- Open access
- Published: 25 August 2015

Visualizing genome and systems biology: technologies, tools, implementation techniques and trends, past, present and future
- Georgios A. Pavlopoulos 1 ,
- Dimitris Malliarakis 2 ,
- Nikolas Papanikolaou 1 ,
- Theodosis Theodosiou 1 ,
- Anton J. Enright 3 &
- Ioannis Iliopoulos 1
GigaScience volume 4 , Article number: 38 ( 2015 ) Cite this article
19k Accesses
53 Citations
32 Altmetric
Metrics details
“Α picture is worth a thousand words.” This widely used adage sums up in a few words the notion that a successful visual representation of a concept should enable easy and rapid absorption of large amounts of information. Although, in general, the notion of capturing complex ideas using images is very appealing, would 1000 words be enough to describe the unknown in a research field such as the life sciences? Life sciences is one of the biggest generators of enormous datasets, mainly as a result of recent and rapid technological advances; their complexity can make these datasets incomprehensible without effective visualization methods. Here we discuss the past, present and future of genomic and systems biology visualization. We briefly comment on many visualization and analysis tools and the purposes that they serve. We focus on the latest libraries and programming languages that enable more effective, efficient and faster approaches for visualizing biological concepts, and also comment on the future human-computer interaction trends that would enable for enhancing visualization further.
Peer Review reports
Introduction
In the current ‘big data’ era [ 1 ], the magnitude of data explosion in life science research is undeniable. The biomedical literature currently includes about 27 million abstracts in PubMed and about 3.5 million full text articles in PubMed Central. Additionally, there are more than 300 established biological databases that store information about various biological entities (bioentities) and their associations. Obvious examples include: diseases, proteins, genes, chemicals, pathways, small molecules, ontologies, sequences, structures and expression data. In the past 250 years, only 1.2 million eukaryotic species (out of the approximately 8.8 million that are estimated to be present on earth) [ 2 ] have been identified and taxonomically classified in the Catalog of Life and the World Register of Marine Species [ 3 ]. The sequencing of the first human genome (2002) took 13 years and cost over $3 million to complete. Although the cost for de novo assembly of a new genome to acceptable coverage is still high, probably at least $40,000, we can now resequence a human genome for $1000 and can generate more than 320 genomes per week [ 4 ]. Notably, few species have been fully sequenced, and a large fraction of their gene function is not fully understood or still remains completely unknown [ 5 ]. The human genome is 3.3 billion base pairs in length and consists of over 20,000 human coding genes organized into 23 pairs of chromosomes [ 6 , 7 ]. Today over 60,000 solved protein structures are hosted in the Protein Data Bank [ 8 ]. Nevertheless, many of the protein functions remain unknown or are partially understood.
Shifting away from basic research to applied sciences, personalized medicine is on the cusp of a revolution allowing the customization of healthcare by tailoring decisions, practices and/or products to the individual patient. To this end, such information should be accompanied by medical history and digital images and should guarantee a high level of privacy. The efficiency and security of distributed cloud computing systems for medical health record organization, storage and handling will be one of the big challenges during the coming years.
Information overload, data interconnectivity, high dimensionality of data and pattern extraction also pose major hurdles. Visualization is one way of coping with such data complexity. Implementation of efficient visualization technologies is necessary not only to present the known but to also reveal the unknown, allowing inference of conclusions, ideas and concepts [ 9 ]. Here we focus on visualization advances in the fields of network and systems biology, present the state-of-the-art tools and provide an overview of the technological advances over time, gaining insights into what to expect in the future of visualization in the life sciences.
In the section on network biology below, we discuss widely used tools related to graph visualization and analysis, we comment on the various network types that often appear in the field of biology and we summarize the strengths of the tools, along with their citation trends over time. In this section we also distinguish between tools for network analysis and tools designed for pathway analysis and visualization. In a section on genomic visualization, we follow the same approach by distinguishing between tools designed for genome browsing and visualization, genome assembly, genome alignments and genome comparisons. Finally, in a section on visualization and analysis of expression data, we distinguish between tree viewers and tools implemented for multivariate analysis.
Network biology visualization
In the field of systems biology, we often meet network representations in which bioentities are interconnected with each other. In such graphs, each node represents a bioentity and edges (connections) represent the associations between them [ 10 ]. These graphs can be weighted, unweighted, directed or undirected. Among the various networks types within the field, some of the most widely used are protein-protein interaction networks, literature-based co-occurrence networks, metabolic/biochemical, signal transduction, gene regulatory and gene co-expression networks [ 11 – 13 ]. As new technological advances and high-throughput techniques come to the forefront every few years, such networks can increase dramatically in size and complexity, and therefore more efficient algorithms for analysis and visualization are necessary. Notably, a network consisting of a hundred nodes and connections is incomprehensible and impossible for a human to visually analyze. For example, techniques such as tandem affinity purification (TAP) [ 14 ], yeast two hybrid (Y2H) [ 15 ] and mass spectrometry [ 16 ] can nowadays generate a significant fraction of the physical interactions of a proteome. As network biology evolves over time, we indicate standard procedures that were developed over the past 20 years and highlight key tools and methodologies that had a crucial role in this maturation process (Fig. 1 ).
Visualization for network biology. a Timeline of the emergence of relevant technologies and concepts. b A simple drawing of an undirected unweighted graph. c A 2D representation of a yeast protein-protein interaction network visualized in Cytoscape ( left ) and potential protein complexes identified by the MCL algorithm from that network ( right ). d A 3D view of a protein-protein interaction network visualized by BiolayoutExpress 3D . e A multilayered network integrating different types of data visualized by Arena3D. f A hive plot view of a network in which nodes are mapped to and positioned on radially distributed linear axes. g Visualization of network changes over time. h Part of lung cancer pathway visualized by iPath. i Remote navigation and control of networks by hand gestures. j Integration and control of 3D networks using VR devices
In the 1990s, two-dimensional (2D) static graph layouts were developed for visualizing networks. Topological analysis, layout and clustering were pre-calculated and results were captured in a single static image. Clustering analysis was performed to detect cliques or highly connected regions within a graph, layout techniques such as Fruchterman-Reingold [ 17 ] were implemented to place nodes in positions where the crossovers between the edges are minimized and topological analysis was used for detecting important nodes of the network such as hubs or nodes with high betweenness centrality. The typical visual encoding consisted of using arrows for directed graphs, adjusting the thickness of an edge to show the importance of a connection, using the same color for nodes that belong to the same cluster or modifying the node’s size to show its topological features, such as its neighbor connectivity. As integrative biology and high-throughput techniques advanced over the years, the necessity to move away from static images and add interactivity and navigation for easier data exploration became clearer.
Bridging between analysis and visualization became necessary, and tools that incorporated both increased the standards in the field. In clustering analysis, for example, new computational methods such as MCL [ 18 ] and variations [ 19 ], Cfinder [ 20 ], MCODE [ 21 ], Clique [ 22 ] and others were applied to biological networks to find highly connected regions of importance. DECAFF [ 23 ], SWEMODE [ 24 ] or STM [ 25 ], for example, were developed to predict protein complexes [ 26 ] incorporating graph annotations, whereas others such as DMSP [ 27 ], GFA [ 28 ] and MATISSE [ 29 ] were focused on gene-expression data. Most of these algorithms were command-line-based and only few tools such as jClust [ 30 ], GIBA [ 31 ], ClusterMaker [ 32 ] or NeAT [ 33 ] have been developed to integrate data in visual environments. These aforementioned techniques along with others are thoroughly discussed elsewhere [ 26 , 34 – 36 ].
