehog signal to be defective.
So what role, then, does Sonic hedgehog play in the syndrome? It turns out that polydactyly is not due to disruptions of Sonic hedgehog function, but of its regulation. In order to make the proper five-digit, pinky-to-thumb pattern, the production of the Sonic hedgehog protein must be restricted to posterior cells. The pattern of the digits depends upon the relative concentration of Sonic hedgehog, which is greatest in the posterior (where the pinky will form) and lowest in the anterior where the thumb will form. Mutations that disrupt Sonic hedgehog regulation such that some protein is made in the anterior of the limb bud cause the formation of an extra thumb.
These mutations were difficult to find in DNA at first because they were not located in the part of the gene that encodes the protein. Rather, they occurred far away in a stretch of DNA sequence that acts like a switch to turn Sonic hedgehog on in the posterior part of the limb bud and to keep the gene off in cells in the anterior part of the limb bud.
A team led by Robert Hill of the Medical Research Council Human Genetics Unit in Edinburgh showed that the Sonic hedgehog switch that controls gene activity in the limb is located about one million base pairs from the part of the gene encoding the Sonic hedgehog signal. On the scale of DNA, finding the mutations so far away from the gene was much like looking for one’s car in a parking lot, and eventually finding it in a vacant field in another town.
Mutations that scramble the switch are responsible for polydactyly in mice and humans. But further work has shown that even slight mutations substituting just a single letter of the DNA sequence can also cause the syndrome, not only in mice and humans, but in Hemingway’s cats.
Mr. Hill’s team analyzed one line of affected Key West cats and found a perfect association between polydactyly and a substitution at one position in the cat Sonic hedgehog gene switch. They also examined other unrelated polydactylous North American cats and found the same substitution, which indicates that all North American polydactylous cats may be descended from one polydactylous ancestor . If that is indeed the case, that ancestor may date back as early as pre-Revolutionary times in New England, and its descendants probably reached Key West by ship long before Hemingway did.
Like Hemingway’s cats and the writer himself, we all have our quirks, some more visible than others. Genetics has made huge strides in understanding the basis of many physical characteristics, like extra toes and fingers. In time, we can look forward to learning more about the genetics of deeper mysteries, like the cause of the profound depression that overtook Hemingway and many of his close relatives or, on the brighter side, perhaps some insights into the source of his great talent.

MRC Mitochondrial Biology Unit
Research at the Medical Research Council (MRC) Mitochondrial Biology Unit (MRC MBU) is focused on the biology of mitochondria and their dysfunction in an ever-increasing range of human diseases. The Unit combines studies exploring the molecular function of the mitochondrial oxidative phosphorylation system, the mitochondrial proteome and genome, and how mitochondria interact with the cell through homeostatic, signalling and execution pathways. Combined with the wealth of clinical, genetic and biochemical data provided by mitochondrial medicine, and the use of model systems with perturbed mitochondrial physiology, the Unit aims to exploit its findings for the development of new therapies to treat human disease.
Find out more about the MRC Mitochondrial Biology Unit .
Last updated: 17 August 2023
This is the website for UKRI: our seven research councils, Research England and Innovate UK. Let us know if you have feedback or would like to help improve our online products and services .
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Perspective
- Published: 13 May 2024
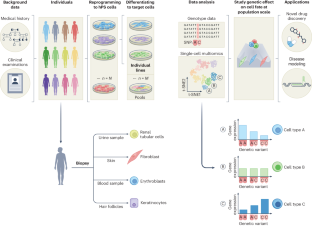
Integrating population genetics, stem cell biology and cellular genomics to study complex human diseases
- Nona Farbehi ORCID: orcid.org/0000-0001-8461-236X 1 , 2 , 3 na1 ,
- Drew R. Neavin ORCID: orcid.org/0000-0002-1783-6491 1 na1 ,
- Anna S. E. Cuomo 1 , 4 ,
- Lorenz Studer ORCID: orcid.org/0000-0003-0741-7987 3 , 5 ,
- Daniel G. MacArthur 4 , 6 &
- Joseph E. Powell ORCID: orcid.org/0000-0002-5070-4124 1 , 3 , 7
Nature Genetics volume 56 , pages 758–766 ( 2024 ) Cite this article
2710 Accesses
13 Altmetric
Metrics details
- Population genetics
- Transcriptomics
Human pluripotent stem (hPS) cells can, in theory, be differentiated into any cell type, making them a powerful in vitro model for human biology. Recent technological advances have facilitated large-scale hPS cell studies that allow investigation of the genetic regulation of molecular phenotypes and their contribution to high-order phenotypes such as human disease. Integrating hPS cells with single-cell sequencing makes identifying context-dependent genetic effects during cell development or upon experimental manipulation possible. Here we discuss how the intersection of stem cell biology, population genetics and cellular genomics can help resolve the functional consequences of human genetic variation. We examine the critical challenges of integrating these fields and approaches to scaling them cost-effectively and practically. We highlight two areas of human biology that can particularly benefit from population-scale hPS cell studies, elucidating mechanisms underlying complex disease risk loci and evaluating relationships between common genetic variation and pharmacotherapeutic phenotypes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
24,99 € / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
195,33 € per year
only 16,28 € per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others

Identifying proteomic risk factors for cancer using prospective and exome analyses of 1463 circulating proteins and risk of 19 cancers in the UK Biobank

