HOME | ABOUT | CONTACT | SHARE Design and developed by MCA Department of Jorhat Engineering College 2023

Candrakanta Glosbe
Google Bing
• Candrakanta abhidhana : Assamese-English dictionary, University of Gauhati (1962)
• Dictionary Assamese and English by Miles Bronson (1867)
• Phrases in English and Assamese by Harriet Cutter, revised by Edward Clark (1877) + 1840 edition
• Brief vocabulary in English and Assamese by Susan Ward (1864)
• Some Assamese proverbs by Philip Richard Gurdon (1896)
• A linguistic study of 24 place names of Assam by Sarat Kumar Phukan, in International Symposium on languages and linguistics (2000)
→ Assamese keyboard (Bengali script with some variants, like the characters r and w )
• Wikipedia : Assamese language
• Dialects or variety of the Assamese language by Manash Jyoti Nirmalia, in Journal of Acharaya Narendra Dev Research Institute (2019)
• Fixing the Assamese language : "Tongue has no bone" (1800-1930) , by Bodhisattva Kar, in Studies in History (2008)
• Language and policies in 19 th -century Assam : war of words , by Madhumita Sengupta, in Indian Historical Review (2012)
• Representing Kamrupi : ideologies of grammar and the question of linguistic boundaries , in A multilingual nation translation and language dynamic in India (2017)
• Translation in Assamese : a brief account , by Biswadip Gogoi, in History of Translation in India (2017)
• Language and nationalism : comprehending the dynamics in 19 th -century Assam (2016)
• Linguistic nationalism in early-colonial Assam : the American Baptist Mission and Orunodoi , by Arnab Dasgupta, in Rupkatha journal on interdisciplinary studies in humanities (2021)
• Derivational morphology of Assamese lexical word categories by Palash Das & Madhumita Barbora, in Indian Journal of Language and Linguistics (2020)
• Assamese: its formation and development by Banikanta Kakati (1941)
• Grammatical notices of the Asamese language by Nathan Brown (1848)
The Assamese language is spoken in the State of Assam, in Northeast India. It's written with the Bengali script.
• Aspects of early Assamese literature by Banikanta Kakati (1953)
• YouVersion : translation of the Bible into Assamese (2019) (+ audio)
• Hymns in Assamese , edited by the Baptist Mission (1850)
জন্মগত ভাবে সকলো মানুহ মৰ্যদা আৰু অধিকাৰত সমান আৰু স্বতন্ত্র। তেওঁলােকৰ বিবেক আছে, বুদ্ধি আছে। তেওঁলােকে প্রত্যেকে প্রত্যেকক ভাতৃ ভাবে ব্যৱহাৰ কৰা উচিত।
• Ethnicity, identity and cartography : possession/dispossession, homecoming/homelessness in contemporary Assam , by Parag Moni Sarma, in Studies of transition states and societies (2011)
• Becoming Hindu : the cultural politics of writing religion in colonial Assam , by Madhumita Sengupta, in Contributions to Indian Sociology (2021)
• The Assam Mission : papers and discussions , American Baptist Missionary Union (1886)
→ Inde
→ Bangladesh
→ languages of India
→ Bengali
→ Meitei
→ Hindi

representation in Dogri डोगरी
- तरजमानी ⇄ Representation
- नमायंदगी ⇄ Representation
representation in Gujarati ગુજરાતી
- અમુક તરીકે વર્ણન કરવું ⇄ representation
- ક હેવું (કે) ⇄ representation
- કૃતિમાં ઉતારવું ⇄ representation
- દર્શ ાવવું ⇄ representation
- ભૂમિકા – નાટક – ભજવવું ⇄ representation
- મન કે આંખ આગળ કશાકની આકૃતિ રજૂ કે ખડી કરવી ⇄ representation
- –નું પ્રતીક હોવું ⇄ representation
- –ને સ્થાને હોવું ⇄ representation
- –નો ભાસ કરાવવો ⇄ representation
representation in Hindi हिन्दी
- दरसाना ⇄ representation
- द्योतित करना ⇄ representation
- निरूपण ⇄ representation
- निस्र्पण ⇄ representation
- नुमाइंदगी ⇄ representation
- प्रतिनिधित्व ⇄ representation
- प्रतिरूप ⇄ representation
- मूत्ति ⇄ representation
- वर्णन ⇄ representation
- वर्णन करना ⇄ representation
- विरोध - पत्र ⇄ representation
representation in Kannada ಕನ್ನಡ
- ಚಿತ್ರ ⇄ representation
- ದರಖಾಸ್ತು ⇄ representation
- ನಿರೂಪಣೆ ⇄ representation
- ಪ್ರದರ್ಶನ ⇄ representation
- ಬಿಮ್ಬ ⇄ representation
- ಮನವಿ ⇄ representation
- ವರ್ಣನೆ ⇄ representation
- ವಿಜ್ಝ್ನ್ಯಾಪನೆ ⇄ representation
- ವಿನಮ್ತಿ ⇄ representation
representation in Kashmiri कॉशुर
- شِکایت ⇄ representation
representation in Malayalam മലയാളം
- ചിത്രീകരണം ⇄ representation
- നിവേദനം ⇄ representation
- മാതൃക ⇄ representation
- സൂചിപ്പിക്കല് ⇄ representation
representation in Marathi मराठी
- निदर्शन ⇄ representation
- निरुपण ⇄ representation
- प्रतिनिधीत्व ⇄ representation
- प्रतिरुपण ⇄ representation
- प्रतिरूपण ⇄ representation
- रूपण ⇄ representation
representation in Sindhi سنڌي
- بيان، تصوير، نقشو، درخواست، عرضي ⇄ Representation
representation in Tamil தமிழ்
- நிகராட்சி ⇄ representation
- வகைக்குறி ⇄ representation
representation in Telugu తెలుగు
- అనగా నాటకము. ⇄ representation
- ఆకారము. మనవి ⇄ representation
- విజ్ఙాపన ⇄ representation
- వృత్తాంతము. ప్రత్యామ్నాయముగా పోవడము. స్త్రీ ప్రతిమ వొకని వలె వేషము కట్టి ఆడడము ⇄ representation
representation in Urdu اُردُو
- صورت حال ⇄ representation
representation in English
- representation ⇄ knowledge baseorrepresentation, the basic store of information required by a computer to solve a problem or perform a task. Ex. Until they acquire ... a knowledge base or knowledge representation, computer systems are going to miss ... many co
- representation ⇄ representation, noun. 1. the act of representing. Ex. the boy's representation of his class on the Student Council. 2. the condition or fact of being represented. Ex. ""Taxation without representation is tyranny."" The United Stat
representation Deals on Amazon
- ›
- assamese-dictionary-translation-meaning-of-representation
Voice speed
Text translation, source text, translation results, document translation, drag and drop.

Website translation
Enter a URL
Image translation
Assamese to English Translation
Translate assamese to english.
- Azerbaijani
- Cantonese (trad)
- Chinese (literary)
- Chinese (simp)
- Chinese (trad)
- English United Kingdom
- French (Canada)
- Haitian Creole
- Inuinnaqtun
- Inuktitut (Latin)
- Kinyarwanda
- Klingon (Latin)
- Kurdish (Kurmanji)
- Kurdish (Sorani)
- Luxembourgish
- Lower Sorbian
- Meiteilon (Manipuri)
- Mongolian (trad)
- Myanmar (Burmese)
- Odia (Oriya)
- Portuguese (Brazil)
- Portuguese (Portugal)
- Quertaro Otomi
- Scots Gaelic
- Serbian (Cyrillic)
- Serbian (Latin)
- Upper Sorbian
- Yucatec Maya

Online Translation
- International
- Today’s Paper
- Premium Stories
- Express Shorts
- Health & Wellness
- Brand Solutions
Explained: Who is Assamese? A proposed definition, and several questions
The question, 'who are the assamese people' has always been contentious. now, a definition has been proposed by a committee that looked into implementation of clause 6 of the assam accord. what is this clause what is the proposed definition, and what are the implications.
The question, “Who is an Assamese?”, has been debated for decades in Assam, whose history has been shaped by people of multiple cultures over the centuries. Now, a report by a government-appointed committee has proposed a definition for “Assamese people”. While this is limited to the purpose of implementing a provision of the 1985 Assam Accord — Clause 6 — it spotlights the complexities at play in Assam.
Why is it a matter of debate?
The Assam Accord was signed at the end of a six-year agitation (1979-85) against illegal migration from Bangladesh. In the context of the Accord, the question of who is Assamese stems from the language of Clause 6: “Constitutional, legislative and administrative safeguards, as may be appropriate, shall be provided to protect, preserve and promote the cultural, social, linguistic identity and heritage of the Assamese people.”

This gives rise to two questions: what will these safeguards be; and, who are the “Assamese people” eligible for these?
Isn’t any resident of Assam, Assamese?
It is not all-encompassing in a state defined by the politics of migration. And yet, the definition of “Assamese” cannot be so narrow as to mean only those who speak Assamese as their first language. Assam has many indigenous tribal and ethnic communities with their own ancestral languages. For Clause 6, it was necessary to expand the definition of “Assamese” beyond the Assamese-speaking population.
Those not eligible for the safeguards under Clause 6 would clearly be from among the migrant populations. But would the entire migrant populations be excluded, or would some of them be eligible for Clause 6 benefits? Hence the debate.