Although most network visualization tools are standalone applications, they guarantee efficient data exploration and the manipulation of visualization with mouse-hovering supporting actions. Such tools are for example the Pajek [ 37 ], Osprey [ 38 ], VisANT [ 39 ] and others. Next-generation visualization tools took advantage of standard file formats such as BioPAX [ 40 , 41 ], SBML [ 42 ], PSI-MI [ 43 ] and CellML [ 44 ]; modern, more sophisticated layouts such as Hive-Plots [ 45 ]; and the available web services and data integration techniques to directly retrieve and handle information from public repositories on the fly. Functional enrichment of genes using the Gene Ontology (GO) repository [ 46 ] is a typical example. Among others, current state-of-the-art tools are the Ondex [ 47 ], Cytoscape [ 48 ] or Gephi [ 49 ], while tools such as iPath [ 50 ], PATIKA [ 51 ], PathVisio [ 52 ] and others [ 53 ] are pathway specific.
As biological networks became larger over time, consisting of thousands of nodes and connections, the so-called ‘hairball’ effect, where many nodes are densely connected with each other became very difficult to cope with. A partial solution to this was to shift from 2D representations to three-dimensional (3D) representations. Tools such as Arena3D [ 54 , 55 ] or BioLayout Express 3D [ 56 ] take advantage of 3D space to show data in a virtual 3D universe. BioLayout Express uses whole 3D space to visualize networks, whereas Arena3D implements a multilayered concept to present 2D networks in a stack. Although a 2D network allows immediate visual feedback, a 3D rendering usually requires the user to interact more with the data in a more explorative mode, but can help reveal interesting features potentially hidden in a 2D representation. Although it is debatable whether 3D rendering is better than 2D visualization, hardware acceleration and performance still need to be taken into account when planning 3D visualizations (Fig. 1 ).
Tables 1 and 2 present currently freely available network and pathway visualization tools and their main characteristics. However, it is not the purpose of this review to perform a deeper comparative analysis of all available 2D and 3D visualization tools, as this is available elsewhere [ 53 , 57 – 59 ]. Nevertheless, as network biology is gaining ground over the years, we sought to investigate the impact of the current tools in the field. To accomplish this, we tracked the tools that appeared after year 2000 and whose respective articles are indexed by Scopus (Fig. 2 ). We chose to keep track of the citations of only the first original publication for each tool. Although the number of citations is a reasonable indicator of popularity, it can sometimes be misleading as several tool versions appear in different articles that we have not yet tracked. Nevertheless, some immediate conclusions can be reached, such as that Cytoscape seems to be by far the biggest player for network visualization, as it comes with more than 200 plugins [ 60 ] implemented by an active module community (Fig. 1b ). Similarly, MapMan [ 61 ] and Reactome SkyPainter [ 62 ] are the most used tools for pathway visualization (Fig. 2b ).
Citation trends and key player tools in network biology. a Citations of network visualization tools based on Scopus. b Citations of pathway visualization tools based on Scopus. The numbers of citations of each tool in 2015 are shown after its name
Over the past 5 years, the data visualization field has become more and more competitive. There is a trend away from standalone applications towards the integration of visualization implementations within web browsers. Therefore, libraries and new programming languages have been dedicated to this task (see the final section below). The greater visibility provided by web implementation means that advanced visualization can more easily become available to non-experts and to the broader community. Finally, one of the biggest visualization challenges today is to capture the dynamics of networks and the way in which topological properties change over time [ 63 ]. For this, motion or other sophisticated ideas, along with new human-computer interaction (HCI) techniques, should be taken into consideration. Although serious efforts on this are on the way [ 54 , 64 , 65 ], there are still much to expect in the future as HCI techniques and virtual reality (VR) devices (such as Oculus Rift) become cheaper and more advanced over time (Fig. 1 ).
Visualization in genomics
There remain many open challenges for advanced visualization for genome assemblies, alignments, polymorphisms, variations, synteny, single nucleotide polymorphisms (SNPs), rearrangements and annotations [ 66 , 67 ]. To better follow progress in the visualization field, we first need to follow the way in which new technologies, questions and trends have been shaped over the years (Fig. 3 ).
Visualization for genome biology. a Timeline of the emergence of relevant technologies and concepts. b A typical normal human karyotype. c Visualization of BLAST hits and alignment of orthologous genes for the human TP53 gene. d The human TP53 gene and its annotations visualized by the UCSC genome browser. e Visualization of a de novo genome assembly from its DNA fragments. f Examples of balanced and unbalanced genomic rearrangements. g Hypothetical visualization of genomic structural variations across time
Up to the 1990s, local and global pairwise and multiple sequence alignment algorithms such as Smith-Waterman [ 68 ], Needleman-Wunsch [ 69 ], FASTA [ 70 ] and BLAST [ 71 ] were the focus of bioinformatics methods development. Multiple sequence alignment tools such as the ClustalW/Clustal X [ 72 ], MUSCLE [ 73 ], T-Coffee [ 74 ] and others [ 75 ] used basic visualization schemes, in which sequences were represented as strings placed vertically in stacks. Colors were used to visually encode base conservation and to indicate matching, non-matching and similar nucleotides [ 76 , 77 ].
Although these tools were successful for small numbers of nucleotide or protein sequences, a question was raised regarding their applicability to whole-genome sequencing and comparison. A few years later (2002), the Sanger (dideoxy) first generation sequencing, particularly capillary approaches, allowed the sequencing of the first whole human genome, consisting of about 3 billion base pairs and over 20,000 human genes [ 78 , 79 ]. Shortly after that, second-generation (Illumina [ 80 ], Roche/454 [ 81 ], Biosystems/SOLiD [ 82 ]) and third-generation techniques (Helicos BioSciences [ 83 ], Pacific Biosciences [ 84 ], Oxford Nanopore [ 85 ] and Complete Genomics [ 86 ]) high-throughput sequencing techniques [ 87 – 91 ] allowed the sequencing of a transcriptome, an exome or a whole genome at a much lower cost and within reasonable timeframes.
Projects such as the 1000 Genomes Project, for comprehensive human genetic variation analysis [ 92 – 94 ], and the International HapMap Project [ 95 – 99 ], for the identification of common genetic variations among people from different countries, are just a few examples of the data explosion that has taken place in the era of comparative genomics, after 2005. Such large-scale genomic datasets necessitate powerful tools to link genomic data to its source genome and across genomes. Therefore, among others [ 66 ], widely used standalone and web-based genome browsers were dedicated to information handling, genome visualization, navigation, exploration and integration with annotations from various repositories. At present, many specialized tools for comparative genomic visualization are available and are widely used.
To follow trends in the field, we summarize the tools into four categories: genome alignment visualization tools (Table 3 ); genome assembly visualization tools (Table 4 ); genome browsers (Table 5 ); and tools to directly compare different genomes with each other for efficient detection of SNPs and genomic variations (Table 6 ). Following the same approach used for network biology (above), we examine the citation progress of the first article that was published for each tool using the Scopus repository (Fig. 4 ). Consed [ 76 ] and Gap [ 100 , 101 ] seem to be the most widely used assembly viewers, while SAMtools tview [ 102 ] is the favorite tool for genomic assembly visualization. In addition, the University of California, Santa Cruz (UCSC) Genome Browser [ 103 ], Artemis [ 104 ] and Ensembl [ 105 , 106 ] seem to be the go-to genome browsers, while Circos [ 107 ], VISTA [ 108 ] and cBio [ 109 ] are the most widely used tools for comparative genomics.