In vitro reconstitution of epigenetic reprogramming in the human germ line
A deep catalogue of protein-coding variation in 983,578 individuals.
Thomson, J. A. Embryonic stem cell lines derived from human blastocysts. Science https://doi.org/10.1126/science.282.5391.1145 (1998).
Takahashi, K. & Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126 , 663–676 (2006).
Article CAS PubMed Google Scholar
Takahashi, K. et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131 , 861–872 (2007).
Liu, G., David, B. T., Trawczynski, M. & Fessler, R. G. Advances in pluripotent stem cells: history, mechanisms, technologies, and applications. Stem Cell Rev. Rep. 16 , 3–32 (2020).
Article PubMed Google Scholar
Efrat, S. Epigenetic memory: lessons from iPS cells derived from human β cells. Front. Endocrinol. 11 , 614234 (2020).
Article Google Scholar
Anderson, R. H. & Francis, K. R. Modeling rare diseases with induced pluripotent stem cell technology. Mol. Cell. Probes 40 , 52–59 (2018).
Article CAS PubMed PubMed Central Google Scholar
Spitalieri, P., Talarico, V. R., Murdocca, M., Novelli, G. & Sangiuolo, F. Human induced pluripotent stem cells for monogenic disease modelling and therapy. World J. Stem Cells 8 , 118–135 (2016).
Article PubMed PubMed Central Google Scholar
Passier, R., Orlova, V. & Mummery, C. Complex tissue and disease modeling using hiPSCs. Cell Stem Cell 18 , 309–321 (2016).
Warren, C. R., Jaquish, C. E. & Cowan, C. A. The NextGen genetic association studies consortium: a foray into in vitro population genetics. Cell Stem Cell 20 , 431–433 (2017).
Visscher, P. M., Brown, M. A., McCarthy, M. I. & Yang, J. Five years of GWAS discovery. Am. J. Hum. Genet. 90 , 7–24 (2012).
Tak, Y. G. & Farnham, P. J. Making sense of GWAS: using epigenomics and genome engineering to understand the functional relevance of SNPs in non-coding regions of the human genome. Epigenetics Chromatin 8 , 57 (2015).
Umans, B. D., Battle, A. & Gilad, Y. Where are the disease-associated eQTLs? Trends Genet. 37 , 109–124 (2021).
Yazar, S. et al. Single-cell eQTL mapping identifies cell type–specific genetic control of autoimmune disease. Science 376 , eabf3041 (2022).
Jerber, J. et al. Population-scale single-cell RNA-seq profiling across dopaminergic neuron differentiation. Nat. Genet. 53 , 304–312 (2021).
Neavin, D. et al. Single cell eQTL analysis identifies cell type-specific genetic control of gene expression in fibroblasts and reprogrammed induced pluripotent stem cells. Genome Biol. 22 , 76 (2021).
Cuomo, A. S. E. et al. Single-cell RNA-sequencing of differentiating iPS cells reveals dynamic genetic effects on gene expression. Nat. Commun. 11 , 810 (2020).
Warren, C. R. et al. Induced pluripotent stem cell differentiation enables functional validation of GWAS variants in metabolic disease. Cell Stem Cell 20 , 547–557 (2017).
Kishore, S. et al. A non-coding disease modifier of pancreatic agenesis identified by genetic correction in a patient-derived iPSC line. Cell Stem Cell 27 , 137–146 (2020).
Magdy, T. et al. RARG variant predictive of doxorubicin-induced cardiotoxicity identifies a cardioprotective therapy. Cell Stem Cell 28 , 2076–2089 (2021).
Bourgeois, S. et al. Towards a functional cure for diabetes using stem cell-derived beta cells: are we there yet? Cells 10 , 191 (2021).
Sharma, A., Sances, S., Workman, M. J. & Svendsen, C. N. Multi-lineage human iPSC-derived platforms for disease modeling and drug discovery. Cell Stem Cell 26 , 309–329 (2020).
Volpato, V. & Webber, C. Addressing variability in iPSC-derived models of human disease: guidelines to promote reproducibility. Dis. Model. Mech. 13 , dmm042317 (2020).
Banovich, N. E. et al. Impact of regulatory variation across human iPSCs and differentiated cells. Genome Res. 28 , 122–131 (2018).
Kilpinen, H. et al. Common genetic variation drives molecular heterogeneity in human iPSCs. Nature 546 , 370–375 (2017).
Panopoulos, A. D. et al. iPSCORE: a resource of 222 iPSC lines enabling functional characterization of genetic variation across a variety of cell types. Stem Cell Rep. 8 , 1086–1100 (2017).
Article CAS Google Scholar
Chen, G., Ning, B. & Shi, T. Single-cell RNA-seq technologies and related computational data analysis. Front. Genet. 10 , 317 (2019).
Elorbany, R. et al. Single-cell sequencing reveals lineage-specific dynamic genetic regulation of gene expression during human cardiomyocyte differentiation. PLoS Genet. 18 , e1009666 (2022).
Ward, M. C., Banovich, N. E., Sarkar, A., Stephens, M. & Gilad, Y. Dynamic effects of genetic variation on gene expression revealed following hypoxic stress in cardiomyocytes. eLife 10 , e57345 (2021).
Shi, Z.-D. et al. Genome editing in hPSCs reveals GATA6 haploinsufficiency and a genetic interaction with GATA4 in human pancreatic development. Cell Stem Cell 20 , 675–688 (2017).
Strober, B. J. et al. Dynamic genetic regulation of gene expression during cellular differentiation. Science 364 , 1287–1290 (2019).
González, F. et al. An iCRISPR platform for rapid, multiplexable, and inducible genome editing in human pluripotent stem cells. Cell Stem Cell 15 , 215–226 (2014).
Barbeira, A. N. et al. Exploiting the GTEx resources to decipher the mechanisms at GWAS loci. Genome Biol. 22 , 49 (2021).
Hamazaki, T., El Rouby, N., Fredette, N. C., Santostefano, K. E. & Terada, N. Concise review: induced pluripotent stem cell research in the era of precision medicine. Stem Cells 35 , 545–550 (2017).
Cuomo, A. S. E. et al. CellRegMap: a statistical framework for mapping context-specific regulatory variants using scRNA-seq. Mol. Syst. Biol. 18 , e10663 (2022).
Cuomo, A. S. E., Nathan, A., Raychaudhuri, S., MacArthur, D. G. & Powell, J. E. Single-cell genomics meets human genetics. Nat. Rev. Genet. 24 , 535–549 (2023).
Mirauta, B. A. et al. Population-scale proteome variation in human induced pluripotent stem cells. eLife 9 , e57390 (2020).
Findley, A. S. et al. Functional dynamic genetic effects on gene regulation are specific to particular cell types and environmental conditions. eLife 10 , e67077 (2021).
Kimura, M. et al. En masse organoid phenotyping informs metabolic-associated genetic susceptibility to NASH. Cell https://doi.org/10.1016/j.cell.2022.09.031 (2022).
Llufrio, E. M., Wang, L., Naser, F. J. & Patti, G. J. Sorting cells alters their redox state and cellular metabolome. Redox Biol. 16 , 381–387 (2018).
Shen, S. et al. Integrating single-cell genomics pipelines to discover mechanisms of stem cell differentiation. Trends Mol. Med. https://doi.org/10.1016/j.molmed.2021.09.006 (2021).
van der Wijst, M. et al. The single-cell eQTLGen consortium. eLife 9 , e52155 (2020).
Soskic, B. et al. Immune disease risk variants regulate gene expression dynamics during CD4 + T cell activation. Nat. Genet. 54 , 817–826 (2022).
Daniszewski, M. et al. Retinal ganglion cell-specific genetic regulation in primary open-angle glaucoma. Cell Genomics 2 , 100142 (2022).
Senabouth, A. et al. Transcriptomic and proteomic retinal pigment epithelium signatures of age-related macular degeneration. Nat. Commun. 13 , 4233 (2022).
Benaglio, P. et al. Mapping genetic effects on cell type-specific chromatin accessibility and annotating complex immune trait variants using single nucleus ATAC-seq in peripheral blood. PLoS Genet. 19 , e1010759 (2023).
Baysoy, A., Bai, Z., Satija, R. & Fan, R. The technological landscape and applications of single-cell multi-omics. Nat. Rev. Mol. Cell Biol. 24 , 695–713 (2023).
Weinshilboum, R. M. & Wang, L. Pharmacogenomics: precision medicine and drug response. Mayo Clin. Proc. 92 , 1711–1722 (2017).
Pirmohamed, M. Personalized pharmacogenomics: predicting efficacy and adverse drug reactions. Annu. Rev. Genom. Hum. Genet. 15 , 349–370 (2014).
Nelson, M. R. et al. The support of human genetic evidence for approved drug indications. Nat. Genet. 47 , 856–860 (2015).
Hay, M., Thomas, D. W., Craighead, J. L., Economides, C. & Rosenthal, J. Clinical development success rates for investigational drugs. Nat. Biotechnol. 32 , 40–51 (2014).
Holmgren, G. et al. Long-term chronic toxicity testing using human pluripotent stem cell-derived hepatocytes. Drug Metab. Dispos. 42 , 1401–1406 (2014).
Kim, J.-H., Kang, M., Jung, J.-H., Lee, S.-J. & Hong, S.-H. Human pluripotent stem cell-derived alveolar epithelial cells as a tool to assess cytotoxicity of particulate matter and cigarette smoke extract. Dev. Reprod. 26 , 155–163 (2022).
Sharma, A. et al. High-throughput screening of tyrosine kinase inhibitor cardiotoxicity with human induced pluripotent stem cells. Sci. Transl. Med. 9 , eaaf2584 (2017).
Han, Y. et al. Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature 589 , 270–275 (2021).
Lam, C. K. & Wu, J. C. Clinical trial in a dish: using patient-derived induced pluripotent stem cells to identify risks of drug-induced cardiotoxicity. Arterioscler. Thromb. Vasc. Biol. 41 , 1019–1031 (2021).
Iwata, R. et al. Mitochondria metabolism sets the species-specific tempo of neuronal development. Science 379 , eabn4705 (2023).
Miller, J. D. et al. Human iPSC-based modeling of late-onset disease via progerin-induced aging. Cell Stem Cell 13 , 691–705 (2013).
Hergenreder, E. et al. Combined small-molecule treatment accelerates maturation of human pluripotent stem cell-derived neurons. Nat. Biotechnol. https://doi.org/10.1038/s41587-023-02031-z (2024).
Fowler, J. L., Ang, L. T. & Loh, K. M. A critical look: challenges in differentiating human pluripotent stem cells into desired cell types and organoids. Wiley Interdiscip. Rev. Dev. Biol. 9 , e368 (2020).
Jiang, S., Feng, W., Chang, C. & Li, G. Modeling human heart development and congenital defects using organoids: how close are we? J. Cardiovasc. Dev. Dis. 9 , 125 (2022).
CAS PubMed PubMed Central Google Scholar
Tremmel, D. M. et al. Validating expression of beta cell maturation-associated genes in human pancreas development. Front. Cell Dev. Biol. 11 , 1103719 (2023).
Washer, S. J. et al. Single-cell transcriptomics defines an improved, validated monoculture protocol for differentiation of human iPSC to microglia. Sci. Rep. 12 , 19454 (2022).
Wolf, F. A., Angerer, P. & Theis, F. J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19 , 15 (2018).
Wilson, S. B. et al. DevKidCC allows for robust classification and direct comparisons of kidney organoid datasets. Genome Med. 14 , 19 (2022).
Subramanian, A. et al. Single cell census of human kidney organoids shows reproducibility and diminished off-target cells after transplantation. Nat. Commun. 10 , 5462 (2019).
Kammers, K. et al. Gene and protein expression in human megakaryocytes derived from induced pluripotent stem cells. J. Thromb. Haemost. 19 , 1783–1799 (2021).
De Sousa, P. A. et al. Rapid establishment of the European Bank for induced Pluripotent Stem Cells (EBiSC)—the Hot Start experience. Stem Cell Res. 20 , 105–114 (2017).
Morrison, M. et al. StemBANCC: governing access to material and data in a large stem cell research consortium. Stem Cell Rev. Rep. 11 , 681–687 (2015).
The GTEx Consortium The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 369 , 1318–1330 (2020).
Article PubMed Central Google Scholar
Mitchell, J. M., Nemesh, J., Ghosh, S. & Handsaker, R. E. Mapping genetic effects on cellular phenotypes with ‘cell villages’. Preprint at bioRxiv https://doi.org/10.1101/2020.06.29.174383 (2020).
Neavin, D. R. et al. A village in a dish model system for population-scale hiPSC studies. Nat. Commun. 14 , 3240 (2023).
Kang, H. M. et al. Multiplexed droplet single-cell RNA-sequencing using natural genetic variation. Nat. Biotechnol. 36 , 89–94 (2018).
Wells, M. F. et al. Natural variation in gene expression and viral susceptibility revealed by neural progenitor cell villages. Cell Stem Cell 30 , 312–332 (2023).
Neavin, D. et al. Demuxafy : improvement in droplet assignment by integrating multiple single-cell demultiplexing and doublet detection methods. Genome Biol. 25 , 94 (2024).
Xu, J. et al. Genotype-free demultiplexing of pooled single-cell RNA-seq. Genome Biol. 20 , 290 (2019).
Heaton, H. et al. Souporcell: robust clustering of single-cell RNA-seq data by genotype without reference genotypes. Nat. Methods 17 , 615–620 (2020).
Huang, Y., McCarthy, D. J. & Stegle, O. Vireo: Bayesian demultiplexing of pooled single-cell RNA-seq data without genotype reference. Genome Biol. 20 , 273 (2019).
Hindson, B. J. et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem. 83 , 8604–8610 (2011).
Dong, X. et al. powerEQTL: an R package and shiny application for sample size and power calculation of bulk tissue and single-cell eQTL analysis. Bioinformatics https://doi.org/10.1093/bioinformatics/btab385 (2021).
Schmid, K. T. et al. scPower accelerates and optimizes the design of multi-sample single cell transcriptomic studies. Nat. Commun. 12 , 6625 (2021).
Camp, J. G., Platt, R. & Treutlein, B. Mapping human cell phenotypes to genotypes with single-cell genomics. Science 365 , 1401–1405 (2019).
Datlinger, P. et al. Pooled CRISPR screening with single-cell transcriptome readout. Nat. Methods 14 , 297–301 (2017).
Dixit, A. et al. Perturb-Seq: dissecting molecular circuits with scalable single-cell RNA profiling of pooled genetic screens. Cell 167 , 1853–1866 (2016).
Rubin, A. J. et al. Coupled single-cell CRISPR screening and epigenomic profiling reveals causal gene regulatory networks. Cell 176 , 361–376 (2019).
Schraivogel, D. et al. Targeted Perturb-seq enables genome-scale genetic screens in single cells. Nat. Methods 17 , 629–635 (2020).
Download references
Acknowledgements
Figures were generated with BioRender.com and further developed by A. Garcia, a scientific illustrator from Bio-Graphics. This research was supported by a National Health and Medical Research Council (NHMRC) Investigator grant (J.E.P., 1175781), research grants from the Australian Research Council (ARC) Special Research Initiative in Stem Cell Science, an ARC Discovery Project (190100825), an EMBO Postdoctoral Fellowship (A.S.E.C.) and an Aligning Science Across Parkinson’s Grant (J.E.P., N.F., D.R.N. and L.S.). J.E.P. is supported by a Fok Family Fellowship.
Author information
These authors contributed equally: Nona Farbehi, Drew R. Neavin.
Authors and Affiliations
Garvan Weizmann Center for Cellular Genomics, Garvan Institute of Medical Research, Sydney, New South Wales, Australia
Nona Farbehi, Drew R. Neavin, Anna S. E. Cuomo & Joseph E. Powell
Graduate School of Biomedical Engineering, University of New South Wales, Sydney, New South Wales, Australia
Nona Farbehi
Aligning Science Across Parkinson’s Collaborative Research Network, Chevy Chase, MD, USA
Nona Farbehi, Lorenz Studer & Joseph E. Powell
Centre for Population Genomics, Garvan Institute of Medical Research, University of New South Wales, Sydney, New South Wales, Australia
Anna S. E. Cuomo & Daniel G. MacArthur
The Center for Stem Cell Biology and Developmental Biology Program, Sloan-Kettering Institute for Cancer Research, New York, NY, USA
Lorenz Studer
Centre for Population Genomics, Murdoch Children’s Research Institute, Melbourne, Victoria, Australia
Daniel G. MacArthur
UNSW Cellular Genomics Futures Institute, University of New South Wales, Sydney, New South Wales, Australia
Joseph E. Powell
You can also search for this author in PubMed Google Scholar
Contributions
All authors conceived the topic and wrote and revised the manuscript.
Corresponding author
Correspondence to Joseph E. Powell .
Ethics declarations
Competing interests.
D.G.M. is a founder with equity in Goldfinch Bio, is a paid advisor to GSK, Insitro, Third Rock Ventures and Foresite Labs, and has received research support from AbbVie, Astellas, Biogen, BioMarin, Eisai, Merck, Pfizer and Sanofi-Genzyme; none of these activities is related to the work presented here. J.E.P. is a founder with equity in Celltellus Laboratory and has received research support from Illumina. The other authors declare no conflict of interest.
Peer review
Peer review information.
Nature Genetics thanks Kelly Frazer, Gosia Trynka and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information.
Supplementary Table 1.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Reprints and permissions
About this article
Cite this article.
Farbehi, N., Neavin, D.R., Cuomo, A.S.E. et al. Integrating population genetics, stem cell biology and cellular genomics to study complex human diseases. Nat Genet 56 , 758–766 (2024). https://doi.org/10.1038/s41588-024-01731-9
Download citation
Received : 24 January 2023
Accepted : 20 March 2024
Published : 13 May 2024
Issue Date : May 2024
DOI : https://doi.org/10.1038/s41588-024-01731-9
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing newsletter — what matters in science, free to your inbox daily.
- U.S. Department of Health & Human Services