But who is a migrant?
In popular conversation, the idea of “indigenous” is taken to mean communities who trace their histories in Assam before 1826, the year when the erstwhile kingdom of Assam was annexed to British India. Large-scale migration from East Bengal took place during British rule, followed by further waves after Independence.
The 1979-85 Assam Movement was triggered by fears that these Bengali Muslim and Bengali Hindu migrants would one day overrun the indigenous population, and dominate the resources and politics of the state. During the agitation, the demand was for the detection and deportation of those who had migrated after 1951. Was this demand accepted?
Not 1951. The Assam Accord was settled at a cut-off of March 24, 1971; anyone who arrived in Assam before that cut-off would be considered a citizen of India. This date was also the basis of the National Register of Citizens ( NRC ), published last year.
Because the Accord legalised additional migrants (1951-71) against the original demand of 1951, Clause 6 was incorporated as a safeguard for the indigenous people. This was the reasoning as explained to The Indian Express last year by Prafulla Kumar Mahanta, who signed the Accord as All Assam Students’ Union president in 1985, and went on to become the Chief Minister.
How has Clause 6 been taken up since?
Because of the complexities involved, previous efforts to work out a framework made little headway. The matter got urgency last year amid protests by the Assamese against the Citizenship Amendment Bill (now an Act) which makes it easier for certain categories of migrants to get Indian citizenship — the key here being Hindus from Bangladesh. The Home Ministry set up a new committee, which submitted its report in February, but the government sat on it for months. This led to four of the committee’ s 14 members making its contents public on Tuesday.
So, what are the recommendations?
For the purpose of implementation of Clause 6, the proposed definition includes indigenous tribals, other indigenous communities, all other citizens of India residing in Assam on or before January 1, 1951 and indigenous Assamese — and their descendants. In short, it covers anyone who can prove their presence (or that of their ancestors) in Assam before 1951.
As for safeguards, the committee has recommended reservations in legislature and jobs for “Assamese people”, and that “land rights be confined” to them.
What are implications of the definition?
- Migrants who entered Assam after 1951 but before March 24, 1971 are not Assamese but are Indian citizens. They would not be eligible, for example, to contest an election in 80-100% of Assam’s seats (if that recommendation is accepted). But they can vote.
- Not just indigenous groups, but East Bengal migrants who entered Assam before 1951, too, would be considered Assamese. “(Migrants of) 1951-71 have been accepted (as citizens)… for their staying here, some people have suffered certain losses that have to be made up,” said committee member Nilay Dutta, Advocate General of Arunachal Pradesh.
📣 Express Explained is now on Telegram . Click here to join our channel (@ieexplained) and stay updated with the latest
What issues does this raise?
Some find it too inclusive. The committee had received some public suggestions that had proposed a base year of 1826 for anyone being considered Assamese, Nilay Dutta said.
Hafizul Ahmed, president of the Sadou Asom Goria-Moria-Deshi Jatiya Parishad that speaks for indigenous Assamese Muslims, told The Indian Express that there should not be a base year for identifying the indigenous people of Assam. The organisation had sought that only communities living in Assam during Ahom rule (pre-1826) be included in the definition, based on their cultural identities.
Others find it exclusionary. The All Assam Minority Students’ Union, which is identified with Bengali Muslims, had been demanding that the 1971 cutoff be used for deciding Clause 6 eligibility too. Its adviser Azizur Rahman said: “How will you prove that a person has been in Assam prior to 1951?” (The 1951 NRC is not available in several parts of the state.)
Sadhan Purkayastha, general secretary of the Citizens’ Right Protection Committee, said lakhs of people in Assam’s Barak Valley stand to lose their rights if the report is implemented. A large number of Bengali Hindus and some Bengali Muslims had migrated from Sylhet to Barak Valley in the 1950s and 1960s.
What is the way forward?
“Several issues come up: for both the state and central government, the key issue is whether it will stand the test of judicial scrutiny because it is bound to be challenged in the courts; and will it stand the test of constitutional validity?” said Sanjoy Hazarika, International Director of the Commonwealth Human Rights Initiative, and a journalist who has written extensively on migration.
- How 1989 elections led to a one-year VP Singh term, Mandir-Mandal
- Riots in France-controlled New Caledonia leave 4 dead: What is happening?
- Why do people make music, what scientists theorise
Among the issues Hazarika raised were:
- Does the definition of an Assamese — a social, historic, ethnic, linguistic, political and cultural and not just religious entity — or a Bengali or a Punjabi or a Tamil also define her/his Indian-ness or Indian citizenship? “This is a key legal and constitutional issue that needs to be considered. It is connected to the NRC process as the Assam Accord cannot be reviewed in isolation of one clause or the other,” he said.
- How will pre-1971 migrants be accommodated? “There isn’t a magic wand to solve the problem which has challenged the core of Assam for over 70 years… I have been an advocate of constitutional reservations and work permits. But we also need to acknowledge the demographic reality in the state and its diversities,” he said.
(Inputs from Abhishek Saha in Guwahati )

What India’s chattering classes don’t understand about American politics Subscriber Only

From 1951-2019: How India voted in Lok Sabha elections Subscriber Only

Artificial General Intelligence: Why are people worried about it? Subscriber Only

What are India's stakes in Iran's Chabahar port? Subscriber Only

UPSC Key | India VIX, LTTE, FLiRT and more Subscriber Only

Why is India seeing a surge in employment? The answer Subscriber Only

Shinde: ‘Uddhav’s neck collar is no longer there, he is Subscriber Only

2 weeks before phase 3 voting, Meta flooded with communal Subscriber Only

Modi's overtures towards Pawar, Uddhav: checking 'sympathy factor' Subscriber Only

Tavleen Singh writes: Modi on the backfoot? Subscriber Only

Why Congress hit ‘400 paar’ in 1984 elections Subscriber Only

What we need is a farmer-friendly agri-export policy Subscriber Only
- Assam Accord
- Explained Politics
- Express Explained

Aam Aadmi Party (AAP) Rajya Sabha member Swati Maliwal Thursday filed a complaint with the Delhi Police in connection with the allegations she first made on Monday, that Chief Minister Arvind Kejriwal’s aide Bibhav Kumar had assaulted her at the AAP chief’s residence.

More Explained

Best of Express

EXPRESS OPINION

May 17: Latest News
- 01 CDC warns of a resurgence of Mpox
- 02 UK school closes amid cryptosporidiosis outbreak, 22 cases confirmed
- 03 Ghatkopar tragedy: BMC , Railway officials hold meet; billboards larger than 40ft ordered to be taken down
- 04 Chembur college bans Hijab for degree students in new uniform policy
- 05 ‘AI can provide most people with equal opportunities’: Google’s Sundar Pichai
- Elections 2024
- Political Pulse
- Entertainment
- Movie Review
- Newsletters
- Web Stories
Academia.edu no longer supports Internet Explorer.
To browse Academia.edu and the wider internet faster and more securely, please take a few seconds to upgrade your browser .
Enter the email address you signed up with and we'll email you a reset link.
- We're Hiring!
- Help Center

Some observational marking of Aspectual Markers in Assamese with the reference of Pragmatics

Related Papers
International Journal of English Linguistics
ZAFAR IQBAL BHATTI
The purpose of the paper is to explore the aspect system in Thali, a language spoken in Thal region, including district Layya, Bhakar and neighboring areas of Jhang, in Punjab province by a large number of people. This research paper presents comparisons and contrasts between Thali aspect system and English aspect system. There are only two aspects in Thali, namely, perfect and progressive. Perfect aspect can be categorized into past perfect and present perfect in terms of time dimensions. Similarly, progressive aspect is also categorized into past progressive and present progressive from time dimensions. All types of aspects in Thali are morphologically marked but aspect system in English is different by using morphological marking as well as several complex constructions like have + past participle, be + present participle, and have + been + present participle for perfect, progressive and perfect progressive, respectively. Thali has only four structures for aspect whereas English ...
Harold Schiffman
Abstract Tamil has a number of verbs, sometimes referred to as 'aspectual verbs' that are added to a main or lexical verb to provide semantic distinctions such as duration, completion, habituality, regularity, continuity, simultaneity, definiteness, expectation of result, remainder of result, current relevance, benefaction, antipathy, and certain other notions. Though these aspectual verbs are found all over the Indian linguistic area (Emeneau 1956) researchers have generally found them difficult to describe in a categorical way, and not until Annamalai 1981 has any attempt been made to treat aspect in Tamil (or for that matter, any Dravidian language) as a variable component of the grammar. In what follows I will summarize what has been discovered about aspect in Tamil and place it in a framework that recognizes both variability and the process of grammaticalization, that is, how a category becomes part of the morphology in a given language. I will show that Tamil aspect is a category that is on the road to grammaticalization, and that it is primarily by the process of metaphoric extension that the semantic and grammatical change takes place. Some aspectual verbs are already, as it were, at their destination, others are proceeding with all deliberate speed toward that goal, but some are straggling, some have gotten lost, and some are only just beginning to pack for the journey. I will show that aspect is a variable category within the grammar of a given speaker, but is also variable across dialects and idiolects, and between Literary Tamil (LT) and Spoken Tamil (ST).
… (Editor) 2009 CSLI …
Jean-Pierre Koenig , Shakthi Poornima Farrar
Indian Linguistics
Sanjukta Ghosh
Walter Bisang
The aim of the paper is twofold. The first aim is to analyse aspect in Thai in the framework of the selection-theory approach developed by Breu and Sasse (1991). The second aim is to study all possible co-occurrenc;es of each of the three aspect markers: lεεw, kamlaƞ, yùu with the four classes of verbs and with the verbs occurring with other strategies within the five classes of states of affairs. It was found that the selectional approach chosen helped explaining the inceptive-stative state of affairs in Thai clearly. It also pointed out that the Thai aspectuality focused on the initial boundary and terminative boundary of the state of affairs. It is here that combinations of the three aspect markers occur. The study shows that they have certain rules of co-occurances.
International Journal of Academic Research in Progressive Education and Development
Francis Onyango
Sunil K Bhatt
Wacana Journal of the Humanities of Indonesia , Philippe Grangé
In this paper, I describe four Indonesian aspect markers, sudah, telah, pernah, and sempat, showing that the main opposition between them relies not only on their aspectual meanings, but also on the various modalities they express. The opposition between the very frequent markers sudah and telah is analysed in detail. The syntactic and semantic survey shows that these two markers are not synonyms in most contexts.
BEST Journals
DELTA [online]
Maria das Graças Salgado , Adriana Lessa
Aspect in the English language has been described through different categories and terminologies, which might lead teachers and students into some misunderstandings. Considering the importance of understanding the systematic representation of this concept in learning a foreign language, we review and compare the various ways aspect is presented in five of the most traditional descriptive English grammar books. We examined whether aspect is explicitly approached; how it is defined; categorized and whether the types of aspect are clearly explained in terms of meaning. Based on that, we contrasted their classification and terminology with an alternative approach, highlighting ambiguities and common grounds. Keywords : aspect; English grammar; classification; comparative analysis.
RELATED PAPERS
Maria Eileen Caballero
Damien Mackey
giorgio marzocchi
Ainul Yakin
American Journal of Islam and Society
Widitra Purba
Global Journal of Human Social Science Research
Burial Taphonomy and Post-Funeral Practices in Pre-Roman Italy. Problems and Perspectives, edited by Martin Guggisberg and
Peter Attema , Paola Catalano
Jael Millán Parra
Tom Fullerton
4th BSME- …
Tazul Islam
Journal of Obstetric, Gynecologic & Neonatal Nursing
SSRN Electronic Journal
Marcel Kahan
Schweizerisches Nationalmuseum (Hrsg.), Menschen. In Stein gemeisselt, Christoph Merian Verlag/Schweizerisches Nationalmuseum, Basel/Zürich
Jacqueline Perifanakis
IEEE Access
SERGIO QUINTERO
Value in Health
Ramiro Gilardino
hoàng nguyễn
The Journal of Agricultural Science
Sandra Paula Alexandre Valentim
The International Review of Retail, Distribution and Consumer Research
Jonathan Reynolds
BMC public health
maryline margueritte
Acta Educationis Generalis
Dinçay KÖKSAL
Mediators of Inflammation
Mohamed Tikly
Journal of Palaeosciences
shreerup Goswami
Mercedes Diaz
Severine Bouard
麦考瑞大学毕业证文凭办理成绩单修改 办理澳洲MQ文凭学位证书学历认证
European Neuropsychopharmacology
Stanislava Vladimirova
European Journal of Anaesthesiology
- We're Hiring!
- Help Center
- Find new research papers in:
- Health Sciences
- Earth Sciences
- Cognitive Science
- Mathematics
- Computer Science
- Academia ©2024
Subscribe Now! Get features like

- Latest News
- Entertainment
- Real Estate
- Lok Sabha Election 2024
- My First Vote
- IPL 2024 Schedule
- IPL Points Table
- IPL Purple Cap
- IPL Orange Cap
- The Interview
- Web Stories
- Virat Kohli
- Mumbai News
- Bengaluru News
- Daily Digest
- Election Schedule 2024

'Gamocha', a symbol of the culture and identity of Assam receives GI tag
Five years after the initial application, the central government has granted gamocha', a representation of assamese culture and identity, the geographical indication (gi) tag..
'Gamocha', a symbol of the culture and identity of Assam, has received the Geographical Indication (GI) tag from the central government, five years after the first application was made. Union Minister of Commerce and Industry Piyush Goyal on Twitter shared the GI registration certificate, issued on Tuesday, bringing cheers to people across the northeastern state. A GI is tagged to primarily agricultural, natural or manufactured products, handicrafts and industrial goods originating from a definite geographical territory. (Also read: By making 'Gamosa', Assamese women keeping state's tradition alive )

Assam Chief Minister Himanta Biswa Sarma said: "A day of pride for Assam, as our #Gamosa gets a Geographical Indication Tag by the Government of India."
Expressing gratitude to Prime Minister Narendra Modi, who is often seen with a 'Gamocha' draped around his neck, Sarma congratulated all the people of Assam for this recognition. Union Shipping, Ports and Waterways Minister Sarbananda Sonowal stated that Assam's pride is "shining bright".
"Due recognition of our heritage & cultural identity. 'Gamosa' gets the GI tag, bringing joy to thousands of weavers of this special item that has become a global symbol of Assam," he added.
Along with Assam ministers Chandra Mohan Patowary, Ajanta Neog, Atul Bora, Pijush Hazarika and Jayanta Malla Baruah, many prominent personalities and hundreds of social media users expressed their happiness over the 'gamocha' getting the GI tag.
The GI tag has been registered in favour of the Directorate of Handloom & Textiles of the Assam government under the Geographical Indication of Goods (Registration and Protection) Act, 1999. The certificate has been awarded for 'Gamocha' as of October 16, 2017 as per the application by the directorate. The process was started in 2017 when an application for a GI tag was filed by the Institute of Handicraft Development of Golaghat district.
The 'gamocha', a handwoven rectangular cotton piece of cloth with red borders and different designs and motifs, is traditionally offered to elders and guests as a mark of respect and honour by Assamese people. It is an integral part of all socio-religious ceremonies in the state and is considered as an Assamese identity and pride.
A 'gamocha' literally means a towel and is commonly used in Assamese households for day-to-day affairs. For specific purposes, it is also made of expensive materials like traditional Assamese 'Pat' silk and in different colours as well.
A 'gamocha' meant for exchange during 'Bihu' festival is known as 'Bihuwan'. This unique scarf, which is found only in Assam, is also used as a signifier of reverence while decorating altars or covering religious books.
Follow more stories on Facebook & Twitter
- Cultural Heritage
IPL 2024 Coverage

Join Hindustan Times
Create free account and unlock exciting features like.