Citation trends and key players in genome biology. a Citations of genome alignment visualization tools based on Scopus. b Citations of genome assembly visualization tools based on Scopus. c Citations of genome browsers based on Scopus. d Citations of comparative genomics visualization tools based on Scopus. The numbers of citations of each tool in 2015 are shown after its name
Although tremendous progress has been made in genomic visualization and very large amounts of money have been invested in such projects, genome browsers [ 110 ] still need to address major problems. One of the biggest challenges is the integration of data in different formats (such as genomic and clinical data) as society enters the personalized medicine era. Furthermore, navigation at different resolution or granularity levels and smooth scaling are necessary as long as simultaneous comparisons across millions of elements [ 111 ] remains a bottleneck. Newer infrastructure and software that allow on-the-fly calculations both in the front end and the back end would definitely be a step forward. Finally, similarly to network biology, time-series data visualization is one of the great challenges. For example, in a hypothetical scenario in which it is required to follow genomic rearrangements over time during tumor development, time-series data visualization would be an invaluable tool. Motion integration and visualization using additional dimensions could be possible solutions. Overall, it would be unrealistic to expect an ideal universal genome browser that serves all the possible purposes in the field.
Visualization and analysis of expression data
Microarrays [ 112 ] and RNA sequencing [ 87 ] are the two main high-throughput techniques for measuring expression levels of large numbers of genes simultaneously. Both methods are revolutionary as one can simultaneously monitor the effects of certain treatments, diseases and developmental stages on gene expression across time (Fig. 5a ) and for multiple transcript isoforms. Although microarrays and RNAseq technologies are comparable to each other [ 113 ], the latter tends to dominate, especially as sequencing technologies have improved, and there now are robust statistics to model the particular noise characteristics of RNAseq, particularly for low expression [ 114 ]. Microarrays are still cheaper and in some contexts may be more convenient as their analysis is still simpler and requires less computing infrastructure.
Multivariate analyses and visualization. a Timeline of the emergence of relevant technologies and concepts. b Visualization of k-means partitional clustering algorithm. c 3D visualization of a principal component analysis. d Visualization of gene-expression measures across time using parallel coordinates. e Visualization of gene-expression clustering across time. f 2D hierarchical clustering to visualize gene expressions against several time points or conditions. g Hypothetical integration of analyses and expression heatmaps and the control of objects by VR devices
In both cases, a typical analysis procedure is first to normalize experimental and batch differences between samples and then to identify up- and downregulated genes based on a fold-change level when comparing across samples, such as between a healthy and a non-healthy tissue. Statistical approaches are used to assess how reliable fold-change measurements are for each transcript of interest by modeling variation across transcripts and experiments. Subsequently, functional enrichment is performed to identify pathways and biological processes in which the up- and downregulated genes may be involved. Although there are numerous functional enrichment suites [ 115 ], David [ 116 ], Panther [ 117 ] and WebGestalt [ 118 ] are among the most widely used.
When gene expression is measured across many time points or conditions so as to observe, for example, the expression patterns following treatment, various analyses can be taken into consideration. Principal component analysis or partitional clustering algorithms such as k-means [ 119 ] can be used to group together genes with similar behavior patterns. Scatter-plotting is the typical visualization to represent such groupings. Thus, each point on a plane represents a gene and the closer two genes appear, the more similar they are (Fig. 5b, c ).
When one wants to categorize genes with similar behavior patterns across time (Fig. 5d ), hierarchical clustering based on expression correlation can be performed. Average linkage, complete linkage, single linkage, neighbor joining [ 120 ] and UPGMA [ 121 ] are the most widely used methods. In such approaches, an all-against-all distance or correlation matrix that shows the similarities between each pair of genes is required and genes are placed as leaves in a tree hierarchy. The two most widely used correlation metrics for expression data are the Spearman and Pearson correlation metrics. A list of tree viewers for hierarchical clustering visualization is presented in Table 7 . A more advanced visualization method is combining trees with heatmaps (Fig. 5e ): genes are grouped together according to their expression patterns in a tree hierarchy and the heat map is a graphical representation of individual gene-expression values represented as colors. Darker colors indicate a higher expression value and vice versa. An even more complex visualization of a 2D hierarchical clustering is shown in Fig. 5f , in which genes are clustered based on their expression patterns across several conditions (vertical tree on the left) and conditions are clustered across genes (horizontal tree). The heatmap shows the correlations between gene groups and conditions by allowing the researcher to come to conclusions about whether a group of genes is affected by a set of conditions or not. Heatmaps do, however, have significant drawbacks with regards to color perception. Perception of the color of a cell in a heatmap is shaped by the color of the surrounding cells, so two cells with identical color can look very different depending on their position in the heatmap.
Although RNAseq analysis is still an active field, microarray analysis has matured a lot over the past 15 years and many suites for analyzing such data are currently available (Table 8 ). To identify the key players in the field of microarray/RNAseq visualization we followed the citation patterns of the available tools from Scopus (Fig. 6 ). MEGA [ 122 ], ARB [ 123 ], NJplot [ 124 ], Dendroscope [ 125 ] and iTOL [ 126 ] are the most widely used tree viewers to visualize phylogenies and hierarchical clustering results. MultiExperiment Viewer [ 127 ], Genesis [ 128 ], GenePattern [ 129 ] and EXPANDER [ 130 ] are advanced suites that can perform various multivariate analyses such as the ones discussed in this section. Nevertheless, the commercial GeneSpring platform and the entire R/BioConductor framework [ 131 , 132 ] are mostly used in such analyses.
Citation trends and tools for gene-expression analysis. a Citations of microarray/RNAseq visualization tools based on Scopus. b Citations of tree viewers based on Scopus. The numbers of citations of each tool in 2015 are shown after its name
Concerning the future of multivariate data visualization, new HCI techniques and VR devices could allow parallel visualizations, analyses and data integration simultaneously (Fig. 5g ).
Programming languages and complementary libraries for building visual prototypes
Although the field of biological data visualization has been active for 25 years, it is still evolving rapidly today, as the complexity and the size of results produced by high-throughput approaches increase. Although most of the current software is offered in the form of standalone distributions, a shift towards web visualization is growing. Important features of modern visualization tools include: interactivity; interoperability; efficient data exploration; quick visual data querying; smart visual adjustment for different devices with different dimensions and resolutions; fast panning; fast zooming in or out; multilayered visualization; visual comparison of data; and smart visual data filtering. As functions and libraries implementing these features for standalone applications become available, similar libraries for web visualizations immediately follow. Therefore, in this section we discuss the latest programming languages, libraries and application program interfaces (APIs) that automate and simplify many of the aforementioned features, enabling higher-quality visualization implementations. It is not in the scope of this review to extensively describe all programming language possibilities for data visualization; therefore, we focus on the five languages that are mostly used for high-throughput biological data. Nevertheless, Table 9 summarizes other languages, along with generic and language-specific libraries (for R, Perl and Python), that target specific problems and make the implementation of biological data visualization more practical.