- Virtual Tour
- Staff Directory
- En Español
You are here
News releases.
News Release
Friday, May 24, 2024
Scientists map networks regulating gene function in the human brain
NIH-funded research details the brain’s cellular and molecular regulatory elements and their impact on brain function.
A consortium of researchers has produced the largest and most advanced multidimensional maps of gene regulation networks in the brains of people with and without mental disorders. These maps detail the many regulatory elements that coordinate the brain’s biological pathways and cellular functions. The research, supported the National Institutes of Health (NIH), used postmortem brain tissue from over 2,500 donors to map gene regulation networks across different stages of brain development and multiple brain-related disorders.
“These groundbreaking findings advance our understanding of where, how, and when genetic risk contributes to mental disorders such as schizophrenia, post-traumatic stress disorder, and depression,” said Joshua A. Gordon, M.D., Ph.D., director of NIH’s National Institute of Mental Health (NIMH). “Moreover, the critical resources, shared freely, will help researchers pinpoint genetic variants that are likely to play a causal role in mental illnesses and identify potential molecular targets for new therapeutics.”
The research is published across 15 papers in Science, Science Advances, and Scientific Reports. The papers report findings along several key themes:
- Population-level analyses that link genetic variants, regulatory elements, and different molecular forms of expressed genes to regulatory networks at the cellular level, in both the developing brain and adult brain
- Single-cell-level maps of the prefrontal cortex from individuals diagnosed with mental disorders and neurodevelopmental disorders
- Experimental analyses validating the function of regulatory elements and genetic variants associated with quantitative trait loci (segments of DNA that are linked with observable traits)
The analyses expand on previous findings, exploring multiple cortical and subcortical regions of the human brain. These brain areas play key roles in a range of essential processes, including decision-making, memory, learning, emotion, reward processing, and motor control.
Approximately 2% of the human genome is composed of genes that code for proteins. The remaining 98% includes DNA segments that help regulate the activity of those genes. To better understand how brain structure and function contribute to mental disorders, researchers in the NIMH-funded PsychENCODE Consortium are using standardized methods and data analysis approaches to build a comprehensive picture of these regulatory elements in the human brain.
In addition to these discoveries, the papers also highlight new methods and tools to help researchers analyze and explore the wealth of data produced by this effort. These resources include a web-based platform offering interactive visualization data from diverse brain cell types in individuals with and without mental disorders, known as PsychSCREEN. Together, these methods and tools provide a comprehensive, integrated data resource for the broader research community.
The papers focus on the second phase of findings from the PsychENCODE Consortium. This effort aims to advance our understanding of how gene regulation impacts brain function and dysfunction.
“These PsychENCODE Consortium findings shed new light on how gene risk maps onto brain function across developmental stages, brain regions, and disorders,” said Jonathan Pevsner, Ph.D., chief of the NIMH Genomics Research Branch. “The work lays a strong foundation for ongoing efforts to characterize regulatory pathways across disorders, elucidate the role of epigenetic mechanisms, and increase the ancestral diversity represented in studies.”
The PsychENCODE papers published in Science and Science Advances are presented as a collection on the Science website.
About the National Institute of Mental Health (NIMH): The mission of the NIMH is to transform the understanding and treatment of mental illnesses through basic and clinical research, paving the way for prevention, recovery and cure. For more information, visit the NIMH website.
About the National Institutes of Health (NIH): NIH, the nation's medical research agency, includes 27 Institutes and Centers and is a component of the U.S. Department of Health and Human Services. NIH is the primary federal agency conducting and supporting basic, clinical, and translational medical research, and is investigating the causes, treatments, and cures for both common and rare diseases. For more information about NIH and its programs, visit www.nih.gov .
NIH…Turning Discovery Into Health ®
Connect with Us
- More Social Media from NIH
- Alzheimer's disease & dementia
- Arthritis & Rheumatism
- Attention deficit disorders
- Autism spectrum disorders
- Biomedical technology
- Diseases, Conditions, Syndromes
- Endocrinology & Metabolism
- Gastroenterology
- Gerontology & Geriatrics
- Health informatics
- Inflammatory disorders
- Medical economics
- Medical research
- Medications
- Neuroscience
- Obstetrics & gynaecology
- Oncology & Cancer
- Ophthalmology
- Overweight & Obesity
- Parkinson's & Movement disorders
- Psychology & Psychiatry
- Radiology & Imaging
- Sleep disorders
- Sports medicine & Kinesiology
- Vaccination
- Breast cancer
- Cardiovascular disease
- Chronic obstructive pulmonary disease
- Colon cancer
- Coronary artery disease
- Heart attack
- Heart disease
- High blood pressure
- Kidney disease
- Lung cancer
- Multiple sclerosis
- Myocardial infarction
- Ovarian cancer
- Post traumatic stress disorder
- Rheumatoid arthritis
- Schizophrenia
- Skin cancer
- Type 2 diabetes
- Full List »
share this!
May 29, 2024
This article has been reviewed according to Science X's editorial process and policies . Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
Research team creates mice that better reflect human genetic variation
by Ben Whitford, Jackson Laboratory