- Terms of use
- Privacy policy
- Weather Today
- HT Newsletters
- Subscription
- Print Ad Rates
- Code of Ethics
- IPL Live Score
- T20 World Cup Schedule
- IPL 2024 Auctions
- T20 World Cup 2024
- Cricket Teams
- Cricket Players
- ICC Rankings
- Cricket Schedule
- T20 World Cup Points Table
- Other Cities
- Income Tax Calculator
- Budget 2024
- Petrol Prices
- Diesel Prices
- Silver Rate
- Relationships
- Art and Culture
- Taylor Swift: A Primer
- Telugu Cinema
- Tamil Cinema
- Board Exams
- Exam Results
- Competitive Exams
- BBA Colleges
- Engineering Colleges
- Medical Colleges
- BCA Colleges
- Medical Exams
- Engineering Exams
- Horoscope 2024
- Festive Calendar 2024
- Compatibility Calculator
- The Economist Articles
- Lok Sabha States
- Lok Sabha Parties
- Lok Sabha Candidates
- Explainer Video
- On The Record
- Vikram Chandra Daily Wrap
- EPL 2023-24
- ISL 2023-24
- Asian Games 2023
- Public Health
- Economic Policy
- International Affairs
- Climate Change
- Gender Equality
- future tech
- Daily Sudoku
- Daily Crossword
- Daily Word Jumble
- HT Friday Finance
- Explore Hindustan Times
- Privacy Policy
- Terms of Use
- Subscription - Terms of Use
- Cambridge Dictionary +Plus
Meaning of representation in English
Your browser doesn't support HTML5 audio
representation noun ( ACTING FOR )
- Defendants have a right to legal representation and must be informed of that right when they are arrested .
- The farmers demanded greater representation in parliament .
- The main opposing parties have nearly equal representation in the legislature .
- The scheme is intended to increase representation of minority groups .
- The members are chosen by a system of proportional representation.
- admissibility
- extinguishment
- extrajudicial
- extrajudicially
- fatal accident inquiry
- federal case
- pay damages
- plea bargain
- plea bargaining
- the Webster ruling
- walk free idiom
- witness to something
representation noun ( DESCRIPTION )
- anti-realism
- anti-realist
- complementary
- confederate
- naturalistically
- non-figurative
- non-representational
- poetic licence
- symbolization
representation noun ( INCLUDING ALL )
- all manner of something idiom
- alphabet soup
- it takes all sorts (to make a world) idiom
- non-segregated
- odds and ends
- of every stripe/of all stripes idiom
- this and that idiom
- variety is the spice of life idiom
- wide choice
representation | Business English
Examples of representation, collocations with representation.
- representation
These are words often used in combination with representation .
Click on a collocation to see more examples of it.
Translations of representation
Get a quick, free translation!

Word of the Day
to try to persuade a customer who is already buying something to buy more, or to buy something more expensive

Searching out and tracking down: talking about finding or discovering things

Learn more with +Plus
- Recent and Recommended {{#preferredDictionaries}} {{name}} {{/preferredDictionaries}}
- Definitions Clear explanations of natural written and spoken English English Learner’s Dictionary Essential British English Essential American English
- Grammar and thesaurus Usage explanations of natural written and spoken English Grammar Thesaurus
- Pronunciation British and American pronunciations with audio English Pronunciation
- English–Chinese (Simplified) Chinese (Simplified)–English
- English–Chinese (Traditional) Chinese (Traditional)–English
- English–Dutch Dutch–English
- English–French French–English
- English–German German–English
- English–Indonesian Indonesian–English
- English–Italian Italian–English
- English–Japanese Japanese–English
- English–Norwegian Norwegian–English
- English–Polish Polish–English
- English–Portuguese Portuguese–English
- English–Spanish Spanish–English
- English–Swedish Swedish–English
- Dictionary +Plus Word Lists
- representation (ACTING FOR)
- representation (DESCRIPTION)
- representation (INCLUDING ALL)
- make representations to sb
- Collocations
- Translations
- All translations
To add representation to a word list please sign up or log in.
Add representation to one of your lists below, or create a new one.
{{message}}
Something went wrong.
There was a problem sending your report.
- More from M-W
- To save this word, you'll need to log in. Log In
representation
Definition of representation
Examples of representation in a sentence.
These examples are programmatically compiled from various online sources to illustrate current usage of the word 'representation.' Any opinions expressed in the examples do not represent those of Merriam-Webster or its editors. Send us feedback about these examples.
Word History
15th century, in the meaning defined at sense 1
Phrases Containing representation
- proportional representation
- self - representation
Dictionary Entries Near representation
representant
representationalism
Cite this Entry
“Representation.” Merriam-Webster.com Dictionary , Merriam-Webster, https://www.merriam-webster.com/dictionary/representation. Accessed 17 May. 2024.
Kids Definition
Kids definition of representation, legal definition, legal definition of representation, more from merriam-webster on representation.
Thesaurus: All synonyms and antonyms for representation
Nglish: Translation of representation for Spanish Speakers
Britannica English: Translation of representation for Arabic Speakers
Britannica.com: Encyclopedia article about representation
Subscribe to America's largest dictionary and get thousands more definitions and advanced search—ad free!


Can you solve 4 words at once?
Word of the day.
See Definitions and Examples »
Get Word of the Day daily email!
Popular in Grammar & Usage
More commonly misspelled words, your vs. you're: how to use them correctly, every letter is silent, sometimes: a-z list of examples, more commonly mispronounced words, how to use em dashes (—), en dashes (–) , and hyphens (-), popular in wordplay, birds say the darndest things, a great big list of bread words, 10 scrabble words without any vowels, 12 more bird names that sound like insults (and sometimes are), 8 uncommon words related to love, games & quizzes.

Gender And Assamese Cinema: The Shifting Pattern

Assamese regional films always ensured to pour cultural richness through their narratives, music and representation through visual narratives. The cinema industry of Assam was formed with the lineage starting from Joymoti directed by Jyoti Prasad Agarwala. In the ’70s and ’80s, this film also led to the growth of films with visually strong narration moving away from the conventional film on entertainment. These films focused more on the richness of culture, politics and the strong use of the visual medium to bring awareness and address different issues of social context through cinema.
Cinema is an expensive trait and one of the strongest mediums to visually represent and expose social reality. This is how the Assamese film wave found its course during the ’60s along with addressing social and cultural discourse. The portrayal of gender roles in Assamese regional cinema has been an essential aspect of the state’s affair of social, cultural and ethical sustainability.
There was also a constant shift seen in the cinema history of Assam with the region always being under the radar due to insurgency. Whereas, many films have actually put light on the serious issues faced by the state for 3 decades of separatism, militancy and insurgency and its conflicting position within its own neighbourhood states. There were also films which are believed to have attempted to dilute the entire cause through their productions.
However, the cause of filmmaking to understand and address social concerns through creative mediums has overall been successful throughout history.
Assamese regional cinema and its growing history
Most of the old literature talks about the Assamese regional film industry heading back to the 1930s when the first film Joymoti was produced. Joymoti was based on the life of an Ahom queen of the same name directed by Agarwala. The film was an instant hit and also the first film to have been produced all over Assam.
In the 1960s and 1970s, a noticeable shift was seen in the techniques of narrative and storytelling. Some of the films also adapted to the storytelling of folk culture and represent historical incidents like Lachit Borphukan and Aranya. These films not only spoke strongly about history but also touched upon the lens of gender and gender roles.

Jahnu Barua, who is a renowned name in the regional film industry of Assam made a solid effort of bringing social interest into parallel cinema. Though, regional cinema has also seen its mighty struggle to keep itself alive due to a lack of knowledge, and support from the government or bureaucrats now and then. However, his films mostly portrayed strong female characters right from the first film Aparoopa .
Assamese regional cinema and gender roles
Assamese regional cinema has always been influenced by the region’s patriarchal society, where men hold a dominant position in both the public and private spheres. The portrayal of gender roles in Assamese regional cinema has often been stereotypical, where women are depicted as passive, submissive, and subservient to men.
In the early days of Assamese regional cinema, women were mostly portrayed as objects of desire or passive characters who played a secondary role to the male protagonist. Female characters were often portrayed as damsels in distress who needed to be rescued by the male hero. However, over the years, there has been a significant shift in the portrayal of female characters in Assamese regional cinema.

The 1980s saw a significant change in the portrayal of female characters in Assamese regional cinema. The emergence of female directors and screenwriters brought a fresh perspective to the narrative, and women’s issues became an essential part of the discourse. Films such as Rati Chakravyuh , Xagoroloi Bohudoor , and Firingoti portrayed women as strong, independent, and capable of making their own decisions. These films also highlighted the challenges and struggles faced by women in a patriarchal society.
In recent years, Assamese regional cinema has continued to explore the nuances of gender roles and break the stereotypes through cinema and creative visual storytelling. Films such as Village Rockstars , Bulbul Can Sing , and Aamis have challenged the traditional notions of gender roles and explored themes such as same-sex relationships, female desire, and the intersection of gender and class.
In the 1990s and 2000s, Assamese regional cinema saw the emergence of a new wave of filmmakers who challenged traditional gender roles and stereotypes. Films such as Hiya Diya Niya , Ramdhenu , and Mon Jaai explored the complexities of relationships and portrayed women as multi-dimensional characters with their desires and aspirations.

Aamis , directed by Bhaskar Hazarika, explores the intersection of gender and desire. The film tells the story of a young woman named Nirmali, who develops a strange and forbidden relationship with a married man. The film explores the complexities of desire and the societal norms restricting women’s agency and autonomy. Aamis was also critically acclaimed and won several awards, including the Best Director Award at the Singapore International Film Festival.
Challenges women filmmakers face
Assamese regional cinema has seen a new wave of films in recent years that explore the complexities of gender roles and stereotypes. These films not only challenge the traditional notions of gender but also provide a platform for the voices of marginalised communities, including women, the LGBTQ+ community, and the working class. One such film is Village Rockstars , directed by Rima Das.
The film also introduces us to the conventional and old method of storytelling by reducing the use of dialogue and furthering the plot with the use of the camera.