‘Processing’ is a programming language and a development platform for writing generative, interactive and animated standalone applications. Basic shapes such as lines, triangles, rectangles and ellipses, inner/outer coloring and basic operations such as transformations, translations, scaling and rotations can be implemented in a single line of code and each shape can be drawn within a canvas of a given dimension and a given refresh rate. It is designed for easier implementations of 2D dynamic visualizations but it also supports 3D rendering, although not optimized. Its core library is now extended by more than 100 other libraries and it is one of the best documented languages in the field. The integrated development environment allows exporting of executable files for all Windows, MacOS and Linux operating systems as well as Java applet .jar files. Finally, it can be used as an excellent educational tool for computer programming fundamentals in a visual context. It is free for download, can easily be plugged in a Java standalone application, and is fully cooperative with the NetBeans and Eclipse environments. Code examples and tutorials can be found at [ 133 ].
Processing.js
Java applets were an easy way to run standalone applications within web browsers. This technology has now mainly been abandoned because of security considerations. To avoid JavaScript’s complexity and compensate for applet limitations, Processing.js was implemented, as the sister project of the popular Processing programming language, to allow interactive web visualization. It is a mediator between HTML5 and Processing and is designed to allow visual prototypes, digital arts, interactive animations, educational graphs and so on to run immediately within any HTML5-compatible browser, such as Firefox, Safari, Chrome, Opera or Internet Explorer. No plugins are required and one can code any visualization directly in the Processing language, include it in a web page, and let Processing.js bridge the two technologies. Processing.js brings the best of visual programming to the web, both for Processing and web developers. Code examples and tutorials can be found at [ 134 ].
D3 is the main competitor of Processing/Processing.js and has gained ground over recent years. It was initially used to generate scalable vector graphics (SVG). Like Processing.js, it is designed for powerful interactive web visualizations and it comes with its own JavaScript-like syntax. It is a JavaScript library for manipulating document object model objects and a programming interface for HTML, XML and SVG. The idea behind this approach is to load data into a browser and then generate document object model elements based on that data. Subsequently, one can apply data-driven transformations on the document. This avoids proprietary representation and affords extraordinary flexibility. With minimal overhead, D3 is extremely fast and supports large datasets and dynamic behaviors for interaction and animation. D3’s functional style allows code reuse through a diverse collection of components and plugins. It is extensively documented and code examples can be found at [ 135 ].
Adobe Flash was once the industry standard for authoring innovative, interactive content. In conjunction with the platform’s programming language, ActionScript, Flash allows designers to implement dynamic visualization, opening up many possibilities for creativity. Some of the most pioneering, best practice visualizations built in Flash can be found with online news and media sites, introducing interactivity to supplement and enhance the presentation of information. Because of the lack of support for Flash across Apple’s suite of devices and the emergence of competing developments, demanding less computational power, including D3 and HTML5, this technology is now fading.
Java 3D is an API, acting as a mediator between OpenGL and Java and enables the creation of standalone 3D graphics applications and internet-based 3D applets. It is easy to use and provides high-level functions for creating and manipulating 3D objects in space and their geometry. Programmers initially create a virtual world and then place any 3D object anywhere in this world. Rotation in three axes, zooming in or out and translation of the whole canvas are functions are offered by default, and the hierarchy of the transformation groups define the 3D transformations that can be applied individually to an object or a set of objects. Java3D code can be compiled under any of the Windows, MacOS and Unix Systems.
The future of biological data visualization
Biological data visualization is a rapidly evolving field. Nevertheless, it is still in its infancy. Hardware acceleration, standardized exchangeable file formats, dimensionality reduction, visual feature selection, multivariate data analyses, interoperability, 3D rendering and visualization of complex data at different resolutions are areas in which great progress has been achieved. Additionally, image processing combined with artificial-intelligence-based pattern recognition, new libraries and programming languages for web visualization, interactivity, visual analytics and visual data retrieval, storing and filtering are still ongoing efforts with remarkable advances over the past years [ 58 , 136 , 137 ]. Today, many of the current visualization tools serve as front ends for very advanced infrastructures dedicated to data manipulation and have driven significant advances in user interfaces. Although the implementation of sophisticated graphical user interfaces is necessary, the effort to minimize back-end calculations is of great importance. Unfortunately, only a limited number of visualization tools today take advantage of libraries designed for parallelization. Multi-threading, for example, allows the distribution of computational tasks in terminals over the network, and CUDA (available on Nvidia graphic cards) allows parallel calculations at multiple graphical processing units.
Despite the fact that multiple screens, light and laser projectors and other technologies partially solve the space limitation problem, HCI techniques are changing the rules of the game and biological data visualization is expected to adjust to these trends in the longer term. 3D control can be achieved without intermediate devices such as mouse, keyboards or touch screens [ 138 ] in modern perceptual input systems. Sony’s EyeToy, Playstation Eye and Artag, for example, use non-spatial computer vision to determine hand gestures. Similarly, the Nintendo Wii and Sony Move devices support object manipulation in 3D space. These actions are mediated through the detection of the position in space of physical devices held by the user or, even more impressively, through immediate tracking of the human body or parts of the human body. Equally impressive is the prospect of ocular tracking, one implementation of which has recently been introduced by the VR startup Fove. The Fove headset tracks eye movement and translates into spatial movement or even other types of action within the simulated 3D space. The recently implemented Molecular Control Toolkit [ 139 ] is a characteristic example of a new API based on the Kinect and Leap Motion devices (which track the human body and human fingers, respectively) to control molecular graphics such as 3D protein structures. Moreover, large screens, tiled arrays or VR environments should be taken into consideration by programmers and designers as they become more and more affordable over time. A great benefit of such technologies is that they allow the representation of complete datasets without the need for algorithms dedicated to dimensionality reduction, which might lead to information loss.
VR environments are expected to bring a revolution in biological data visualization as one could integrate metabolomics networks and gene expression in virtual worlds, as in MetNet3D [ 140 ], or create virtual universes of living systems such as a whole cell [ 59 , 141 – 144 ]. A visual representation of the whole cell with its components in an immense environment in which users can visually explore the location of molecules and their interaction in space and time could lead to a better understanding of the biological systems. Oculus Rift (which promoted the reemergence of VR devices), Project Morpheus, Google Cardboard, Sony Smart Eyeglass, HTC Vive, Samsung Gear VR, Avegant Glyph, Razer OSVR, Archos VR Headset and Carl Zeiss VR One are state-of-the-art commercial devices that offer VR experiences. All of them overlay the user’s eyesight with some kind of screen and aim to replace the field of view with a digital 3D alternative. Between them, those devices use many technologies and new ideas such as the monitoring of the position of the head (allowing for more axes of movement), the substitution of the VR screen with smartphones (thus harnessing efficient modern smartphone processors), eye tracking and projection of images straight onto the retina.
Approaching the problem from a different angle, Google Glass, HoloLens and Magic Leap offer an augmented reality experience (the latter is rumored to achieve that by projecting a digital light field into the user’s eye). Augmented reality can facilitate the learning process of the biological systems because it builds on exploratory learning. This allows scientists to visualize existing knowledge, whereas the unstructured nature of augmented reality could allow them to construct knowledge themselves by making connections between information and their own experiences or intuition and thus offer novel insights to the studied biological system [ 145 ]. Efforts such as the Visible Cell [ 141 ] and CELLmicrocosmos have already begun. The Visible Cell project aims to inform advanced in silico studies of cell and molecular organization in 3D using the mammalian cell as a unitary example of an ordered complex system; the CELLmicrocosmos integrative cell modeling and stereoscopic 3D visualization project is a typical example of the use of 3D vision.