The great majority of how we understand human disease, and attempt to cure it, derives from mice genetically fashioned to be prolific breeders, docile and easy to handle—all qualities that have made it the favorite tool of large-scale biomedical research. However, this human-imposed selection of these behavioral and reproductive traits has come at a hefty cost. By weeding out undesirable traits in the highly standardized laboratory mouse, researchers have also placed unseen constraints on what is possible to know and learn from them.
Beth Dumont, an evolutionary biologist at The Jackson Laboratory, and colleagues are looking to remedy this situation. By collecting mice in their natural habitats, in fields, barns and forests, and in divergent ecological niches (polar, tropical, and arid climates) across North and South America, Dumont and colleagues have developed 10 new laboratory -grade research mouse strains with genomes packed with information that had been stamped out of classical mice.
These new mouse strains, detailed recently in PLoS Genetics , will provide an important new resource for researchers worldwide. Their genomes introduce millions of novel genetic variants compared to classical in-bred strains, including predicted versions of a gene that—on average—decreases the fitness of the organism carrying it, and gene-spanning structural variants, including loss of DNA, duplicated DNA, and detached chromosomes that reattach in the opposite direction—all genetic profiles that better reflect human genetic variation .
"Since the early 1900s, we've actively removed a lot of genetic information from laboratory mice that is incredibly relevant to human health," said Dumont. "We've bred out traits that relate to anxiety, aggressiveness, and infertility, for instance, and that means we're missing out on a wealth of potentially transformative biomedical research , making it much harder to determine how effectively new treatments or pharmaceuticals will work in humans."
Dumont and her colleague, Professor Michael W. Nachman at the University of California, Berkeley and teams of graduate students went to work. They collected—with some difficulty—wild mice across five locations in Canada, the United States, and Brazil. These wild mice, which all belonged to a single mouse subspecies (M. musculus domesticus; important because they can all interbreed, which is not necessarily the case with all genetically diverse mice) looked very different from laboratory mice, having experienced adaptive pressures to survive and thrive in their respective environments.
For example, mice from Canada were bigger and had high metabolic activity to stay warm in a colder climate; mice from Brazil were smaller and their metabolism was radically different, indicating that mice collected in Brazil were uniquely adapted to a hot climate.
Dumont, Nachman and colleagues meticulously inbred these mice for 20 generations to eliminate deleterious genes while imposing very minimal selection for docile behavior and/or reproductive output. Dumont then re-derived these mice via IVF to prevent the introduction of wild pathogens, making them now suitable for laboratory use—if you can catch them. The new strains are also noticeably feistier.
"They're fast, and they don't want you to hold them," said Dumont. "I haven't been bitten, but working with them does require quick reflexes."
The wild-derived laboratory strains showed variation in phenotypic traits beyond size and speed. The introduction of millions of genetic variants captured broad variations across many biological domains. Their biochemical, neurobehavioral, physiological, morphological, and metabolic traits differed much more broadly than those in inbred laboratory mice, more accurately modeling the complex genetic basis of human disease -related phenotypes.
"Tapping into wild strains has the potential to establish a powerful suite of resources for the modeling of human traits and diseases, enabling important discoveries across pretty much every disease area," said Dumont. "As researchers start to recognize that, we'll see increasing interest in wild-derived lab mice ."
Explore further
Feedback to editors

Improving cell therapy by creating T-cell 'super soldiers'
11 hours ago

Study shows more than just social media use may be causing depression in young adults


More out-of-state patients seek abortions in Washington state

Researchers develop microneedle patch that can detect skin cancer early
Blood flow makes waves across the surface of the mouse brain
12 hours ago

Study finds that memory complaints can predict biological changes in the brain

Research shows robotic ultrasound systems can aid doctors during surgery
13 hours ago

Researchers uncover surprising role of opioid receptors in gut development
Could a medicated foam make gene therapies more accessible?
14 hours ago

Novel method combines nano informatics and AI for advances in cancer prediction
Related stories.

City mouse or country mouse? Biologist collects mice from homes to study how they got so good at urban living
Mar 16, 2024

Unraveling the diversity of the wild house mouse
Jun 14, 2022

Eastern and Western house mice took parallel evolutionary paths after colonizing US
Apr 29, 2021

New study reveals details across 20 diverse inbred mouse strains
Apr 5, 2023
Researchers make mice a more powerful tool to study a wide range of human diseases
Apr 3, 2024
Gut bacteria from wild mice boost health in lab mice
Oct 19, 2017
Recommended for you

CRISPR-based mapping uncovers 'switches' for immune genes central to health
16 hours ago
Antibody–peptide inhibitor conjugates: A new path for cancer therapy
Grow the skin you're in: In vivo generation of chimeric skin grafts
15 hours ago
The use of new technologies expands understanding of brain tumors in children
Let us know if there is a problem with our content.
Use this form if you have come across a typo, inaccuracy or would like to send an edit request for the content on this page. For general inquiries, please use our contact form . For general feedback, use the public comments section below (please adhere to guidelines ).
Please select the most appropriate category to facilitate processing of your request
Thank you for taking time to provide your feedback to the editors.
Your feedback is important to us. However, we do not guarantee individual replies due to the high volume of messages.
E-mail the story
Your email address is used only to let the recipient know who sent the email. Neither your address nor the recipient's address will be used for any other purpose. The information you enter will appear in your e-mail message and is not retained by Medical Xpress in any form.
Newsletter sign up
Get weekly and/or daily updates delivered to your inbox. You can unsubscribe at any time and we'll never share your details to third parties.
More information Privacy policy
Donate and enjoy an ad-free experience
We keep our content available to everyone. Consider supporting Science X's mission by getting a premium account.
E-mail newsletter