The film tells the story of a young girl named Dhunu, who dreams of owning a guitar and forming a rock band with her friends. The film explores the challenges faced by a young girl growing up in a patriarchal society and her struggle to break free from gender stereotypes. Village Rockstars was not only critically acclaimed but also won several national and international awards, including the National Film Award for Best Feature Film.
Another film that explores the complexities of gender and sexuality is Bulbul Can Sing , also directed by Rima Das . The film tells the story of a young girl named Bulbul, who grows up in a remote village in Assam and discovers her sexuality in a conservative and homophobic society. The film explores the challenges faced by a young girl coming to terms with her sexuality and the discrimination faced by the LGBTQ+ community in India. Bulbul Can Sing was also critically acclaimed and won several awards, including the Best Asian Film Award at the Pingyao International Film Festival.
Paradigm shift
Ever since its inception, in 1930, Assamese regional film has explored and crossed diverse paths throughout its intersection with respect to caste, class, gender, race, sexuality and so on. The emergence of female directors and screenwriters has brought a fresh perspective to the discourse, and women’s issues have become an integral part of the narrative.

New films such as Village Rockstars , Bulbul Can Sing , and Aamis have challenged the traditional notions of gender and explored themes such as same-sex relationships, female desire, and the intersection of gender and class. These films not only provide a platform for marginalised communities but also challenge the societal norms that restrict women’s agency and autonomy.
As the industry continues to evolve, it is essential to give space to diverse voices and perspectives and ensure that the discourse remains relevant and representative of the state’s social and cultural values.

Ritwik is a northeastern Bahujan who has been a journalist for 2 years. He has also been a UN fellow for the year 2021-22. He has worked on several papers like writing about reportage in broadcast media on women impacted by floods in the northeast. He is currently working on queer representation in the regional press of Assam. Most of the work focused on gender, sexuality, intersectionality, climate change, marginalised and vulnerable communities and data journalism. He has been a contributing writer at Youth ki Awaaz, Feminism in India and Swaddle for the last 4 years.
Related Posts

Women On And Off Camera In Amazon MiniTV’s ‘Amber Girls School’
By Deepti Bholanda
Consent Or Coercion: Navigating Dating Spaces As A Heterosexual Woman
By Anjana Anilkumar

‘Ripley’: A Grey Character Shown In Stunning Black And White
By Dr. Guni Vats
- India Today
- Business Today
- Reader’s Digest
- Harper's Bazaar
- Brides Today
- Cosmopolitan
- Aaj Tak Campus
- India Today Hindi
Definition of Assamese must include those living here for years: Himanta Sarma
Himanta biswa sarma has emphasised the need for an inclusive definition of "assamese," recognising changing demographics and advocating a mass movement to preserve the community's identity..
Listen to Story

- Himanta Sarma calls for mass movement to preserve Assamese identity
- Suggests redefinition of Assamese to include Hindi speakers
- Sarma blames demographic change in Assam on Bangladeshi infiltration
The definition of Assamese has undergone change over the years, and it must include people who have lived in Assam for centuries, such as Hindi speakers and the tea tribes, Chief Minister Himanta Biswa Sarma said on Saturday.
He said that a mass movement is needed to convince the Assamese people that the identity of the community can be preserved through "quality".
Sarma was speaking at a programme in which rehabilitation grants were distributed under the ULFA Peace Accord to surrendered cadres.
"The situation in Assam is not due to some policy of the central government but due to infiltration from Bangladesh, which changed the demography of the state. When the census report comes, Assamese people will be only about 40 per cent of the population," he claimed.
Sarma said that the definition of Assamese has undergone change over the years, and it must include communities such as the tea tribes and Hindi speakers who have been living here for over a century.
Though the Assamese people may be dwindling in numbers, they can come together to assert their identity, he maintained.
"A mass movement is needed so that we can convince the people that though we do not have the numbers, with quality we can keep our Assamese identity alive," the CM said.
Praising the ULFA leadership and cadres for returning to the mainstream, Sarma said they should urge those who are still pursuing armed struggle to join them.
"I always ask Paresh Barua to come and spend 10 days in Assam. After that he will not want to return to Myanmar or China," he said, referring to the chief of the ULFA (I) which has refused to come forward for peace talks.
Sarma maintained that being "rational rather than emotional" is the need of the hour to take forward the state in the path of development. Published By: Vani Mehrotra Published On: Mar 2, 2024 ALSO READ | 'Decisions taken in dining room of one family': Himanta Sarma jabs Congress ALSO READ | BJP to contest 11 seats from Assam in Lok Sabha polls: Himanta Biswa Sarma
Dictionary English - Assamese
Translations from dictionary english - assamese, definitions, grammar.
In Glosbe you will find translations from English into Assamese coming from various sources. The translations are sorted from the most common to the less popular. We make every effort to ensure that each expression has definitions or information about the inflection.
In context translations English - Assamese, translated sentences
Glosbe dictionaries are unique. In Glosbe you can check not only English or Assamese translations. We also offer usage examples showing dozens of translated sentences. You can see not only the translation of the phrase you are searching for, but also how it is translated depending on the context.
Translation memory for English - Assamese languages
The translated sentences you will find in Glosbe come from parallel corpora (large databases with translated texts). Translation memory is like having the support of thousands of translators available in a fraction of a second.
Pronunciation, recordings
Often the text alone is not enough. We also need to hear what the phrase or sentence sounds like. In Glosbe you will find not only translations from the English-Assamese dictionary, but also audio recordings and high-quality computer readers.
Picture dictionary
A picture is worth more than a thousand words. In addition to text translations, in Glosbe you will find pictures that present searched terms.
Automatic English - Assamese translator
Do you need to translate a longer text? No problem, in Glosbe you will find a English - Assamese translator that will easily translate the article or file you are interested in.
It's nice to welcome you to the Glosbe Community. How about adding entries to the dictionary?
Add translation
Help us to build the best dictionary.
Glosbe is a community based project created by people just like you.
Please, add new entries to the dictionary.
Recent changes
Statistics of the english - assamese dictionary, language english, language assamese.

NORTHEAST NOW
NORTHEAST NEWS
Assam CM pushes for inclusive definition of Assamese identity