Finally, starting from a living entity, the process of digitizing it, visualizing it, placing it in virtual worlds or even recreating it as a physical object using 3D printing is no longer the realm of science fiction. Data visualization and biological data visualization are rapidly developing in parallel with advances in the gaming industry and HCI. These efforts are complementary and there are already strong interactions developing between these fields, something that is expected to become more obvious in the future.
Howe D, Costanzo M, Fey P, Gojobori T, Hannick L, Hide W, et al. Big data: the future of biocuration. Nature. 2008;455:47–50.
Article CAS PubMed PubMed Central Google Scholar
Liolios K, Mavromatis K, Tavernarakis N, Kyrpides NC. The Genomes On Line Database (GOLD) in 2007: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res. 2008;36:D475–9.
Article CAS PubMed Google Scholar
Census of Marine Life. How many species on Earth? About 8.7 million, new estimate says. ScienceDaily. 24 August 2011. http://www.sciencedaily.com/releases/2011/08/110823180459.htm . Accessed 27 July 2015.
May M. Life Science Technologies: Big biological impacts from big data. Science. 2014; doi: 10.1126/science.opms.p1400086 .
Reddy TB, Thomas AD, Stamatis D, Bertsch J, Isbandi M, Jansson J, et al. The Genomes OnLine Database (GOLD) v. 5: a metadata management system based on a four level (meta)genome project classification. Nucleic Acids Res. 2015;43:D1099–1106.
International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–45.
Article CAS Google Scholar
Ezkurdia I, Juan D, Rodriguez JM, Frankish A, Diekhans M, Harrow J, et al. Multiple evidence strands suggest that there may be as few as 19,000 human protein-coding genes. Hum Mol Genet. 2014;23:5866–78.
Berman H, Henrick K, Nakamura H. Announcing the worldwide Protein Data Bank. Nat Struct Biol. 2003;10:980.
Pavlopoulos GA, Iacucci E, Iliopoulos I, Bagos PG. Interpreting the Omics 'era' data. Multimedia Services in Intelligent Environments vol. 25. Heidelber: Springer; 2013. p. 79–100.
Book Google Scholar
Pavlopoulos GA, Secrier M, Moschopoulos CN, Soldatos TG, Kossida S, Aerts J, et al. Using graph theory to analyze biological networks. BioData Min. 2011;4:10.
Article PubMed PubMed Central Google Scholar
Moschopoulos CN, Pavlopoulos GA, Likothanassis SD, Kossida S. Analyzing protein-protein interaction networks with web tools. Curr Bioinform. 2011;6:389–97.
Papanikolaou N, Pavlopoulos GA, Theodosiou T, Iliopoulos I. Protein-protein interaction predictions using text mining methods. Methods. 2015;74:47–53.
Pavlopoulos GA, Promponas VJ, Ouzounis CA, Iliopoulos I. Biological information extraction and co-occurrence analysis. Methods Mol Biol. 2014;1159:77–92.
Article PubMed Google Scholar
Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, et al. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–29.
Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci USA. 2001;98:4569–74.
Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–7.
Fruchterman T, Reingold E. Graph drawing by force-directed placement. Softw Pract Exp. 1991;21:1129–64.
Article Google Scholar
Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–84.
Moschopoulos CN, Pavlopoulos GA, Likothanassis SD, Kossida S. An enhanced Markov clustering method for detecting protein complexes. 8th IEEE International Conference on Bioinformatics and Bioengineering. 2008. doi: 10.1109/BIBE.2008.4696656 .
Adamcsek B, Palla G, Farkas IJ, Derenyi I, Vicsek T. CFinder: locating cliques and overlapping modules in biological networks. Bioinformatics. 2006;22:1021–3.
Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4:2.
Spirin V, Mirny LA. Protein complexes and functional modules in molecular networks. Proc Natl Acad Sci USA. 2003;100:12123–8.
Li XL, Foo CS, Ng SK. Discovering protein complexes in dense reliable neighborhoods of protein interaction networks. Comput Syst Bioinformatics Conf. 2007;6:157–68.
Lubovac Z, Gamalielsson J, Olsson B. Combining functional and topological properties to identify core modules in protein interaction networks. Proteins. 2006;64:948–59.
Cho YR, Hwang W, Ramanathan M, Zhang A. Semantic integration to identify overlapping functional modules in protein interaction networks. BMC Bioinformatics. 2007;8:265.
Article PubMed PubMed Central CAS Google Scholar
Moschopoulos CN, Pavlopoulos GA, Iacucci E, Aerts J, Likothanassis S, Schneider R, et al. Which clustering algorithm is better for predicting protein complexes? BMC Res Notes. 2011;4:549.
Maraziotis IA, Dimitrakopoulou K, Bezerianos A. Growing functional modules from a seed protein via integration of protein interaction and gene expression data. BMC Bioinformatics. 2007;8:408.
Feng J, Jiang R, Jiang T. A max-flow-based approach to the identification of protein complexes using protein interaction and microarray data. IEEE/ACM Trans Comput Biol Bioinform. 2011;8:621–34.
Ulitsky I, Shamir R. Identification of functional modules using network topology and high-throughput data. BMC Syst Biol. 2007;1:8.
Pavlopoulos GA, Moschopoulos CN, Hooper SD, Schneider R, Kossida S. jClust: a clustering and visualization toolbox. Bioinformatics. 2009;25:1994–6.
Moschopoulos CN, Pavlopoulos GA, Schneider R, Likothanassis SD, Kossida S. GIBA: a clustering tool for detecting protein complexes. BMC Bioinformatics. 2009;10 Suppl 6:S11.
Morris JH, Apeltsin L, Newman AM, Baumbach J, Wittkop T, Su G, et al. clusterMaker: a multi-algorithm clustering plugin for Cytoscape. BMC Bioinformatics. 2011;12:436.
Brohee S, Faust K, Lima-Mendez G, Sand O, Janky R, Vanderstocken G, et al. NeAT: a toolbox for the analysis of biological networks, clusters, classes and pathways. Nucleic Acids Res. 2008;36:W444–51.
Li X, Wu M, Kwoh CK, Ng SK. Computational approaches for detecting protein complexes from protein interaction networks: a survey. BMC Genomics. 2010;11 Suppl 1:S3.
Brohee S, Faust K, Lima-Mendez G, Vanderstocken G, van Helden J. Network analysis tools: from biological networks to clusters and pathways. Nat Protoc. 2008;3:1616–29.
Brohee S, van Helden J. Evaluation of clustering algorithms for protein-protein interaction networks. BMC Bioinformatics. 2006;7:488.
Batagelj V, Mrvar A. Pajek - Program for large network analysis. Connections. 1998;21:47–57.
Google Scholar
Breitkreutz BJ, Stark C, Tyers M. Osprey: a network visualization system. Genome Biol. 2003;4:R22.
Hu Z, Hung JH, Wang Y, Chang YC, Huang CL, Huyck M, et al. VisANT 3.5: multi-scale network visualization, analysis and inference based on the gene ontology. Nucleic Acids Res. 2009;37:W115–21.
Luciano JS, Stevens RD. e-Science and biological pathway semantics. BMC Bioinformatics. 2007;8 Suppl 3:S3.
Luciano JS. PAX of mind for pathway researchers. Drug Discov Today. 2005;10:937–42.