- Schools & departments
Professor Chris Ponting
Group Leader

- Institute of Genetics and Cancer
Contact details
- Email: [email protected]
- MRC Human Genetics Unit
Professor Chris Ponting is Chair of Medical Bioinformatics and a Principal Investigator at the MRC Human Genetics Unit, Institute of Genetics and Cancer. Chris started his research in particle physics before moving via biophysics to bioinformatics and genomics. Aside from one year at the National Centre for Biotechnology Information (NIH, Bethesda, MD), he pursued his research at the University of Oxford before moving to Edinburgh in 2016. His research group has made substantial contributions to protein science, evolutionary biology, genetics and genomics. Early in his career he discovered many important protein domain families. He then provided the first evolutionary analyses for mammalian genomes whilst leading protein analysis teams for the human and mouse genome sequencing projects. More recently, his research established that 8.2% of the human genome is constrained, and thus is likely functional.
Chris has been on Editorial Boards of Genome Research, Genome Biology, Human Molecular Genetics, Annual Review of Genomics and Human Genetics, and Trends in Genetics, and was a Senior Editor of eLife until 2015. He served as Program Committee member for the CSHL Biology of Genomes, American Society of Human Genetics and Genome Science conferences. He was Head of the UK Node of ELIXIR and Chair of EMBL-EBI’s External Training Advisory Group and founded CGAT ( www.cgat.org ), an MRC-funded training centre. Professor Ponting is a Fellow of the Academy of Medical Sciences and a Member of the European Molecular Biology Organisation.
Qualifications
Bachelor 1986, B.A. (2:1, Hons., physics), University of Oxford, UK
- Masters 1988, M.Sc. (1, physics), University of British Columbia, Canada
- Doctoral 1991, Doctor of Philosophy (biophysics), DPhil, University of Oxford
Publications
- GWAS and meta-analysis identifies 49 genetic variants underlying critical Covid-19 (30 pages) 17 May 2023 In: Nature, vol. 617, pp. 763-793 DOI : https://doi.org/10.1038/s41586-023-06034-3 Research output: Contribution to Journal › Article (E-pub ahead of print)
- Human Genetics Seen Through An Evolutionary Lens 10 May 2023 In: Cell Genomics, vol. 3 DOI : https://doi.org/10.1016/j.xgen.2023.100323 Research output: Contribution to Journal › Article (Published)
- An ependymal cell census identifies heterogeneous and ongoing cell maturation in the adult mouse spinal cord that changes dynamically on injury 19 Jan 2023 In: Developmental Cell, vol. 58, pp. 239-255.e10 DOI : https://doi.org/10.1016/j.devcel.2023.01.003 Research output: Contribution to Journal › Article (Published)
- OAF: a new member of the BRICHOS family (5 pages) 24 Nov 2022 In: Bioinformatics Advances, vol. 2 DOI : https://doi.org/10.1093/bioadv/vbac087 Research output: Contribution to Journal › Article (Published)
- Genome-Wide Analysis of Human Long Noncoding RNAs: A Provocative Review 31 Aug 2022 In: Annual review of genomics and human genetics, vol. 23, pp. 153-172 DOI : https://doi.org/10.1146/annurev-genom-112921-123710 Research output: Contribution to Journal › Review article (Published)
- DecodeME: Community recruitment for a large genetics study of myalgic encephalomyelitis / chronic fatigue syndrome 19 Jul 2022 In: Bmc neurology DOI : https://doi.org/10.1186/s12883-022-02763-6 Research output: Contribution to Journal › Article (E-pub ahead of print)
- Whole genome sequencing reveals host factors underlying critical Covid-19 7 Jul 2022 In: Nature, vol. 607, pp. 97-103 DOI : https://doi.org/10.1038/s41586-022-04576-6 Research output: Contribution to Journal › Article (Published)
- The genetics of ME: a commentary on Hajdarevic et al 14 Jun 2022 In: Brain, Behavior, and Immunity DOI : https://doi.org/10.1016/j.bbi.2022.06.008 Research output: Contribution to Journal › Comment/debate (E-pub ahead of print)
- Refining the domain architecture model of the replication origin firing factor Treslin/TICRR 1 May 2022 In: Life Science Alliance, vol. 5 DOI : https://doi.org/10.26508/lsa.202101088 Research output: Contribution to Journal › Article (Published)
- Mapping the developing human cardiac endothelium at single cell resolution identifies MECOM as a regulator of arteriovenous gene expression 25 Feb 2022 In: Cardiovascular Research DOI : https://doi.org/10.1093/cvr/cvac023 Research output: Contribution to Journal › Article (Published)
View all 360 publications on Research Explorer

Transforming the understanding and treatment of mental illnesses.
Información en español
Celebrating 75 Years! Learn More >>
- Science News
- Meetings and Events
- Social Media
- Press Resources
- Email Updates
- Innovation Speaker Series
Scientists Map Networks Regulating Gene Function in the Human Brain
NIH-funded research details the brain’s cellular and molecular regulatory elements and their impact on brain function
May 23, 2024 • Press Release
A consortium of researchers has produced the largest and most advanced multidimensional maps of gene regulation networks in the brains of people with and without mental disorders. These maps detail the many regulatory elements that coordinate the brain’s biological pathways and cellular functions. The research, supported the National Institutes of Health (NIH), used postmortem brain tissue from over 2,500 donors to map gene regulation networks across different stages of brain development and multiple brain-related disorders.
“These groundbreaking findings advance our understanding of where, how, and when genetic risk contributes to mental disorders such as schizophrenia, post-traumatic stress disorder, and depression,” said Joshua A. Gordon, M.D., Ph.D., director of NIH’s National Institute of Mental Health (NIMH). “Moreover, the critical resources, shared freely, will help researchers pinpoint genetic variants that are likely to play a causal role in mental illnesses and identify potential molecular targets for new therapeutics.”
The research is published across 15 papers in Science , Science Advances , and Scientific Reports . The papers report findings along several key themes:
- Population-level analyses that link genetic variants, regulatory elements, and different molecular forms of expressed genes to regulatory networks at the cellular level, in both the developing brain and adult brain
- Single-cell-level maps of the prefrontal cortex from individuals diagnosed with mental disorders and neurodevelopmental disorders
- Experimental analyses validating the function of regulatory elements and genetic variants associated with quantitative trait loci (segments of DNA that are linked with observable traits)
The analyses expand on previous findings , exploring multiple cortical and subcortical regions of the human brain. These brain areas play key roles in a range of essential processes, including decision-making, memory, learning, emotion, reward processing, and motor control.
Approximately 2% of the human genome is composed of genes that code for proteins. The remaining 98% includes DNA segments that help regulate the activity of those genes. To better understand how brain structure and function contribute to mental disorders, researchers in the NIMH-funded PsychENCODE Consortium are using standardized methods and data analysis approaches to build a comprehensive picture of these regulatory elements in the human brain.
In addition to these discoveries, the papers also highlight new methods and tools to help researchers analyze and explore the wealth of data produced by this effort. These resources include a web-based platform offering interactive visualization data from diverse brain cell types in individuals with and without mental disorders, known as PsychSCREEN . Together, these methods and tools provide a comprehensive, integrated data resource for the broader research community.
The papers focus on the second phase of findings from the PsychENCODE Consortium. This effort aims to advance our understanding of how gene regulation impacts brain function and dysfunction.
“These PsychENCODE Consortium findings shed new light on how gene risk maps onto brain function across developmental stages, brain regions, and disorders,” said Jonathan Pevsner, Ph.D., chief of the NIMH Genomics Research Branch. “The work lays a strong foundation for ongoing efforts to characterize regulatory pathways across disorders, elucidate the role of epigenetic mechanisms, and increase the ancestral diversity represented in studies.”
The PsychENCODE papers are presented as a collection on the Science website .
MH116438 , MH116488 , MH116492 , MH116529 , MH117406 , MH116489 , MH117291 , MH117292 , MH117293 , MH129817 , MH116442 , MH121521 , MH122590 , MH122591 , MH122592 , MH122678 , MH122681 , MH126393 , MH122509 , MH125516 , MH126459 , MH129301
About the National Institute of Mental Health (NIMH): The mission of the NIMH is to transform the understanding and treatment of mental illnesses through basic and clinical research, paving the way for prevention, recovery and cure. For more information, visit the NIMH website .
About the National Institutes of Health (NIH) : NIH, the nation's medical research agency, includes 27 Institutes and Centers and is a component of the U.S. Department of Health and Human Services. NIH is the primary federal agency conducting and supporting basic, clinical, and translational medical research, and is investigating the causes, treatments, and cures for both common and rare diseases. For more information about NIH and its programs, visit the NIH website .
NIH…Turning Discovery Into Health ®
Prediction of colorectal cancer risk based on profiling with common genetic variants
Affiliations.
- 1 School of Public Health, Zhejiang University, Hangzhou, China.
- 2 Centre for Global Health, Usher Institute, University of Edinburgh, Edinburgh, UK.
- 3 Colon Cancer Genetics Group, Cancer Research UK Edinburgh Centre and Medical Research Council Human Genetics Unit, Medical Research Council Institute of Genetics and Molecular Medicine, University of Edinburgh, Edinburgh, UK.
- 4 Cancer Research UK Edinburgh Centre, Medical Research Council Institute of Genetics and Molecular Medicine, University of Edinburgh, Edinburgh, UK.
- 5 Danish Institute for Advanced Study (DIAS), Department of Public Health, University of Southern Denmark, Odense, Denmark.
- 6 Centre for Population Health Sciences, Usher Institute, University of Edinburgh, Edinburgh, UK.
- 7 Division of Genetics and Epidemiology, The Institute of Cancer Research, London, UK.
- PMID: 32638365
- DOI: 10.1002/ijc.33191
Increasing numbers of common genetic variants associated with colorectal cancer (CRC) have been identified. Our study aimed to determine whether risk prediction based on common genetic variants might enable stratification for CRC risk. Meta-analysis of 11 genome-wide association studies comprising 16 871 cases and 26 328 controls was performed to capture CRC susceptibility variants. Genetic prediction models with several candidate polygenic risk scores (PRSs) were generated from Scottish CRC case-control studies (6478 cases and 11 043 controls) and the score with the best performance was then tested in UK Biobank (UKBB) (4800 cases and 20 287 controls). A weighted PRS of 116 CRC single nucleotide polymorphisms (wPRS 116 ) was found with the best predictive performance, reporting a c-statistics of 0.60 and an odds ratio (OR) of 1.46 (95% confidence interval [CI] = 1.41-1.50, per SD increase) in Scottish data set. The predictive performance of this wPRS 116 was consistently validated in UKBB data set with c-statistics of 0.61 and an OR of 1.49 (95% CI = 1.44-1.54, per SD increase). Modeling the levels of PRS with age and sex in the general UK population shows that employing genetic risk profiling can achieve a moderate degree of risk discrimination that could be helpful to identify a subpopulation with higher CRC risk due to genetic susceptibility.
Keywords: colorectal cancer; genetic prediction; genome-wide association study; polygenic risk score.
© 2020 UICC.
Publication types
- Meta-Analysis
- Research Support, Non-U.S. Gov't
- Case-Control Studies
- Colorectal Neoplasms / epidemiology*
- Colorectal Neoplasms / genetics
- Genetic Predisposition to Disease
- Genome-Wide Association Study / methods*
- Models, Genetic
- Multifactorial Inheritance
- Polymorphism, Single Nucleotide*
Grants and funding
- 12076/CRUK_/Cancer Research UK/United Kingdom
- MC_PC_17228/MRC_/Medical Research Council/United Kingdom
- C1298/A25514/CRUK_/Cancer Research UK/United Kingdom
- U127527198/MRC_/Medical Research Council/United Kingdom
- 18927/CRUK_/Cancer Research UK/United Kingdom
- 27327/CRUK_/Cancer Research UK/United Kingdom
- C6199/A16459/CRUK_/Cancer Research UK/United Kingdom
- C31250/A22804/CRUK_/Cancer Research UK/United Kingdom
- C348/A12076/CRUK_/Cancer Research UK/United Kingdom
- MC_UU_00007/1/MRC_/Medical Research Council/United Kingdom
- MC_PC_U127527198/MRC_/Medical Research Council/United Kingdom
- MC_U127527198/MRC_/Medical Research Council/United Kingdom
- MR/K018647/1/MRC_/Medical Research Council/United Kingdom
- MC_QA137853/MRC_/Medical Research Council/United Kingdom
- 22804/CRUK_/Cancer Research UK/United Kingdom