Share this:
- Google News

Guwahati: Assam Chief Minister Himanta Biswa Sarma has emphasized the evolving nature of Assamese identity.
Sarma spoke on Saturday in Guwahati at a programme distributing rehabilitation grants to surrendered ULFA cadres under the peace accord.
Sarma highlighted the need for a broader definition that encompasses communities like Hindi speakers and tea tribes who have called Assam home for generations.
He attributed the potential decline in the percentage of Assamese speakers to “infiltration from Bangladesh” impacting the state’s demographics.
While acknowledging a possible decrease in numbers, Sarma stressed the importance of preserving Assamese identity through “quality.” He called for a “mass movement” to convince people that their unique character can endure.
The Chief Minister commended the ULFA leadership for embracing peace and urged remaining militants to follow suit.
He expressed confidence that experiencing Assam firsthand would convince them to abandon the armed struggle.
Sarma concluded by advocating for a “rational” approach to propel the state towards development.
NE NOW NEWS
Northeast Now is a multi-app based hyper-regional bilingual news portal. Mail us at: [email protected] More by NE NOW NEWS
- Open access
- Published: 15 May 2024
Readsynth: short-read simulation for consideration of composition-biases in reduced metagenome sequencing approaches
- Ryan Kuster 1 &
- Margaret Staton 1
BMC Bioinformatics volume 25 , Article number: 191 ( 2024 ) Cite this article
8 Altmetric
Metrics details
The application of reduced metagenomic sequencing approaches holds promise as a middle ground between targeted amplicon sequencing and whole metagenome sequencing approaches but has not been widely adopted as a technique. A major barrier to adoption is the lack of read simulation software built to handle characteristic features of these novel approaches. Reduced metagenomic sequencing (RMS) produces unique patterns of fragmentation per genome that are sensitive to restriction enzyme choice, and the non-uniform size selection of these fragments may introduce novel challenges to taxonomic assignment as well as relative abundance estimates.
Through the development and application of simulation software, readsynth, we compare simulated metagenomic sequencing libraries with existing RMS data to assess the influence of multiple library preparation and sequencing steps on downstream analytical results. Based on read depth per position, readsynth achieved 0.79 Pearson’s correlation and 0.94 Spearman’s correlation to these benchmarks. Application of a novel estimation approach, fixed length taxonomic ratios , improved quantification accuracy of simulated human gut microbial communities when compared to estimates of mean or median coverage.
Conclusions
We investigate the possible strengths and weaknesses of applying the RMS technique to profiling microbial communities via simulations with readsynth. The choice of restriction enzymes and size selection steps in library prep are non-trivial decisions that bias downstream profiling and quantification. The simulations investigated in this study illustrate the possible limits of preparing metagenomic libraries with a reduced representation sequencing approach, but also allow for the development of strategies for producing and handling the sequence data produced by this promising application.
Peer Review reports
Since its first application, reduced metagenomic sequencing (RMS) has remained a niche approach to profiling microbial communities. First coined by Liu et al., RMS is the application of reduced-representation sequencing (RRS) to metagenomic libraries, adopting steps modified from the original ddRADseq protocol [ 1 , 2 ]. Metagenomic profiling of human samples comparing RMS and whole genome sequencing (WGS) yielded similar microbial profiles, and concerns of GC content bias weren’t detected. Three further studies have shown the potential for RMS to yield similar results or even outperform the traditional 16S and WGS approaches [ 3 , 4 , 5 ].
Although traditional approaches of amplicon and WGS can be effective for studying community structure, they exist on extreme ends of sequencing efforts and the benefits of each may exist in a middle ground. The conserved gene regions used in amplicon sequencing, often the 16S or ITS gene, can create bias towards some community members due to the primers selected and often these marker gene targets lack the resolution to consistently identify one species or strain from another [ 6 ]. WGS increases resolution but is comparatively expensive for deep sampling of rare taxa. Continuous, overlapping reads originating from closely related taxa may be computationally challenging to assign. Comparable to genotyping by sequencing (GBS), RMS reduces a genome into sampled DNA fragments by using one or more restriction enzymes [ 7 ]. These subsetted fragments represent predictable, targeted loci within a genome and function as hundreds of markers within each microbial genome allowing for species and even strain-level identification not found in amplicon sequencing alone. RRS increases per-locus sequencing depth for these fragments and allows for more samples to be analyzed in the same sequencing run, improving sequencing accuracy and lowering the per-sample cost of sequencing [ 1 , 8 , 9 ]. Unlike the original application of reduced representation sequencing for GBS, which produces fragments from the genome of an individual, RMS creates fragments from many unknown source genomes into a single pooled sequencing run.
Despite the potential benefits of RMS, the ability of this approach to accurately quantify at the species and strain level has not been tested at scale, possibly hindering adoption by metagenomics researchers. A major caveat to the RMS approach is that restriction enzyme motifs are enriched in a taxa-dependent manner [ 3 ]. Genome size, which can range over an order of magnitude in bacteria, as well as the varying degrees of sequence conservation among closely related microbes may complicate RMS quantification [ 10 ]. For example, genomic loci yielding RMS fragments may be variable even among closely related individuals, as a single base mutation altering a restriction cut site can lead to “allelic dropout” [ 11 ]. The degree to which this variation affects taxonomic identification and determination of relative abundance of community members has not been studied deeply. Every community member comprising the metagenomic sample will yield a unique distribution of fragment lengths.
Further complicating RMS profiling, sequencing effort (i.e. total read count) and size selection constraints may significantly impact the taxa included in the final sequencing library. Both PCR amplification and fragment size selection steps will affect the probability of a given DNA fragment surviving into the final sequencing reaction in a size-dependent manner with possible PCR biases dependent on template length [ 12 , 13 ]. Gel or bead mediated size selection is a critical step in RRS library design that helps to remove adapter dimers and optimize flow cell performance, and so remains an unavoidable library preparation step in most cases. An alternative RRS approach using isolength (type IIB) restriction enzymes has been proposed as a potential solution to this issue of variable fragment length [ 14 , 15 ]. Using a single enzyme that produces constant fragment lengths frequently across the genome may create detailed fingerprints of genomic communities without size selection steps, assuming adapter dimers can be reduced. Normalizing for the non-uniform read depths associated with a single organism is a task that existing metagenome profiling software aren’t designed to handle.
In applications such as RMS, where existing short read approaches are being used in novel ways, it may be difficult to gain traction because the features of the data are so understudied for the intended application. Spending money to develop new experimental techniques can be a highly risky endeavor, and it also requires the development of custom software tools and statistical approaches necessary to analyze this new form of data. In instances such as these, simulation can be useful to predict and overcome artifacts from library prep that could not be anticipated or otherwise measured without a ground truth. Capturing the nuances of RMS behavior under different conditions is a necessary step in understanding its application, therefore, we introduce the software readsynth as a simulation aid to researchers in considering these promising alternative approaches to sequencing metagenomes. Readsynth simulates read count constrained metagenomic sequence data based on pre-assembled genomes and user-defined community compositions along with multiple library preparation parameters.
In this study, both real and simulated RMS libraries are investigated to understand the influence of non-uniform fragment sizes and possible molecular and computational considerations that might be used to overcome these biases. Bias in fragment lengths may be occurring due to the biology of the individual community members being sampled and/or the technical library preparation. These factors prevent the total numbers of reads mapped to reference genomes from being directly used for calculating relative abundance. Our proposed solution uses the fixed length taxonomic ratio (FLTR) of read depths occurring within a non-uniform fragment length distribution. Barring the presence of strong GC bias, the ratio of two taxa should remain constant within all fragment lengths where both taxa are present. We first benchmark readsynth’s ability to faithfully capture features of mock community data. To demonstrate the utility of readsynth simulations, we then investigate this novel FLTR quantification approach that may overcome some of the challenges produced by non-uniform fragment distributions. Upon finding the library conditions that produce the best resolution, we consider the ability of these approaches when the search space includes a full database of all available reference genomes.
Implementation
Simulation software overview.
Readsynth was developed to simulate Illumina short read libraries to assess the compositional abundance of highly custom communities under multiple reduced sequencing conditions. Readsynth is a command line software package written in Python and C++ that uses commonly maintained statistical packages and consists of a digestion, size-selection, and read-writing stages (Fig. 1 ). The software was written to be highly customizable across three categories: (1) microbial community composition, defined by input genomes and their relative abundance; (2) experimental parameters, including reduced library approach, enzyme digestion rates, expected fragment length distributions, and custom adapter design, and (3) sequencing parameters, such as total read number, read length, and base quality value profiles.
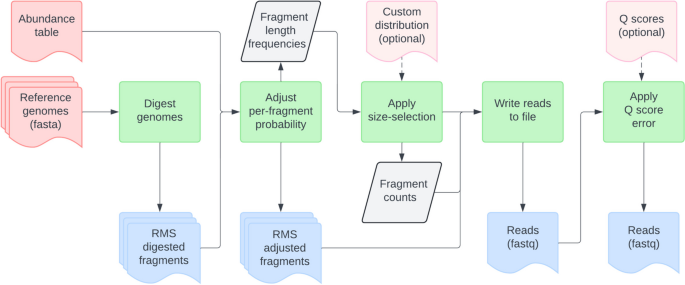
Overview of readsynth inputs and flow of data. Necessary input files (shown in pink) are a collection of genome files and a corresponding abundance table for each. Output files include per fragment count estimates and the final, paired-end read fastq files
Readsynth first reads each input genome assembly individually to capture the set of possible fragments and calculate the probability of each sequence fragment surviving to the final library. Given a user input set of IUPAC restriction site motifs, overlap-tolerant regular expression (regex) searches are performed to exhaustively detect all possible cleavage sites and define fragments within the expected size-selection distribution. Fragments resulting from any combination of palindromic restriction enzyme motifs are modeled probabilistically to account for partial enzyme digestion. The probability of a fragment remaining at the end of digestion is calculated based on the probability of an enzyme cut producing the necessary forward and reverse adapter-boundary sites, adjusted accordingly for fragments harboring internal cut sites.
The per-fragment probability is a function of enzyme cleavage occurring at both ends of a sequence based on a user defined enzyme cut efficiency (c). Sequences that harbor greater numbers of internal cut sites (i) are less frequently represented as a sequenced read. The probabilities for each fragment length are then summed, approximating the expected fragments given a single genome copy for each genome in consideration.
Size selection
To simulate size selection, each fragment’s post-digestion probability is adjusted based on multiple additional factors affecting its representation. First, the expected fragment counts for each genome are scaled by their proportional abundance, as defined in the abundance table. The combined distribution of digested fragment lengths for all input genomes then undergoes size selection. The counts of the digestion distribution are used to scale a Gaussian probability density function at a given length, x, and this intersection of sample spaces defines the final size selection distribution (Fig. 2 ). This approach follows the size variability expected in gel-based size-selection equipment (e.g., SageScience BluePippin) at the narrow and broad range selection techniques while preventing artificial inflation of reads in lower abundance than produced by the Gaussian curve [ 1 ]. To simulate the hardware-imposed limitation on the composition of metagenomic fragments, the input read number (n) is divided evenly amongst the resulting size-selected distribution of the digested metagenome.
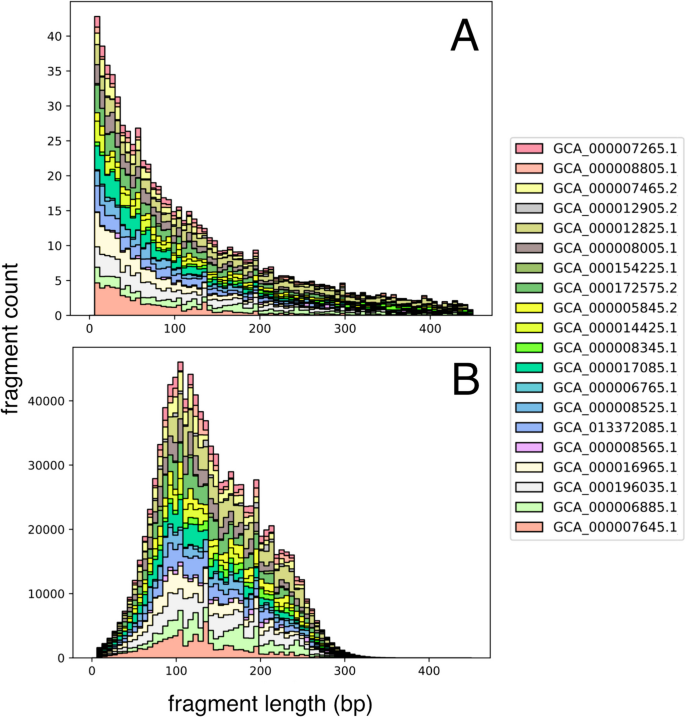
Digestion distribution of reduced metagenome sequencing (RMS) fragment lengths (bp) A after simulated enzyme fragmentation, counts represent expected fragment frequency with 1X genome coverage; B final read counts of size-selected fragments from the intersection of a Gaussian normal (μ: 150 bp, σ: 50) and A, scaled to the count at length 100 bp from A. Bar colors indicate individual bacterial genomes present in the simulated community
Error modeling
Readsynth applies a straightforward substitution error model to every read using randomly sampled Q scores from any existing fastq file, with several publicly available profiles to select from. Phred-like error probability rates from the sampled Q scores are used to mutate each nucleotide base to a non-self modification using pseudo-random number generation. Simulated fragments that are shorter than the simulated read length resulted in expected adapter contamination in data output (Supplementary Fig. 1 ), and users may provide any number of custom-designed adapters with specific overhangs.
Software benchmarking
In order to benchmark the simulation accuracy of readsynth, we use existing RMS data as a ground truth. The loci-specific enrichment of reads as well as the similarity in taxonomic profiles between real and simulated reads are used as performance metrics. Two previously sequenced mock community data sets produced with RMS approaches were considered. These communities used standardized concentrations of each community member, making them ideal for comparison with simulated reads. The first dataset from Snipen et al. consists of a Human Microbiome Project mock community of 20 bacterial strains (BEI HM-782D) digested using the restriction enzymes EcoRI and MseI [ 5 ]. To simulate the abundance of each taxon, the input abundance table used the reciprocal of the ribosomal copy number determined from the ribosomal RNA operon copy number database [ 16 ]. The second dataset from Sun et al. used a separate community of 20 bacterial strains (ATCC MSA-1002), which was assumed to have even genomic copy number [ 14 ]. This dataset was created using the type IIB restriction enzyme BcgI which produces short fragments with no length variation.
Extracting sequencing features from real and simulated mock communities
Sequence reads from the existing mock communities were adapter trimmed using Cutadapt 4.1 [ 17 ]. Reads were then mapped against each of the community member reference genomes using BWA MEM (Burrows Wheeler Aligner 0.7.17) [ 18 ]. Samtools 1.15.1 was used to select only paired end reads with the appropriate orientation. The Samtools stats and depth commands were used to summarize the read lengths as well as the per-position depth of all aligned reads [ 19 ]. Using positional information from these reads, a custom fragment length distribution was estimated for use in simulation. The count ratio of fully digested fragments to the larger fragments immediately encompassing them (r) was used to estimate the cut efficiency (c) of enzyme digestion except for the case of complete digestion in which c = 1. In cases of incomplete digestion, following the per-fragment probability model, fragments containing internal cut sites will occur less frequently than their contained fragments such that r must be greater than 1.