Hucka M, Finney A, Sauro HM, Bolouri H, Doyle JC, Kitano H, et al. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19:524–31.
Hermjakob H, Montecchi-Palazzi L, Bader G, Wojcik J, Salwinski L, Ceol A, et al. The HUPO PSI's molecular interaction format--a community standard for the representation of protein interaction data. Nat Biotechnol. 2004;22:177–83.
Lloyd CM, Halstead MD, Nielsen PF. CellML: its future, present and past. Prog Biophys Mol Biol. 2004;85:433–50.
Krzywinski M, Birol I, Jones SJ, Marra MA. Hive plots--rational approach to visualizing networks. Brief Bioinform. 2012;13:627–44.
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat Genet. 2000;25:25–9.
Kohler J, Baumbach J, Taubert J, Specht M, Skusa A, Ruegg A, et al. Graph-based analysis and visualization of experimental results with ONDEX. Bioinformatics. 2006;22:1383–90.
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504.
Bastian M, Heymann S, Jacomy M. Gephi: an open source software for exploring and manipulating networks. In: International AAAI Conference on Weblogs and Social Media. 2009. https://www.aaai.org/ocs/index.php/ICWSM/09/paper/view/154 . Accessed 27 July 2015.
Letunic I, Yamada T, Kanehisa M, Bork P. iPath: interactive exploration of biochemical pathways and networks. Trends Biochem Sci. 2008;33:101–3.
Dogrusoz U, Erson EZ, Giral E, Demir E, Babur O, Cetintas A, et al. PATIKAweb: a web interface for analyzing biological pathways through advanced querying and visualization. Bioinformatics. 2006;22:374–5.
van Iersel MP, Kelder T, Pico AR, Hanspers K, Coort S, Conklin BR, et al. Presenting and exploring biological pathways with PathVisio. BMC Bioinformatics. 2008;9:399.
Bader GD, Cary MP, Sander C. Pathguide: a pathway resource list. Nucleic Acids Res. 2006;34:D504–6.
Secrier M, Pavlopoulos GA, Aerts J, Schneider R. Arena3D: visualizing time-driven phenotypic differences in biological systems. BMC Bioinformatics. 2012;13:45.
Pavlopoulos GA, O'Donoghue SI, Satagopam VP, Soldatos TG, Pafilis E, Schneider R. Arena3D: visualization of biological networks in 3D. BMC Syst Biol. 2008;2:104.
Freeman TC, Goldovsky L, Brosch M, van Dongen S, Maziere P, Grocock RJ, et al. Construction, visualisation, and clustering of transcription networks from microarray expression data. PLoS Comput Biol. 2007;3:2032–42.
Gehlenborg N, O'Donoghue SI, Baliga NS, Goesmann A, Hibbs MA, Kitano H, et al. Visualization of omics data for systems biology. Nat Methods. 2010;7 Suppl 3:S56–68.
Pavlopoulos GA, Wegener AL, Schneider R. A survey of visualization tools for biological network analysis. BioData Min. 2008;1:12.
Suderman M, Hallett M. Tools for visually exploring biological networks. Bioinformatics. 2007;23:2651–9.
Saito R, Smoot ME, Ono K, Ruscheinski J, Wang PL, Lotia S, et al. A travel guide to Cytoscape plugins. Nat Methods. 2012;9:1069–76.
Thimm O, Blasing O, Gibon Y, Nagel A, Meyer S, Kruger P, et al. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004;37:914–39.
Matthews L, Gopinath G, Gillespie M, Caudy M, Croft D, de Bono B, et al. Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res. 2009;37:D619–22.
Klein C, Marino A, Sagot MF, Vieira Milreu P, Brilli M. Structural and dynamical analysis of biological networks. Brief Funct Genomics. 2012;11:420–33.
Secrier M, Schneider R. PhenoTimer: software for the visual mapping of time-resolved phenotypic landscapes. PloS One. 2013;8:e72361.
Secrier M, Schneider R. Visualizing time-related data in biology, a review. Brief Bioinform. 2014;15:771–82.
Nielsen CB, Cantor M, Dubchak I, Gordon D, Wang T. Visualizing genomes: techniques and challenges. Nat Methods. 2010;7 Suppl 3:S5–15.
Procter JB, Thompson J, Letunic I, Creevey C, Jossinet F, Barton GJ. Visualization of multiple alignments, phylogenies and gene family evolution. Nat Methods. 2010;7 Suppl 3:S16–25.
Smith TF, Waterman MS. Identification of common molecular subsequences. J Mol Biol. 1981;147:195–7.
Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48:443–53.
Lipman DJ, Pearson WR. Rapid and sensitive protein similarity searches. Science. 1985;227:1435–41.
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10.
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8.
Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7.
Notredame C, Higgins DG, Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–17.
Daugelaite J, O' Driscoll A, Sleator R. An overview of multiple sequence alignments and cloud computing in bioinformatics. ISRN Biomathematics. 2013. doi: 10.1155/2013/615630 .
Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:195–202.
Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–85.
Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921.
Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, et al. The sequence of the human genome. Science. 2001;291:1304–51.
Bennett S. Solexa Ltd. Pharmacogenomics. 2004;5:433–8.
Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–80.
CAS PubMed PubMed Central Google Scholar
Barabási A-L, Gulbahce N, Loscalzo J. Network medicine: a network-based approach to human disease. Nat Rev Genet. 2011;12:56–68.
Xie C, Tammi MT. CNV-seq, a new method to detect copy number variation using high-throughput sequencing. BMC Bioinformatics. 2009;10:80.
Keravala A, Lee S, Thyagarajan B, Olivares EC, Gabrovsky VE, Woodard LE, et al. Mutational derivatives of PhiC31 integrase with increased efficiency and specificity. Mol Ther. 2009;17:112–20.
Chiang DY, Getz G, Jaffe DB, O'Kelly MJ, Zhao X, Carter SL, et al. High-resolution mapping of copy-number alterations with massively parallel sequencing. Nat Methods. 2009;6:99–103.
Kim TM, Luquette LJ, Xi R, Park PJ. rSW-seq: algorithm for detection of copy number alterations in deep sequencing data. BMC Bioinformatics. 2010;11:432.
Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63.
Morin R, Bainbridge M, Fejes A, Hirst M, Krzywinski M, Pugh T, et al. Profiling the HeLa S3 transcriptome using randomly primed cDNA and massively parallel short-read sequencing. Biotechniques. 2008;45:81–94.
Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet. 2010;11:31–46.
Hall N. Advanced sequencing technologies and their wider impact in microbiology. J Exp Biol. 2007;210:1518–25.
Li H, Homer N. A survey of sequence alignment algorithms for next-generation sequencing. Brief Bioinform. 2010;11:473–83.
Durbin RM, Abecasis GR, Altshuler DL, Auton A, Brooks LD, Gibbs RA, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73.
Karchin R. Next generation tools for the annotation of human SNPs. Brief Bioinform. 2009;10:35–52.
Medvedev P, Stanciu M, Brudno M. Computational methods for discovering structural variation with next-generation sequencing. Nat Methods. 2009;6 Suppl 11:S13–20.
Buchanan CC, Torstenson ES, Bush WS, Ritchie MD. A comparison of cataloged variation between International HapMap Consortium and 1000 Genomes Project data. J Am Med Inform Assoc. 2012;19:289–94.