Blood- and brain-based genome-wide association studies of smoking
- Find this author on Google Scholar
- Find this author on PubMed
- Search for this author on this site
- For correspondence: [email protected]
- Info/History
- Supplementary material
- Preview PDF
Background Self-reported smoking is often incorporated into disease prediction tools but suffers from recall bias and does not capture passive exposure. Blood-based DNA methylation (DNAm) is an objective way to assess smoking. However, studies have not fully explored tissue-specificity or epigenome-wide coverage beyond array data. Here, we update the existing biomarkers of smoking and conduct a detailed analysis of the associations between blood DNAm and self-reported smoking.
Methods and Findings A blood-based Bayesian epigenome-wide association study (EWAS) of smoking was carried out in 17,865 Generation Scotland individuals at ∼850k CpG sites (Illumina EPIC array). For 24 pairs of smokers and non-smokers a high-resolution approach was implemented (∼4 million sites, TWIST methylome panel). A DNAm-derived biomarker of smoking (mCigarette) was tested in the independent Lothian Birth Cohort 1936 (n=882, Illumina 450k array) and in the ALSPAC parents and offspring at four time points (range n=496–1,207). To explore tissue specific signals, EWASs of smoking were run across five brain regions for 14 individuals using DNAm from the EPIC array. Lastly, genome-wide association studies (GWASs) of smoking pack years and an epigenetic score for smoking (GrimAge DNAm pack years) were conducted (n=17,105). The primary EWAS analyses identified two novel genome-wide significant loci, mapping to genes related to addiction and carcinogenesis. Associations with CpG sites which are currently absent from methylation arrays were identified by the high resolution EWAS of smoking (n=48). The mCigarette pack years biomarker showed excellent discrimination across all smoking categories (current, former, never), and outperformed existing predictors in associations with pack years in an external test dataset (Pearson r=0.75). Several CpGs showed near-perfect discrimination of smoking status in both blood and brain, but these loci did not overlap across tissues. The GWAS of DNAm (but not self-reported) pack years identified novel and established smoking-related loci. However, the self-reported phenotype GWAS had a higher genetic correlation with a large meta-analysis GWAS of self-reported pack years. Among the study shortcomings are its potential lack of generalizability to non-Europeans and the absence of serum cotinine data.
Conclusion A multi-tissue, multi-cohort analysis of the relationship between smoking, DNA and DNAm (assessed via arrays and targeted sequencing) has improved our understanding of the biological consequences of smoking.
Competing Interest Statement
R.E.M has received a speaker fee from Illumina and is an advisor to the Epigenetic Clock Development Foundation. R.F.H. has received consultant fees from Illumina. R.E.M and R.F.H. have received consultant fees from Optima partners. All other authors declare no competing interests.
Funding Statement
Generation Scotland: Generation Scotland received core support from the Chief Scientist Office of the Scottish Government Health Directorates (CZD/16/6) and the Scottish Funding Council (HR03006). Genotyping and DNA methylation profiling of the Generation Scotland samples was carried out by the Genetics Core Laboratory at the Edinburgh Clinical Research Facility, Edinburgh, Scotland and was funded by the Medical Research Council UK and the Wellcome Trust (Wellcome Trust Strategic Award STratifying Resilience and Depression Longitudinally (STRADL; Reference 104036/Z/14/Z). The DNA methylation data assayed for Generation Scotland was partially funded by a 2018 NARSAD Young Investigator Grant from the Brain & Behavior Research Foundation (Ref: 27404; awardee: Dr David M Howard) and by a JMAS SIM fellowship from the Royal College of Physicians of Edinburgh (Awardee: Dr Heather C Whalley). LBC1936: The LBC1936 is supported by the BBSRC, and the Economic and Social Research Council [BB/W008793/1] (which supports S.E.H.), Age UK (Disconnected Mind project), the Milton Damerel Trust, the Medical Research Council (MR/M01311/1), and the University of Edinburgh. Methylation typing of LBC1936 was supported by the Centre for Cognitive Ageing and Cognitive Epidemiology (Pilot Fund award), Age UK, The Wellcome Trust Institutional Strategic Support Fund, The University of Edinburgh, and The University of Queensland. Genotyping was funded by the BBSRC (BB/F019394/1). S.R.C. is supported by a Sir Henry Dale Fellowship jointly funded by the Wellcome Trust and the Royal Society (Grant Number 221890/Z/20/Z). ALSPAC: The UK Medical Research Council and Wellcome (Grant ref: 217065/Z/19/Z) and the University of Bristol provide core support for ALSPAC. This publication is the work of the authors and they will serve as guarantors for the contents of this paper. A comprehensive list of grants funding is available on the ALSPAC website ( http://www.bristol.ac.uk/alspac/external/documents/grant-acknowledgements.pdf ). Funding for ALSPAC DNAm measurements were supported by the Wellcome (102215/2/13/2); the University of Bristol; the UK Economic and Social Research Council (ES/N000498/1); the UK Medical Research Council (MC_UU_12013/1, MC_UU_12013/2); the Biotechnology and Biological Sciences Research Council (BBI025751/1 and BB/I025263/1); and the John Templeton Foundation (60828). P.Y. and M.S. work is supported by the National Institute for Health and Care Research Bristol Biomedical Research Centre, the Medical Research Council Integrative Epidemiology Unit at the University of Bristol (MC_UU_00032/3, MC_UU_00032/4, MC_UU_00032/6), and Cancer Research UK [C18281/A29019, EDDISA-Jan22\100003]. A.D.C. is supported by a Medical Research Council PhD Studentship in Precision Medicine with funding from the Medical Research Council Doctoral Training Program and the University of Edinburgh College of Medicine and Veterinary Medicine. R.F.H is supported by an MRC IEU Fellowship. E.B. and R.E.M. are supported by Alzheimer's Society major project grant AS-PG-19b-010. This research was funded in whole, or in part, by the Wellcome Trust (104036/Z/14/Z, 108890/Z/15/Z, 220857/Z/20/Z, and 221890/Z/20/Z). For the purpose of open access, the author has applied a CC BY public copyright licence to any Author Accepted Manuscript version arising from this submission.
Author Declarations
I confirm all relevant ethical guidelines have been followed, and any necessary IRB and/or ethics committee approvals have been obtained.
The details of the IRB/oversight body that provided approval or exemption for the research described are given below:
All components of Generation Scotland received ethical approval from the NHS Tayside Committee on Medical Research Ethics (REC Reference Number: 05/S1401/89). All participants provided broad and enduring written informed consent for biomedical research. Generation Scotland has also been granted Research Tissue Bank status by the East of Scotland Research Ethics Service (REC Reference Number: 15/0040/ES), providing generic ethical approval for a wide range of uses within medical research. This study was performed in accordance with the Helsinki declaration. Ethical approval for the LBC1936 study was obtained from the Multi-Centre Research Ethics Committee for Scotland (MREC/01/0/56) and the Lothian Research Ethics committee (LREC/1998/4/183; LREC/2003/2/29). All participants provided written informed consent. These studies were performed in accordance with the Helsinki declaration. Ethical approval for the ALSPAC study was obtained from the ALSPAC Ethics and Law Committee and the Local Research Ethics Committees. Consent for biological samples has been collected in accordance with the Human Tissue Act (2004). Informed consent for the use of data collected via questionnaires and clinics was obtained from participants following the recommendations of the ALSPAC Ethics and Law Committee at the time.
I confirm that all necessary patient/participant consent has been obtained and the appropriate institutional forms have been archived, and that any patient/participant/sample identifiers included were not known to anyone (e.g., hospital staff, patients or participants themselves) outside the research group so cannot be used to identify individuals.
I understand that all clinical trials and any other prospective interventional studies must be registered with an ICMJE-approved registry, such as ClinicalTrials.gov. I confirm that any such study reported in the manuscript has been registered and the trial registration ID is provided (note: if posting a prospective study registered retrospectively, please provide a statement in the trial ID field explaining why the study was not registered in advance).
I have followed all appropriate research reporting guidelines, such as any relevant EQUATOR Network research reporting checklist(s) and other pertinent material, if applicable.
7. Availability of data and materials
According to the terms of consent for Generation Scotland participants, access to data must be reviewed by the Generation Scotland Access Committee. Applications should be made to access{at}generationscotland.org .
Lothian Birth Cohort data are available on request from the Lothian Birth Cohort Study, University of Edinburgh ( https://www.ed.ac.uk/lothian-birth-cohorts/data-access-collaboration ). Lothian Birth Cohort data are not publicly available due to them containing information that could compromise participant consent and confidentiality.
ALSPAC data are available on request from bona fide researchers. The study website contains details of all the data that is available through a fully searchable data dictionary and variable search tool ( http://www.bristol.ac.uk/alspac/researchers/our-data/ ).
All custom R (version 4.3.1), Python (version 3.9.7), and bash code is available with open access at the following GitHub repository: https://github.com/aleksandra-chybowska/Smoking_EpiScore/
GWAS and EWAS summary statistics will be made available on Edinburgh DataShare on publication.
View the discussion thread.
Supplementary Material
Thank you for your interest in spreading the word about medRxiv.
NOTE: Your email address is requested solely to identify you as the sender of this article.