Only fragment lengths in the range of 100 bp to 450 bp were used to estimate cut efficiency, as these were observed to be less constrained by size selection and therefore more reliable in preserving true read ratios (Supplementary Fig. 2 ). Aligning community mixtures of many taxa against individual reference genomes in BWA MEM returned many reads that aligned to multiple genomes (Supplementary Fig. 3 ). Further, variability between the published reference sequence and the real sequence data resulted in the rare presence of RMS fragments not reproducible in simulation. In order to make meaningful comparisons between the real and simulated data, custom Python scripts were used to extract positional information from high quality read alignments in the sam-formatted file in order to preserve the fragment size distribution while removing duplicate alignments.
Simulation was performed using fragment size distribution, enzyme cut efficiency, and Q score profile derived from the real sequence data. Although we expect these derived inputs to most closely resemble the real data, additional simulations were run to test the importance of fragment size distribution and enzyme cut efficiency. Three fragment length distributions were considered to measure the impact of size selection. The first was created using the exact fragment counts extracted as a custom.json file from the extracted sequence alignments. The second simulated size selection using the mean and standard deviation from the samtools summary statistics to define the distribution shape. The third size selection distribution used identical standard deviation values, but with the mean fragment size increased by 100 bp. BWA MEM and Samtools were again used to map simulated reads to the reference genomes and the read depth at every expected fragment position was counted. Cumulative read depth across every position in the mock community was compared between real and simulated sequences using both Pearson and Spearman correlation.
Assessing compositional-biases using simulation
Simulations of RMS sequencing reads were informed using the mock community and two representative gut metagenomic communities based on sequencing efforts from complex microbial samples. Mock community sequences from Snipen et al. (replicates SRR10199716, SRR10199724, and SRR10199725) were again used as a baseline to quantify taxonomic abundances from a mixture of 20 bacterial taxa at known relative abundance. Taxa were assumed to be approximately equimolar in ribosomal operon count as described in the product specifications. A single human stool sample (SRR5298272) prepared with RMS using NlaIII and HpyCH4IV was used to establish a microbial community with greater richness for simulation [ 2 ]. These biological sample reads were assigned putative taxonomic labels using Kraken2 and Bracken (Standard plus protozoa, fungi & plant database ‘PlusPFP’ June 2022) and resulting taxonomic identification numbers were used to download representative genomes from each of the 691 non-host hits [ 20 , 21 ]. Finally, the OTU profile resulting from gut samples from 2,084 individuals from the Healthy Life in an Urban Setting (HELIUS) study served as a basis for simulating microbial communities with authentic taxonomic abundances [ 22 ]. Of the 744 OTUs, 610 unique RefSeq genome references from the genus and species level served as the basis for simulation. When multiple OTUs from the same genus level were encountered, multiple species from this genus were selected based on genomes with full, major releases in GenBank, which included many highly similar strains to be simulated in the same community. All simulations were performed using the software readsynth (0.1.0; commit 88d8bb1).
To assess the sensitivity of RMS to capture rare taxa, metagenomes based on HELIUS communities were simulated using a series of increasing total read counts (1 × 10 5 , 1 × 10 6 , 1 × 10 7 , and 1 × 10 8 paired end reads) and four combinations of restriction enzyme double digests. The combinations of restriction enzymes selected (EcoRI/AgeI, EcoRI/MseI, HhaI/AgeI, and HhaI/MseI) were chosen for the diversity of the cut site frequency and GC content of the recognition motifs (Table 1 ). Additionally, a type IIB enzyme “BcgI” digest was simulated to assess the application of the resulting isolength fragments.
Paired end reads from each of the real and simulated datasets were aligned to a combined reference genome concatenated from all known members in the metagenome using BWA MEM. Mapped reads with a MAPQ score of zero were removed to avoid reads that map closely to multiple reference genomes. Custom Python scripts (available at github.com/ryandkuster/readsynth_analysis) were used to recreate the original genomic fragments corresponding with each read pair based on the simulated start and end positions for each fragment produced in simulation. Only those fragments harboring no internal cut sites were kept as incomplete digests are expected to unpredictably affect quantification and assessment of existing RMS data found these fragments to be rare. The observed count and corresponding taxonomic assignment were stored for each fragment.
We also assess the performance of RMS data when the reference database is not curated to only those references in the ground truth set, as is the case in most practical applications of metagenomic profiling. Reads simulated using taxa derived from the Liu et al. RMS stool sample dataset were queried against the Kraken2 ‘PlusPFP’ database. We wanted to see if mapping and recreating fragments using an inclusive database would produce taxa count and abundance comparable with the input 691 taxa simulated at even relative abundance.
Fixed length taxonomic ratios
The frequency of fragments was analyzed individually across the range of observed fragment lengths. Within each discrete fragment length, the ratios of the observed counts between all taxa present were calculated. The ratio was calculated iteratively by dividing the individual fragment count by the average fragment count for each of the other taxa present at that length (Fig. 3 ). These taxonomic ratios were then averaged over all fragment lengths to produce an n x n matrix of all n taxa. Because many taxonomic ratios were missing, all columns in the taxonomic ratio matrix were scaled to the taxon with the greatest number of relationships to all other taxa. The row average of this scaled matrix was used to predict relative taxonomic abundance for all taxa.
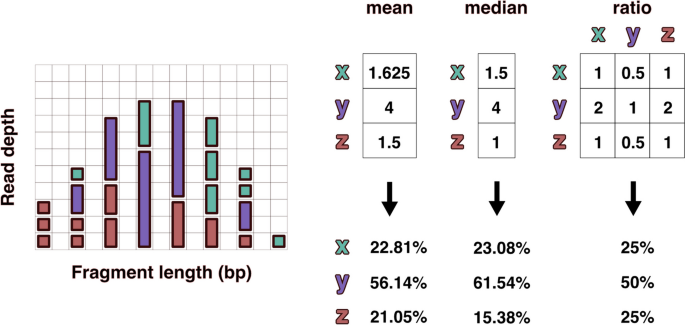
Visual representation of read depths originating from hypothetical taxa x, y, and z present in a 1:2:1 ratio. The left distribution shows fragments resulting from reduced sequencing under size selection or PCR fragment length biases. The differences in relative abundance estimates produced using mean and median read depths is compared with the taxonomic ratio approach (FLTR) introduced here. The ratio table on the right displays the averaged pairwise read depth ratios calculated individually within each fragment length. Even assuming no bias within each fragment length, the mean and median read depth estimates produce relative abundances that don’t account for variance in the size distribution, which is often not normal or uniform
Performance of software
A hypothetical gut microbiome was established to assess the computational performance of readsynth on larger metagenomic communities. Reference genomes derived from human gut samples (Liu et al. [ 2 ]) were downloaded and simulated under various input settings using a single core on a AMD EPYC 7F72 24-Core Processor (x86_64, 3028.149 MHz) (Supplementary Table 1 ). The cumulative size of the metagenome reference sequences influenced the time to simulate, but choice of enzymes had the largest impact on time and performance. Frequent, 4-base cutters produced many potential fragments to process, and use of less frequent motifs performed more efficiently. The most time consuming task tested, two 4-base cutters against 691 microbial genomes and the human genome, took 204 min; all other tasks took less than two hours.
Benchmarking of simulation accuracy
Spearman correlation captured a monotonic relationship between real and simulated read depths across the length of all 20 reference genomes in the mock community, and therefore was sensitive to differences in fragment presence or absence. The reads simulated using 100 percent enzyme cut efficiency corresponded closely with the real data (Fig. 4 group E; r = 0.92 to 0.94), comparable to the covariance measured between replicates of the real data. Simulation with a lower enzyme cut efficiency of 80 percent produced lower correlation with the real data, possibly resulting from increased novel loci surviving the fragmentation process (Fig. 4 group D, r = 0.66 to 0.68).
Spearman correlation coefficient of metagenome-wide, position-specific read depth for each of three replicates of real and simulated mock community sequencing. Real1-3 are the depth correlations of the real sequencing data replicates. Mapped read depth correlations from readsynth simulations are shown in A – E : A custom (.json) dictionary of fragment lengths derived from real sequence data and cut efficiency derived from real sequence data; B normal distribution of fragment lengths and cut efficiency derived from real sequence data; C normal distribution of fragment lengths with fragment mean increased by 100 bp and cut efficiency derived from real sequence data; D normal distribution of fragment lengths and cut efficiency reduced to 0.8; E normal distribution of fragment lengths and cut efficiency increased to 1
Pearson correlation was sensitive for comparing fragment read depths and was affected by noise in the data, as can be seen by variation within the replicates of the real sequencing datasets. The simulation that produced the highest read depth correlation coefficients between 0.76 to 0.79 used exact fragment size distributions informed by the real dataset (Supplementary Fig. 4 , group A). Using a normal distribution produced coefficients between 0.71 and 0.76 (groups B, D, E), suggesting the real data was approximated by applying readsynth’s normal distribution approach. The distribution with mean fragment lengths shifted only 100 bp longer affected read depth considerably with coefficients of 0.67 to 0.7 across replications (Supplementary Fig. 4 , group C). Simulations of the isolength dataset produced lower Pearson correlation (r = 0.56) but higher Spearman correlation (r = 0.97). The use of type IIB enzymes avoids variability in fragment length and while simulations with readsynth aligned closely with sequence presence-absence, simulation did not capture variability in the per-fragment depth.
Comparing Kraken2/Bracken profiles of real and simulated mock communities
The adapter-trimmed reads from the previously described mock community sequencing efforts were profiled using custom databases with Kraken2 (2.1.2) and Bracken (2.7). Raw reads mapping to each species-level identification was used as a metric of performance between the real and simulated reads. Generally, simulations captured trends in the distribution of real sequence data accurately (Fig. 5 ). The simulations with increased fragment size selection and decreased cut efficiency (Fig. 5 , groups C and D) caused larger shifts in predicted distributions. These changes are largely due to taxa-specific patterns of fragmentation where some community members, such as Phocaeicola vulgatus and Staphylococcus epidermidis , contain disproportionate read counts originating from either long or short fragments (Supplementary Fig. 5 ).
Kraken2/Bracken percent abundance profiles based on read assignments of real and simulated mock community sequencing. Real1-3 are the relative abundances of mapped reads per species produced from the real sequencing data. Mapped read depth correlations from readsynth simulations are shown in A – E : A custom (.json) dictionary of fragment lengths; B normal distribution and cut rate; C normal distribution and cut rate with fragment mean increased by 100 bp; D normal distribution and cut rate with cut rate reduced to 0.8; E normal distribution and cut rate with cut rate increased to 1. All fragment distributions and cut rates estimated from real sequence data unless specified
Impact of simulation parameters and quantification method
The FLTR approach is an alternative to read median or mean depth for profiling that measures the average relationships in read depth ratios between the taxa present at a given length. To test the FLTR approach, we first compared it to mean depth and median depth using the Snipen et al. 2021 mock community samples. Across the three replicates, using ratios produced relative abundance estimates similar to those produced using either the median or mean read depths, with Pearson correlation to the ground truth relative abundances ranging between 0.809 and 0.852 (Supplementary Fig. 6 ). Estimates of relative abundance across the three replicates show consistent, taxa-specific patterns of over- and underabundance in several of the 20 mock community members (Supplementary Fig. 7 ). These differences may be the result of sequence-specific amplification biases or slight deviations from the reported proportions expected from the mock community standard.
Data simulated from the 610 HELIUS gut microbial taxa were used to assess differences in profiling performance for the new FLTR approach versus previously published methods under varying library preparation conditions. Across all simulated HELIUS datasets, FLTR estimates of relative abundance outperformed the mean and median read depth in every instance (Supplementary Table 2 ). In some extreme cases, relying on the mean and median depths fall far from the target relative abundances (Fig. 6 ). Reads simulated from the HELIUS data captured several general trends not captured with the mock community analyses. Most notably, we see a strong interaction between the characteristics of restriction enzymes selected and the signal of the taxa simulated. Use of two frequent restriction enzymes, HhaI and MseI, each with 4 bp recognition motifs, required 100 million reads in order to detect > 90% of the input taxa. Interestingly, the use of two infrequent restriction enzymes, EcoRI and AgeI, was able to identify a higher percentage of taxa at lower sequencing efforts relative to the other treatments considered (Fig. 7 ). At 10 million reads, the combination of EcoRI and MseI captured 96.1% of taxa. The majority of simulated datasets performed moderately well at 10 million reads, and this was used as the basis for additional simulations. Lowering the mean fragment size for the EcoRI/MseI simulations by 100 bp reduced the number of identifiable taxa to 89%, and an increase by 100 bp marginally improved detection to 96.7%. Simulating the isolength, BcgI fragmentation of the HELIUS data returned only 56.1% of the 610 taxa, compared with the 66.7% of taxa identified by HhaI/MseI at the same sequencing effort (Supplementary Fig. 8 ). Across all simulations at 10 million reads, between 16 and 20% of reads were discarded per simulation due to the multi-mapping criterion described in the methods.
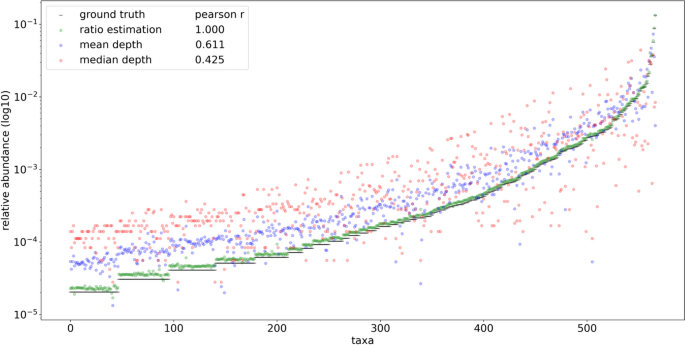
Comparisons of ground truth relative abundance (black) vs. results obtained using taxonomic ratio approach (green), mean depth (blue), and median depth (red) for 567 taxa returned using HELIUS simulated reads digested with HhaI and AgeI using 1 × 10 8 total reads
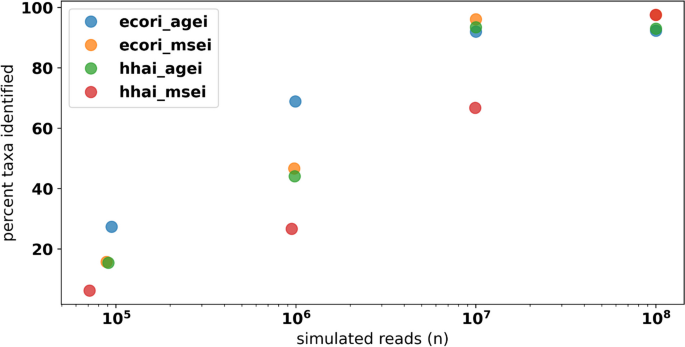
The relationship between the input simulated reads target and the percent of taxa identified across 4 combinations of restriction enzyme double digests. Of the expected 610 unique taxa, many are not captured at lower levels of coverage. Combination of rare cutters (EcoRI and AgeI) performed better at these lower levels of coverage