Tanaka T. [International HapMap project]. Nihon Rinsho. 2005;63 Suppl 12:29–34.
PubMed Google Scholar
Thorisson GA, Smith AV, Krishnan L, Stein LD. The International HapMap Project web site. Genome Res. 2005;15:1592–3.
International HapMap Consortium. Integrating ethics and science in the International HapMap Project. Nat Rev Genet. 2004;5:467–75.
International HapMap Consortium. The International HapMap Project. Nature. 2003;426:789–96.
Bonfield JK, Smith K, Staden R. A new DNA sequence assembly program. Nucleic Acids Res. 1995;23:4992–9.
Dear S, Staden R. A sequence assembly and editing program for efficient management of large projects. Nucleic Acids Res. 1991;19:3907–11.
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9.
Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006.
Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream M-A, et al. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–5.
Flicek P, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, et al. Ensembl 2012. Nucleic Acids Res. 2012;40:D84–90.
Hubbard T, Barker D, Birney E, Cameron G, Chen Y, Clark L, et al. The Ensembl genome database project. Nucleic Acids Res. 2002;30:38–41.
Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–45.
Mayor C, Brudno M, Schwartz JR, Poliakov A, Rubin EM, Frazer KA, et al. VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics. 2000;16:1046–7.
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4.
Wang J, Kong L, Gao G, Luo J. A brief introduction to web-based genome browsers. Brief Bioinform. 2013;14:131–43.
Article PubMed CAS Google Scholar
Pavlopoulos GA, Oulas A, Iacucci E, Sifrim A, Moreau Y, Schneider R, et al. Unraveling genomic variation from next generation sequencing data. BioData Min. 2013;6:13.
Quackenbush J. Computational analysis of microarray data. Nat Rev Genet. 2001;2:418–27.
Mantione KJ, Kream RM, Kuzelova H, Ptacek R, Raboch J, Samuel JM, et al. Comparing bioinformatic gene expression profiling methods: microarray and RNA-Seq. Medical science monitor basic research. 2014;20:138–42.
MAQC Consortium, Shi L, Reid LH, Jones WD, Shippy R, Warrington JA, et al. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol. 2006;24:1151–61.
Article PubMed Central CAS Google Scholar
da Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13.
da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57.
Mi H, Muruganujan A, Thomas PD. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2013;41:D377–86.
Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33:W741–8.
MacQueen J. Some methods for classification and analysis of multivariate observations. In: Proceedings of the Fifth Berkeley Symposium on Mathematical Statistics and Probability, Volume 1. Berkeley: University of California Press; 1967. p. 281–97.
Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–25.
CAS PubMed Google Scholar
Li Y, Xu L. Unweighted multiple group method with arithmetic mean. 2010 IEEE Fifth International Conference on Bio-Inspired Computing: Theories and Applications (BIC-TA). 2010;830–4.
Kumar S, Tamura K, Jakobsen IB, Nei M. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics. 2001;17:1244–5.
Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–71.
Perriere G, Gouy M. WWW-query: an on-line retrieval system for biological sequence banks. Biochimie. 1996;78:364–9.
Huson DH, Richter DC, Rausch C, Dezulian T, Franz M, Rupp R. Dendroscope: an interactive viewer for large phylogenetic trees. BMC Bioinformatics. 2007;8:460.
Letunic I, Bork P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–8.
Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–8.
Sturn A, Quackenbush J, Trajanoski Z. Genesis: cluster analysis of microarray data. Bioinformatics. 2002;18:207–8.
Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat Genet. 2006;38:500–1.
Shamir R, Maron-Katz A, Tanay A, Linhart C, Steinfeld I, Sharan R, et al. EXPANDER--an integrative program suite for microarray data analysis. BMC Bioinformatics. 2005;6:232.
R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing; 2008.
Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods. 2015;12:115–21.
Fry B, Reas C. Processing. 2015. http://processing.org . Accessed 30 July 2015.
Fry B, Reas C, Resig, J. Processing.js. 2015. http://processingjs.org . Accessed 30 July 2015.
Bostock M. Data-Driven Documents. 2015. http://d3js.org/ . Accessed 30 July 2015.
O'Donoghue SI, Gavin AC, Gehlenborg N, Goodsell DS, Heriche JK, Nielsen CB, et al. Visualizing biological data-now and in the future. Nat Methods. 2010;7 Suppl 3:S2–4.
Thomas J, Cook KA. Illuminating the path: the research and development agenda for visual analytics. National Visualization and Analytics Center. 2005. http://vis.pnnl.gov/pdf/RD_Agenda_VisualAnalytics.pdf . Accessed 27 July 2015.
Karam M, Schraefel. A taxonomy of gestures in human computer interactions. In: Electronics and Computer Science. Southampton: University of Southampton; 2005. p. 1–45.
Sabir K, Stolte C, Tabor B, O'Donoghue SI. The molecular control toolkit: controlling 3D molecular graphics via gesture and voice. IEEE Symposium on Biological Data Visualization (BioVis). 2013;2013:49–56. doi: 10.1109/BioVis.2013.6664346 .
Yang Y, Engin L, Wurtele ES, Cruz-Neira C, Dickerson JA. Integration of metabolic networks and gene expression in virtual reality. Bioinformatics. 2005;21:3645–50.
Burrage K, Hood L, Ragan MA. Advanced computing for systems biology. Brief Bioinform. 2006;7:390–8.
McComb T, Cairncross O, Noske AB, Wood DL, Marsh BJ, Ragan MA. Illoura: a software tool for analysis, visualization and semantic querying of cellular and other spatial biological data. Bioinformatics. 2009;25:1208–10.
Loew LM, Schaff JC. The Virtual Cell: a software environment for computational cell biology. Trends Biotechnol. 2001;19:401–6.
McClean P, Johnson C, Rogers R, Daniels L, Reber J, Slator BM, et al. Molecular and cellular biology animations: development and impact on student learning. Cell Biol Educ. 2005;4:169–79.
Kaufmann H. Collaborative Augmented Reality in Education. Imagina Conference 2003;TUW-137414.
Garcia-Garcia J, Guney E, Aragues R, Planas-Iglesias J, Oliva B. Biana: a software framework for compiling biological interactions and analyzing networks. BMC Bioinformatics. 2010;11:56.
Theocharidis A, van Dongen S, Enright AJ, Freeman TC. Network visualization and analysis of gene expression data using BioLayout Express(3D). Nat Protoc. 2009;4:1535–50.
Baitaluk M, Sedova M, Ray A, Gupta A. BiologicalNetworks: visualization and analysis tool for systems biology. Nucleic Acids Res. 2006;34:W466–71.
Kozhenkov S, Sedova M, Dubinina Y, Gupta A, Ray A, Ponomarenko J, et al. BiologicalNetworks--tools enabling the integration of multi-scale data for the host-pathogen studies. BMC Syst Biol. 2011;5:7.
Sirava M, Schafer T, Eiglsperger M, Kaufmann M, Kohlbacher O, Bornberg-Bauer E, et al. BioMiner--modeling, analyzing, and visualizing biochemical pathways and networks. Bioinformatics. 2002;18 Suppl 2:S219–30.
Nagasaki M, Saito A, Jeong E, Li C, Kojima K, Ikeda E, et al. Cell Illustrator 4.0: a computational platform for systems biology. In Silico Biol. 2010;10:5–26.