Citation Manager Formats
- EndNote (tagged)
- EndNote 8 (xml)
- RefWorks Tagged
- Ref Manager
- Tweet Widget
- Facebook Like
- Google Plus One
Subject Area
- Genetic and Genomic Medicine
- Addiction Medicine (325)
- Allergy and Immunology (633)
- Anesthesia (168)
- Cardiovascular Medicine (2409)
- Dentistry and Oral Medicine (291)
- Dermatology (208)
- Emergency Medicine (382)
- Endocrinology (including Diabetes Mellitus and Metabolic Disease) (854)
- Epidemiology (11816)
- Forensic Medicine (10)
- Gastroenterology (705)
- Genetic and Genomic Medicine (3781)
- Geriatric Medicine (351)
- Health Economics (638)
- Health Informatics (2414)
- Health Policy (941)
- Health Systems and Quality Improvement (909)
- Hematology (343)
- HIV/AIDS (789)
- Infectious Diseases (except HIV/AIDS) (13358)
- Intensive Care and Critical Care Medicine (770)
- Medical Education (371)
- Medical Ethics (105)
- Nephrology (403)
- Neurology (3535)
- Nursing (199)
- Nutrition (531)
- Obstetrics and Gynecology (683)
- Occupational and Environmental Health (671)
- Oncology (1838)
- Ophthalmology (541)
- Orthopedics (222)
- Otolaryngology (287)
- Pain Medicine (234)
- Palliative Medicine (67)
- Pathology (449)
- Pediatrics (1041)
- Pharmacology and Therapeutics (427)
- Primary Care Research (425)
- Psychiatry and Clinical Psychology (3204)
- Public and Global Health (6197)
- Radiology and Imaging (1297)
- Rehabilitation Medicine and Physical Therapy (753)
- Respiratory Medicine (833)
- Rheumatology (380)
- Sexual and Reproductive Health (375)
- Sports Medicine (325)
- Surgery (407)
- Toxicology (51)
- Transplantation (173)
- Urology (148)
- School of Medicine Columbia
- Location Location
- Contact Contact
- Colleges and Schools
Genetic Counseling

There’s never been a better time to be a genetic counselor. Opportunities abound in any number of university, hospital, laboratory, research and industry settings. With a master's in genetic counseling, you’ll be ready for the workforce and a challenging, rewarding health professional career in one of the most dynamic areas of medicine.
Our Program
Learn about who we are as a program. Learn about our mission, our vision, our values, our goals and our history.
The people that make it happen. Meet our incredible faculty, staff, students and alumni.
Through coursework, fieldwork and community engagement, our curriculum cultivates a well-rounded genetic counselor.
Application
Find information, forms and deadlines on how to apply for our program, here.
Tuition, Scholarships and Financial Aid
Learn more about our program's tuition for both South Carolina residents and non-residents, as well as scholarship and financial aid opportunities, here.
Preparing for Graduate School
We know how important it is to be certain of your chosen profession and prepared for graduate education. We've created several opportunities that will help you explore a career in genetic counseling.
Program Events and Special Lectures
Our program is uniquely positioned in an academic, research and medical school setting allowing for special curricular events outside of the classroom.
Diversity, Equity, Inclusion and Justice
The USC Genetic Counseling Program strives to consistently foster an environment of inclusion for students, and we recognize that more can be done to improve diversity, equity, inclusion and justice (DEIJ) in our program and in the genetic counseling profession.
Challenge the conventional. Create the exceptional. No Limits.
Internet Explorer Alert
It appears you are using Internet Explorer as your web browser. Please note, Internet Explorer is no longer up-to-date and can cause problems in how this website functions This site functions best using the latest versions of any of the following browsers: Edge, Firefox, Chrome, Opera, or Safari . You can find the latest versions of these browsers at https://browsehappy.com
- Publications
- HealthyChildren.org
Shopping cart
Order Subtotal
Your cart is empty.
Looks like you haven't added anything to your cart.
Career Resources
- Philanthropy
- About the AAP
- Infant Feeding for Persons Living With and at Risk for HIV in the United States: Clinical Report
- Research on HIV transmission through breastfeeding spurs update to AAP guidance
- Breastfeeding for People with HIV: AAP Policy Explained
- American Academy of Pediatrics Shifts Position on Breastfeeding for People with Human Immunodeficiency Virus
- News Releases
- Policy Collections
- The State of Children in 2020
- Healthy Children
- Secure Families
- Strong Communities
- A Leading Nation for Youth
- Transition Plan: Advancing Child Health in the Biden-Harris Administration
- Health Care Access & Coverage
- Immigrant Child Health
- Gun Violence Prevention
- Tobacco & E-Cigarettes
- Child Nutrition
- Assault Weapons Bans
- Childhood Immunizations
- E-Cigarette and Tobacco Products
- Children’s Health Care Coverage Fact Sheets
- Opioid Fact Sheets
- Advocacy Training Modules
- Subspecialty Advocacy Report
- AAP Washington Office Internship
Online Courses
- Live and Virtual Activities
- National Conference and Exhibition
- Prep®- Pediatric Review and Education Programs
- Journals and Publications
- NRP LMS Login
- Patient Care
- Practice Management
- AAP Committees
- AAP Councils
- AAP Sections
- Volunteer Network
- Join a Chapter
- Chapter Websites
- Chapter Executive Directors
- District Map
- Create Account
- News from the AAP
- Latest Studies in Pediatrics
- Pediatrics OnCall Podcast
- AAP Voices Blog
- Campaigns and Toolkits
- Spokesperson Resources
- Join the AAP
- Exclusive for Members
- Membership FAQs
- AAP Membership Directory
- Member Advantage Programs
- Red Book Member Benefit
- My Membership
- Join a Council
- Join a Section
- National Election Center
- Medical Students
- Pediatric Residents
- Fellowship Trainees
- Planning Your Career
- Conducting Your Job Search
Making Career Transitions
- COVID-19 State-Level Data Reports
- Children and COVID-19 Vaccination Trends
- Practice Research in the Office Setting (PROS)
- Pediatrician Life and Career Experience Study (PLACES)
- Periodic Survey
- Annual Survey of Graduating Residents
- Child Population Characteristics Trends
- Child Health Trends
- Child Health Care Trends
- Friends of Children Fund
- Tomorrow’s Children Endowment
- Disaster Recovery Fund
- Monthly Giving Plans
- Honor a Person You Care About
- Donor-Advised Funds
- AAP in Your Will
- Become a Corporate Partner
- Employment Opportunities
- Equity and Inclusion Efforts
- RFP Opportunities
- Board of Directors
- Senior Leadership Team
- Constitution & By-Laws
- Strategic Plan

Log In to Your Account
Access your account, the AAP Member Directory, transcripts and more. Already logged in? Find your Account here.

Collaborate
Connect with other AAP members and explore the AAP communities you belong to.