In order to compare the profiling performance of RMS reads when mapping to a known set of reference genomes and a fully inclusive database, 691 taxa based on the Liu et al. dataset were simulated using even abundance. Using even relative abundance allows qualitative assessment of profiling quantification efforts independent from taxonomic assignment, which expectedly contain naming and identification discrepancies between the input reference genome naming and the resulting assignments. Unlike the HELIUS results, aligning these reads to the known set of references was able to uniquely identify every input genome (Supplementary Fig. 9 ). The same simulated reads were then assigned to the species level of the ‘PlusPFP’ Kraken2 database using Bracken, which includes the target genomes and an additional 28,763 non-target genomes at the species level or lower. The resulting number of taxonomic hits was inflated from 691 to 1534 total taxa. To explore the basis of these false positive taxonomic assignments and potential methods of reduction, the reference genomes from these preliminary hits were then used for BWA MEM alignment and fragment recreation in place of the curated, known reference approach described above. Upon selecting reads whose source fragments aligned to the expected cut positions and removing multi mapped reads, 93.8% were able to map precisely to fragments expected in 749 of the 1,534 taxa from the Kraken2/Bracken profile, greatly reducing the false positives. Visual inspection of the recreated fragment length distribution captured the expected profile (Supplementary Fig. 10 ). Applying the FLTR approach to these reads yielded broadly even estimates of relative abundance matching the expected community composition. Some of the estimates have much lower than even representation, but comparatively these results further support the fallibility of using mean and median read depths to estimate abundances using RMS (Fig. 8 ).
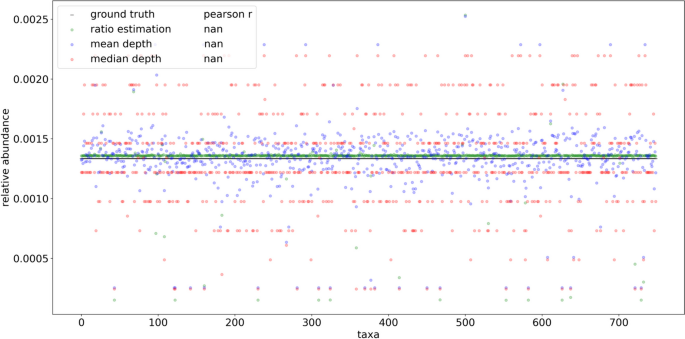
Comparisons of ground truth vs. results obtained using taxonomic ratio approach, mean depth, and median depths by mapping simulated reads naively to a fully inclusive, pre-made Kraken2 database (‘PlusPFP’) before fragment recreation. A total of 749 taxa were returned using this approach. Reads from 691 taxa were simulated based on the metagenomic taxa from Liu et al. [ 2 ] sequence SRR5298272 using even representation and 1/749 was used as ground truth to reflect the assumption of equal abundance. In silico reference genomes were digested with NlaIII and HpyCH4IV producing ~ 3.3 × 10 6 total reads
Simulation is a meaningful way to measure the behavior of bioinformatics approaches, but its utility hinges on its ability to faithfully capture features found in real sequencing data. The applications of short reads are highly variable. Often the impacts of library prep are overlooked in the production of bioinformatics tools, and more tools need to be considerate of these possible nuances. Readsynth simulations of RMS mock communities produced realistic taxonomic distributions of genomic fragments and the output sequence profiles corresponded closely to those found in the limited RMS mock community sequences that exist. Each taxon present in the simulated RMS mock community produces a unique digest profile at varying relative abundance, and readsynth simulations faithfully captured these influential patterns in read depth across all combined loci in the metagenome. These simulations also demonstrate the influence that library preparation can have on profiling efforts. The fragment origin and read depth were sensitive to even minor differences in simulated library parameters, particularly size selection and enzyme efficiency.
Restriction enzyme digests of metagenomic communities produce irregular fragment length distributions that cannot be readily modeled assuming features of an underlying distribution. Therefore, calculating relative proportions in the context of a mixture of individuals prepared this way is a challenge that has not been properly addressed to account for the influences of library preparation. The FLTR methodology proposed here may benefit efforts in metagenomic quantification when fragment or target length differs between organisms. It may also have useful applications outside of metagenomics where fragment size bias affects abundance estimates using read depth, such as genotyping approaches using reduced representation sequencing. We found that using the FLTR approach appears to be a more stable metric than either mean or median depth, often because RMS fragments are not unimodally distributed within an individual genome. Removal of multi-mapped reads may compound this effect by leaving only a small number of reads representing a genome, and these reads may exist in regions of the fragment length distribution that are influenced by size-selection and PCR length biases. For RMS approaches to overcome the confounding influence of variable fragment lengths, it is a necessary prerequisite to first recreate fragments in order to know fragment lengths. Fortunately, if a reference genome assembly is reliable enough to produce a strong hit to RMS reads, it should be able to provide a framework for simulating the expected fragments so long as the cut sites are preserved. Conversely, reads that remain unassigned or ambiguous cannot be interpreted as proportional due to the uneven contribution of community members, the removal of which may result in inflated estimates of relative abundance. It may be possible to retain multi-mapped reads using estimated abundances of uniquely aligned kmers, such as the KrakenUniq approach to assignment. Such approaches have not been tested in this study.
Simulated assessments of RMS’s capability to capture rare taxa suggest that it may be possible but is highly dependent on the library preparation methods. Given a finite set of sequence reads, the restriction enzymes selected and size sampling protocols will determine which taxa produce enough signal to be detected. Fragmenting a set of highly diverse genomes with a frequent cutting enzyme may produce hundreds of thousands or even millions of potential fragments, but when these reads are distributed between taxa whose relative abundances differ by orders of magnitude, the signal for rare taxa may be lost. It is likely that enzyme digests that produce shorter fragments will create more multi-mapping collisions, as the shorter read lengths reduce mapping confidence. Here we found that digests using HhaI/MseI or the BcgI isolength enzyme produced the greatest number of potential fragments, but based on the sensitivity of the profiling approach employed, few were actually informative because the short fragments they produce often lack resolution against a highly inclusive database. This mapping-based phenomenon may also explain the success of infrequent cutters, which captured a greater percent of the taxa present using a fraction of the overall fragments as the frequent cutters. Increasing sequencing effort may allow for greater resolution when many fragments are produced; however, using upwards of 100 million reads per sample to capture rare taxa may reduce the cost benefits of RRS. Therefore, it is recommended to consider each community’s complexity when considering the necessary read coverage, as has been similarly proposed for WGS profiling [ 23 ].
When two closely related taxa exist within a community, simulations indicate that it may become difficult to estimate the relative abundance of each using the RMS approach. This is because most fragments originating from both organisms will be identical and only a small fraction of loci will be uniquely informative to a taxon. In instances when only a small number of fragments may be used for quantification, abundance estimates using read depths may become highly sensitive to per-fragment biases originating from size-selection or PCR. The development of ddRADseq protocols for GBS were based around comparing conserved regions of the genome between closely related species. With RMS, conservation between these fragments may be counterproductive when trying to parse the individual members of a complex community. The taxonomic ratio methodology described here does not rely on normalization, but it is also not designed to handle redundancies in multi-mapping fragments, as evidenced by the set of unidentified taxa in the simulated HELIUS community. Eliminating fragments based on similarity may ultimately eliminate all useful, identifying markers if multiple, nearly identical strains are present. Retaining these fragments requires a means of identifying distinct taxa when aligning against a fully inclusive database of potential matches, and the results may be highly dependent on the database as well as the behavior of the aligning tool used [ 24 ]. We also recommend future investigations into profiling software choice and handling ambiguously assigned reads.
While simulation cannot be expected to capture all the nuances of real sequencing data, it can help find the edge cases where existing tools might fail to perform as intended. Using a set of largely pre-existing bioinformatics tools, our assessments here of simulated RMS data may be successful in some instances and very underpowered in others. While RMS offers promising applications, profiling benchmarks have not been widely tested on mixed samples including viral, protist, or non-fungal eukaryotic members and instead focus largely on prokaryotic and fungal taxa. Simulation could be a useful means in determining whether such mixed communities contain enzyme site biases preventing meaningful profiling accuracy. RMS may provide a fast and affordable profiling technique for communities that are relatively simple in structure. It may also be fast and economical in instances where detecting rare taxa is not critical. One of the largest obstacles preventing community use of RMS is the lack of bioinformatic tools developed to handle the data it produces, and applying existing profiling tools will not work out of the box. Both developers and users of new tools should be cognizant of the intersection between sample preparation and downstream analytical tools selected.
Availability and requirements
Project name: Readsynth
Project home page: github.com/ryandkuster/readsynth
Operating system(s): Linux/MacOS,
Programming language: Python3, C++
Other requirements: Python packages: numpy, pandas, and seaborn
License: Apache-2.0
Any restrictions to use by non-academics: none.
Availability of data and materials
The code and parameters used in all simulation and analytical steps used in this study are available on GitHub at https://github.com/ryandkuster/readsynth_analysis and raw data are stored at https://doi.org/ https://doi.org/10.5061/dryad.nzs7h44zk . The freely available package readsynth can be downloaded at https://github.com/ryandkuster/readsynth . The mock RMS and isolength sequencing datasets analyzed for this study can be found in the NCBI BioProject PRJNA574678 https://www.ncbi.nlm.nih.gov/bioproject/PRJNA574678 and figshare https://doi.org/ https://doi.org/10.6084/m9.figshare.12272360.v8 . The RMS gut metagenome data is available in NCBI BioProject PRJNA377403 https://www.ncbi.nlm.nih.gov/bioproject/PRJNA377403 . The Kraken2/Bracken PlusPFP database (6/7/2022) and indices used in this study were downloaded from https://benlangmead.github.io/aws-indexes/k2 .
Abbreviations
Reduced metagenomic sequencing
Reduced representation sequencing
Double-digest restriction-site associated DNA sequencing
Genotyping by sequencing
Polymerase chain reaction
Operational taxonomic unit
Peterson BK, Weber JN, Kay EH, Fisher HS, Hoekstra HE. Double digest RADseq: an inexpensive method for De novo SNP discovery and genotyping in model and non-model species. PLoS ONE. 2012;7(5):11.
Article Google Scholar
Liu M, Worden P, Monahan LG, DeMaere MZ, Burke CM, Djordjevic SP, et al. Evaluation of ddRAD seq for reduced representation metagenome sequencing. PeerJ. 2017;5:9.
Ravi A, Avershina E, Angell IL, Ludvigsen J, Manohar P, Padmanaban S, et al. Comparison of reduced metagenome and 16S rRNA gene sequencing for determination of genetic diversity and mother-child overlap of the gut associated microbiota. J Microbiol Methods. 2018;149:44–52.
Article CAS PubMed Google Scholar
Hess MK, Rowe SJ, Van Stijn TC, Henry HM, Hickey SM, Brauning R, et al. A restriction enzyme reduced representation sequencing approach for low-cost, high-throughput metagenome profiling. PLoS ONE. 2020;15(4):18.
Snipen L, Angell IL, Rognes T, Rudi K. Reduced metagenome sequencing for strain-resolution taxonomic profiles. Microbiome. 2021;9(1):19.
Goodrich JK, Di Rienzi SC, Poole AC, Koren O, Walters WA, Caporaso JG, et al. Conducting a microbiome study. Cell. 2014;158(2):250–62.
Article CAS PubMed PubMed Central Google Scholar
Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, et al. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE. 2011;6(5):10.
Miller MR, Dunham JP, Amores A, Cresko WA, Johnson EA. Rapid and cost-effective polymorphism identification and genotyping using restriction site associated DNA (RAD) markers. Genome Res. 2007;17(2):240–8.
Baird NA, Etter PD, Atwood TS, Currey MC, Shiver AL, Lewis ZA, et al. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS ONE. 2008;3(10):7.
Ochman H, Caro-Quintero A. Genome size and structure, bacterial. In: Kliman RM, editor. Encyclopedia of evolutionary biology [Internet]. Oxford: Academic Press; 2016. p. 179–85. Available from: https://www.sciencedirect.com/science/article/pii/B9780128000496002353
Andrews KR, Good JM, Miller MR, Luikart G, Hohenlohe PA. Harnessing the power of RADseq for ecological and evolutionary genomics. Nat Rev Genet. 2016;17(2):81–92.
Davey JW, Cezard T, Fuentes-Utrilla P, Eland C, Gharbi K, Blaxter ML. Special features of RAD sequencing data: implications for genotyping. Mol Ecol. 2013;22(11):3151–64.
DaCosta JM, Sorenson MD. Amplification biases and consistent recovery of loci in a double-digest RAD-seq protocol. PLoS ONE. 2014;9(9):14.
Sun Z, Huang S, Zhu P, Tzehau L, Zhao H, Lv J, et al. Species-resolved sequencing of low-biomass or degraded microbiomes using 2bRAD-M. Genome Biol. 2022;23(1):36.
Sun Z, Liu J, Zhang M, Wang T, Huang S, Weiss ST, et al. Removal of false positives in metagenomics-based taxonomy profiling via targeting type IIB restriction sites. Nat Commun. 2023;14(1):5321.
Stoddard SF, Smith BJ, Hein R, Roller BRK, Schmidt TM. rrnDB: improved tools for interpreting rRNA gene abundance in bacteria and archaea and a new foundation for future development. Nucleic Acids Res. 2015;43(D1):D593–8.
Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal Vol 17 No 1 Gener Seq Data Anal [Internet]. 2011; Available from: https://journal.embnet.org/index.php/embnetjournal/article/view/200
Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinforma Oxf Engl. 2010;26(5):589–95.
Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, et al. Twelve years of SAMtools and BCFtools. GigaScience. 2021;10(2):giab008.
Article PubMed PubMed Central Google Scholar
Lu J, Breitwieser FP, Thielen P, Salzberg SL. Bracken: estimating species abundance in metagenomics data. Peerj Comput Sci. 2017;5:14082.
Google Scholar
Wood DE, Lu J, Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019;20(1):13.
Deschasaux M, Bouter KE, Prodan A, Levin E, Groen AK, Herrema H, et al. Depicting the composition of gut microbiota in a population with varied ethnic origins but shared geography. Nat Med. 2018;24(10):1526–31.
Rodriguez-R LM, Konstantinidis KT. Estimating coverage in metagenomic data sets and why it matters. ISME J. 2014;8(11):2349–51.
Anyansi C, Straub TJ, Manson AL, Earl AM, Abeel T. Computational methods for strain-level microbial detection in colony and metagenome sequencing data. Front Microbiol. 2020;11:17.
Download references
Acknowledgements
We acknowledge Bode Olukolu for the initiation of research into genotyping by sequencing for microbiome exploration. We would like to thank Aaron Onufrak for software testing and the Hadziabdic lab for guidance on amplicon sequencing methodology.
Financial support was provided by the Department of Entomology and Plant Pathology, University of Tennessee Institute of Agriculture and the Scholarly and Research Incentive Funds from the Graduate School, University of Tennessee, Knoxville.
Author information
Authors and affiliations.
Department of Entomology and Plant Pathology, University of Tennessee, Knoxville, TN, USA
Ryan Kuster & Margaret Staton
You can also search for this author in PubMed Google Scholar
Contributions
RK designed the research and carried out the experiments. MS supervised the project, provided software guidance, and contributed to the manuscript. All authors contributed to the article and approved the submitted version.
Corresponding author
Correspondence to Ryan Kuster .
Ethics declarations
Ethics approval.
Not applicable.
Consent for publication
Competing interests.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Additional information
Publisher's note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file1 (docx 21675 kb), rights and permissions.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ . The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
Reprints and permissions
About this article
Cite this article.
Kuster, R., Staton, M. Readsynth: short-read simulation for consideration of composition-biases in reduced metagenome sequencing approaches. BMC Bioinformatics 25 , 191 (2024). https://doi.org/10.1186/s12859-024-05809-3
Download citation
Received : 09 January 2024
Accepted : 10 May 2024
Published : 15 May 2024
DOI : https://doi.org/10.1186/s12859-024-05809-3
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Reduced-representation
BMC Bioinformatics
ISSN: 1471-2105
- General enquiries: [email protected]

IMAGES
VIDEO
COMMENTS
assamesedictionary.in is an Assamese English Online Dictionary developed by Departmrnt of Computer Application Jorhat Engineering College (JEC), Assam. It has a library of more than 32,000 assamese words (oxomia xobdo). It also provides meaning of the assamese word with an english description and pronounciation of the word with sound. The words of 'Assamese Dictionary' have been collected ...
Assamese অসমীয়া. Assamese. অসমীয়া. • Candrakanta abhidhana: Assamese-English dictionary, University of Gauhati (1962) • Dictionary Assamese and English by Miles Bronson (1867) • Phrases in English and Assamese by Harriet Cutter, revised by Edward Clark (1877) + 1840 edition. • Brief vocabulary in English ...
Translations from dictionary Assamese - English, definitions, grammar. In Glosbe you will find translations from Assamese into English coming from various sources. The translations are sorted from the most common to the less popular. We make every effort to ensure that each expression has definitions or information about the inflection.
Check 'representation' translations into Assamese. Look through examples of representation translation in sentences, listen to pronunciation and learn grammar.
representation meaning in Assamese অসমীয়া is a translation of representation in Assamese অসমীয়া dictionary. Click for meanings of representation, including synonyms, antonyms.
Google's service, offered free of charge, instantly translates words, phrases, and web pages between English and over 100 other languages.
Assamese to English Translation tool includes online translation service, English text-to-speech service, English spell checking tool, on-screen keyboard for major languages, back translation, email client and much more. The most convenient translation environment ever created. Featured Language Translations.
And yet, the definition of "Assamese" cannot be so narrow as to mean only those who speak Assamese as their first language. Assam has many indigenous tribal and ethnic communities with their own ancestral languages. For Clause 6, it was necessary to expand the definition of "Assamese" beyond the Assamese-speaking population.
After reading the narratives of which has been taken in this research paper, the issue of women in Assamese folktales can be seen a matter of study and research, taking into consideration the ...
Accordingly, based on this definition I am writing this paper as an attempt to analyse the Aspectual Markers in Assamese with the reference of Pragmatics. As per now we come to know Assamese has three basic aspectual markers - '-is', '-i- s' and '-ith k'. They covey different meaning in different context.
In My Opinion Misrepresentation as applies any action has equal and opposite to reaction. It doesn't only affect the misrepresented culture and it's people but also the factions of people who have misrepresented it and all the people related to them. So let's just not mis represent any culture be it Bodo, Assamese,Garo,Rabha any.
Hi guys, This video is help you to learn about assamese meaning of evidence, evident, evidential and evidently. all words are translated in English and assam...
The name "Assamese" is of British colonial coinage of the 19th and 20th century. Assamese is an English word meaning "of Assam" —though most Assamese people live in Assam, not all the people of Assam today are Assamese people. Definition Plain Assamese tribal communities (depicting from top to down: Ahom, Chutia, Hojai Kachari)
nawot loi guchi gol." (ibid: 8) mean that the merchant took away the lonely beautiful girl whom he found crying on the river shore. This provides clear evidence of the merchant‟s intention as a man. "ghar pai mudoie tar dujoni ghoiniyekor aagote taiko ejoni xoru ghoini pati lole." (ibid: 8) mean that immediately upon reaching
It is an integral part of all socio-religious ceremonies in the state and is considered as an Assamese identity and pride. A 'gamocha' literally means a towel and is commonly used in Assamese ...
REPRESENTATION definition: 1. a person or organization that speaks, acts, or is present officially for someone else: 2. the…. Learn more.
The Assamese alphabet (Assamese: অসমীয়া বৰ্ণমালা, romanized: Ôxômiya Bôrnômala) is a writing system of the Assamese language and is a part of the Bengali-Assamese script.This script was also used in Assam and nearby regions for Sanskrit as well as other languages such as Bodo (now Devanagari), Khasi (now Roman), Mising (now Roman), Jaintia (now Roman) etc.
representation: [noun] one that represents: such as. an artistic likeness or image. a statement or account made to influence opinion or action. an incidental or collateral statement of fact on the faith of which a contract is entered into. a dramatic production or performance. a usually formal statement made against something or to effect a ...
Assamese regional films always ensured to pour cultural richness through their narratives, music and representation through visual narratives. The cinema industry of Assam was formed with the lineage starting from Joymoti directed by Jyoti Prasad Agarwala. In the '70s and '80s, this film also led to the growth of films with visually strong ...
When the census report comes, Assamese people will be only about 40 per cent of the population," he claimed. Sarma said that the definition of Assamese has undergone change over the years, and it must include communities such as the tea tribes and Hindi speakers who have been living here for over a century.
Translations from dictionary English - Assamese, definitions, grammar. In Glosbe you will find translations from English into Assamese coming from various sources. The translations are sorted from the most common to the less popular. We make every effort to ensure that each expression has definitions or information about the inflection.
Sarma highlighted the need for a broader definition that encompasses communities like Hindi speakers and tea tribes who have called Assam home for generations. He attributed the potential decline in the percentage of Assamese speakers to "infiltration from Bangladesh" impacting the state's demographics.
Hold mock elections in your class to elect four class representatives. Hold the election in three different ways: ± Each student can give one vote. The four highest vote getters are elected. ± Each student has four votes and can give them all to one candidate or split the votes among different candidates.
A sparse representation-based local occlusion recognition method for athlete expressions is proposed to address the problems of large mean square error, low recall rate, and poor recognition performance. We calculate the gradient direction and size of ...
In order to win an election and become a county councillor, candidates are required to meet a quota of votes. This is calculated by dividing the number of votes casted on voting day in that ...
Applying the FLTR approach to these reads yielded broadly even estimates of relative abundance matching the expected community composition. Some of the estimates have much lower than even representation, but comparatively these results further support the fallibility of using mean and median read depths to estimate abundances using RMS (Fig. 8).