Become a Member
Not a member yet? Learn more about the benefits of AAP membership.
What's New at the AAP
Research on hiv transmission through breastfeeding spurs update to aap guidance, 33rd edition of the red book is now available. order today, register for the 2024 national conference and exhibition, aap announces candidates for president-elect, learning opportunities.
Continuing Medical Education from AAP helps you stay current in practice and provides tools and resources for every stage of your career.
Live & Virtual Events
Neonatal resuscitation program®.
From your time as a medical student to becoming a pediatric resident and beyond, there are resources and opportunities only available through AAP.
Physician Health & Wellness

Equity, Diversity, and Inclusion
From the beginning, the American Academy of Pediatrics has been guided by its mission to ensure the health and well-being of all children. This includes promoting nurturing, inclusive environments and actively opposing intolerance, bigotry, bias, and discrimination.
Our Equity Agenda
Global Health
We work for every child's future. The AAP's global mission is to attain optimal physical, mental, and social health and well-being for all children around the world.
Our Global Initiatives

You have the power to change a life. The AAP is dedicated to the health of all children and the pediatric professionals who care for them. Your gift today makes that possible.
More AAP Resources
Journals & publications.
The American Academy of Pediatrics is the leading publisher, globally, in the field and practice of Pediatrics.
shopAAP is the official store of the American Academy of Pediatrics. All purchases directly benefit and support the health and well-being of all infants, children, adolescents, and young adults. Find a new look and feel now on shopAAP!

IMAGES
VIDEO
COMMENTS
The MRC Human Genetics Unit discovers how changes in our DNA impact our lives. The human genome contains all the information (DNA) molecules, cells and tissues need to develop and function. Our mission is to understand how our genome works and how changes in DNA influence human health and cause disease. We combine the latest computational and ...
The mission of the Medical Research Council (MRC) Human Genetics Unit at the University of Edinburgh (MRC HGU) is to explore the mechanisms underlying human genetic disease and normal development and physiology. The Unit's research seeks to identify the genetic risk factors in common disease in isolated populations, develop animal model systems to study human genetic disease and mammalian ...
The MRC Human Genetics Unit combine the latest computational and experimental technologies to investigate how our genomes work to control the function of molecules, cells and tissues in people and populations. For more than half a century our research has been dedicated to understanding human genetic disease. Today we continue to apply our ...
The Medical Research Council (UK) Human Genetics Unit is situated at the Western General Hospital in Edinburgh. It is one of the largest MRC research establishments, housing over two hundred scientists, support staff, research fellows, PhD students, and visiting workers.
Undertakes research to obtain a molecular and cellular understanding of genetic factors implicated in human disease and normal and abnormal development.
The Medical Research Council Human Genetics Unit is at the forefront of research into human genetics. Its role is to advance the understanding of genetic factors implicated in human disease and normal and abnormal development and physiology. ... Dive into the research topics where MRC Human Genetics Unit is active. These topic labels come from ...
MRC Human Genetics Unit. Institute of Genetics and Cancer. The University of Edinburgh. Western General Hospital. Crewe Road, Edinburgh EH4 2XU. Tel: +44 (0)131 651 8500
The Institute of Genetics and Cancer at the University of Edinburgh is a strategic partnership between the Medical Research Council (MRC) and University of Edinburgh, joining the MRC Human Genetics Unit with the Centre for Genomic and Experimental Medicine and the Edinburgh Cancer Research Centre, under the Directorship of Professor Margaret Frame.
The MRC Human Genetics Unit is at the forefront of research into human genetics. Its role is to advance the understanding of genetic factors implicated in human disease and normal and abnormal development and physiology. It is situated at the Western General Hospital campus in Edinburgh. mrc-human-genetics-unit
Medical Research Council Human Genetics Unit Comparing redheads to people with brown or black hair, they identified eight previously unknown genetic differences that are associated with red hair. The team also looked at the functions of the genes they identified and found that some of them work by controlling when MC1R is switched on or off.
A team led by Robert Hill of the Medical Research Council Human Genetics Unit in Edinburgh showed that the Sonic hedgehog switch that controls gene activity in the limb is located about one ...
18 Medical Research Council Human Genetics Unit, Institute of Genetics and Cancer, University of Edinburgh, Edinburgh, UK. 19 Human Genetics Center, Department of Epidemiology, Human Genetics, and Environmental Sciences, School of Public Health, The University of Texas Health Science Center at Houston, Houston, TX, USA.
4 Colon Cancer Genetics Group, Medical Research Council Human Genetics Unit, Medical Research Council Institute of Genetics & Molecular Medicine, Western General Hospital, The University of Edinburgh, Edinburgh, UK. 5 Danish Institute for Advanced Study (DIAS), Department of Public Health, University of Southern Denmark, Odense, Denmark.
Medical Research Council (MRC) Human Genetics Unit, MRC Institute of Genetics and Molecular Medicine, University of Edinburgh, Edinburgh EH4 2XU, UK. [3]. 2 1] MRC Integrative Epidemiology Unit, University of Bristol, Bristol BS8 2BN, UK. [2] Queensland Brain Institute Centre, University of Queensland, Brisbane, Queensland 4072, Australia. [3]
Medical Research Council (UKRI) | mrc · MRC Human Genetics Unit. Contact. Connect with experts in your field. Join ResearchGate to contact this researcher and connect with your scientific community.
Colon Cancer Genetics Group, Medical Research Council Human Genetics Unit, Medical Research Council Institute of Genetics & Molecular Medicine, Western General Hospital, The University of Edinburgh, Edinburgh, United Kingdom.
Research at the Medical Research Council (MRC) Mitochondrial Biology Unit (MRC MBU) is focused on the biology of mitochondria and their dysfunction in an ever-increasing range of human diseases. The Unit combines studies exploring the molecular function of the mitochondrial oxidative phosphorylation system, the mitochondrial proteome and genome, and how mitochondria interact with the cell ...
This research was supported by a National Health and Medical Research Council (NHMRC) Investigator grant (J.E.P., 1175781), research grants from the Australian Research Council (ARC) Special ...
About the National Institutes of Health (NIH): NIH, the nation's medical research agency, includes 27 Institutes and Centers and is a component of the U.S. Department of Health and Human Services. NIH is the primary federal agency conducting and supporting basic, clinical, and translational medical research, and is investigating the causes ...
Research team creates mice that better reflect human genetic variation. by Ben Whitford, Jackson Laboratory. Credit: Pixabay/CC0 Public Domain. The great majority of how we understand human ...
Additionally, P.R. (Icahn School of Medicine at Mount Sinai and James J. Peters VA Medical Center) and J.B. (Icahn School of Medicine at Mount Sinai) were supported by the National Institute of Mental Health (NIMH), NIH grants RF1-MH128970, R01-MH125246, and R01-MH109897, as well as the National Institute on Aging, NIH grants R01-AG050986, R01 ...
Professor Chris Ponting is Chair of Medical Bioinformatics and a Principal Investigator at the MRC Human Genetics Unit, Institute of Genetics and Cancer. Chris started his research in particle physics before moving via biophysics to bioinformatics and genomics. Aside from one year at the National Centre for Biotechnology Information (NIH ...
About the National Institutes of Health (NIH): NIH, the nation's medical research agency, includes 27 Institutes and Centers and is a component of the U.S. Department of Health and Human Services.NIH is the primary federal agency conducting and supporting basic, clinical, and translational medical research, and is investigating the causes, treatments, and cures for both common and rare diseases.
Maryland. Howard Hughes Medical Institute. J. Craig Venter Institute. Kennedy Krieger Institute. National Human Genome Research Institute. USC Institute Of Translational Genomics. Massachusetts. Broad Institute ( Massachusetts Institute of Technology and Harvard University) Dana-Farber Cancer Institute.
Affiliations 1 School of Public Health, Zhejiang University, Hangzhou, China.; 2 Centre for Global Health, Usher Institute, University of Edinburgh, Edinburgh, UK.; 3 Colon Cancer Genetics Group, Cancer Research UK Edinburgh Centre and Medical Research Council Human Genetics Unit, Medical Research Council Institute of Genetics and Molecular Medicine, University of Edinburgh, Edinburgh, UK.
In 1964, the World Medical Association passed the Declaration of Helsinki, a set of ethical principles for the medical community regarding human experimentation. In 1966, the United States National Institutes of Health (NIH) Office for Protection of Research Subjects (OPRR) was created.
Genotyping and DNA methylation profiling of the Generation Scotland samples was carried out by the Genetics Core Laboratory at the Edinburgh Clinical Research Facility, Edinburgh, Scotland and was funded by the Medical Research Council UK and the Wellcome Trust (Wellcome Trust Strategic Award STratifying Resilience and Depression Longitudinally ...
Diversity, Equity, Inclusion and Justice. The USC Genetic Counseling Program strives to consistently foster an environment of inclusion for students, and we recognize that more can be done to improve diversity, equity, inclusion and justice (DEIJ) in our program and in the genetic counseling profession.
Overview. Our mission is to assist Pennsylvanians in leading safe, healthy, and productive lives through equitable, trauma-informed, and outcome-focused services while being an accountable steward of commonwealth resources. DHS Executive Leadership.
The American Academy of Pediatrics (AAP) is dedicated to improving the health and well-being of children. Explore our comprehensive resources, evidence-based guidelines, and expert insights on pediatric care. Discover the latest research, educational materials, and advocacy initiatives aimed at promoting child health. Join the AAP community and access valuable tools, training, and networking ...