Hoops S, Sahle S, Gauges R, Lee C, Pahle J, Simus N, et al. COPASI--a COmplex PAthway SImulator. Bioinformatics. 2006;22:3067–74.
Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–2.
Ramsey S, Orrell D, Bolouri H. Dizzy: stochastic simulation of large-scale genetic regulatory networks. J Bioinform Comput Biol. 2005;3:415–36.
Kauffman J, Kittas A, Bennett L, Tsoka S. DyCoNet: a Gephi plugin for community detection in dynamic complex networks. PloS One. 2014;9:e101357.
Westenberg MA, van Hijum SAFT, Kuipers OP, Roerdink JBTM. Visualizing genome expression and regulatory network dynamics in genomic and metabolic context. Comput Graph Forum. 2008;27:887–94.
Baker C, Carpendale S, Prusinkiewicz P, Surette M. GeneVis: simulation and visualization of genetic networks. Information Visualization. 2003;2:201–17.
Csardi G, Nepusz T. The igraph software package for complex network research. InterJournal Complex Syst. 2006;1695.
Hooper SD, Bork P. Medusa: a simple tool for interaction graph analysis. Bioinformatics. 2005;21:4432–3.
Pavlopoulos GA, Hooper SD, Sifrim A, Schneider R, Aerts J. Medusa: a tool for exploring and clustering biological networks. BMC Res Notes. 2011;4:384.
Brown KR, Otasek D, Ali M, McGuffin MJ, Xie W, Devani B, et al. NAViGaTOR: network analysis. Visualization and Graphing Toronto Bioinformatics. 2009;25:3327–9.
Djebbari A, Ali M, Otasek D, Kotlyar M, Fortney K, Wong S, et al. NAViGaTOR: large scalable and interactive navigation and analysis of large graphs. Internet Math. 2011;7:314–47.
Kao HL, Gunsalus KC. Browsing multidimensional molecular networks with the generic network browser (N-Browse). Curr Protoc Bioinf. 2008: Chapter 9:Unit 9.11.
Nikitin A, Egorov S, Daraselia N, Mazo I. Pathway studio--the analysis and navigation of molecular networks. Bioinformatics. 2003;19:2155–7.
Orlev N, Shamir R, Shiloh Y. PIVOT: protein interacions visualizatiOn tool. Bioinformatics. 2004;20:424–5.
Krumsiek J, Friedel CC, Zimmer R. ProCope--protein complex prediction and evaluation. Bioinformatics. 2008;24:2115–16.
Iragne F, Nikolski M, Mathieu B, Auber D, Sherman D. ProViz: protein interaction visualization and exploration. Bioinformatics. 2005;21:272–4.
Forman JJ, Clemons PA, Schreiber SL, Haggarty SJ. SpectralNET--an application for spectral graph analysis and visualization. BMC Bioinformatics. 2005;6:260.
Auber D. A huge graph visualization framework. In: Mutzel P, Jünger M, editors. Graph Drawing Software (Mathematics and Visualization). Heidelberg: Springer; 2004. p. 105–26.
Chapter Google Scholar
Brinkrolf C, Janowski SJ, Kormeier B, Lewinski M, Hippe K, Borck D, et al. VANESA - a software application for the visualization and analysis of networks in system biology applications. J Integr Bioinform. 2014;11:239.
Junker BH, Klukas C, Schreiber F. VANTED: a system for advanced data analysis and visualization in the context of biological networks. BMC Bioinformatics. 2006;7:109.
Prieto C, De Las Rivas J. APID: agile protein interaction DataAnalyzer. Nucleic Acids Res. 2006;34:W298–302.
Villeger AC, Pettifer SR, Kell DB. Arcadia: a visualization tool for metabolic pathways. Bioinformatics. 2010;26:1470–1.
Berger SI, Iyengar R, Ma'ayan A. AVIS: AJAX viewer of interactive signaling networks. Bioinformatics. 2007;23:2803–5.
Myers CL, Robson D, Wible A, Hibbs MA, Chiriac C, Theesfeld CL, et al. Discovery of biological networks from diverse functional genomic data. Genome Biol. 2005;6:R114.
Florez LA, Lammers CR, Michna R, Stulke J. Cell Publisher: a web platform for the intuitive visualization and sharing of metabolic, signalling and regulatory pathways. Bioinformatics. 2010;26:2997–9.
Huttenhower C, Mehmood SO, Troyanskaya OG. Graphle: interactive exploration of large, dense graphs. BMC Bioinformatics. 2009;10:417.
Reimand J, Tooming L, Peterson H, Adler P, Vilo J. GraphWeb: mining heterogeneous biological networks for gene modules with functional significance. Nucleic Acids Res. 2008;36:W452–9.
Lin CY, Chin CH, Wu HH, Chen SH, Ho CW, Ko MT. Hubba: hub objects analyzer--a framework of interactome hubs identification for network biology. Nucleic Acids Res. 2008;36:W438–43.
Kalaev M, Smoot M, Ideker T, Sharan R. NetworkBLAST: comparative analysis of protein networks. Bioinformatics. 2008;24:594–6.
Luo W, Brouwer C. Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics. 2013;29:1830–1.
Wu J, Vallenius T, Ovaska K, Westermarck J, Makela TP, Hautaniemi S. Integrated network analysis platform for protein-protein interactions. Nat Methods. 2009;6:75–7.
Pitkanen E, Akerlund A, Rantanen A, Jouhten P, Ukkonen E. ReMatch: a web-based tool to construct, store and share stoichiometric metabolic models with carbon maps for metabolic flux analysis. J Integr Bioinform. 2008;5. doi: 10.2390/biecoll-jib-2008-102 .
Minguez P, Gotz S, Montaner D, Al-Shahrour F, Dopazo J. SNOW, a web-based tool for the statistical analysis of protein-protein interaction networks. Nucleic Acids Res. 2009;37:W109–14.
Kuhn M, Szklarczyk D, Franceschini A, Campillos M, von Mering C, Jensen LJ, et al. STITCH 2: an interaction network database for small molecules and proteins. Nucleic Acids Res. 2010;38:D552–6.
von Mering C, Jensen LJ, Kuhn M, Chaffron S, Doerks T, Kruger B, et al. STRING 7--recent developments in the integration and prediction of protein interactions. Nucleic Acids Res. 2007;35:D358–62.
Curtis RE, Yuen A, Song L, Goyal A, Xing EP. TVNViewer: an interactive visualization tool for exploring networks that change over time or space. Bioinformatics. 2011;27:1880–1.
Yip KY, Yu H, Kim PM, Schultz M, Gerstein M. The tYNA platform for comparative interactomics: a web tool for managing, comparing and mining multiple networks. Bioinformatics. 2006;22:2968–70.
Hu Z, Mellor J, Wu J, DeLisi C. VisANT: an online visualization and analysis tool for biological interaction data. BMC Bioinformatics. 2004;5:17.
Gerasch A, Faber D, Küntzer J, Niermann P, Kohlbacher O, Lenhof H-P, et al. BiNA: a visual analytics tool for biological network data. PloS One. 2014;9:e87397.
Longabaugh WJ, Davidson EH, Bolouri H. Computational representation of developmental genetic regulatory networks. Dev Biol. 2005;283:1–16.
Streit M, Lex A, Kalkusch M, Zatloukal K, Schmalstieg D. Caleydo: connecting pathways and gene expression. Bioinformatics. 2009;25:2760–1.
Funahashi A, Matsuoka Y, Akiya J, Morohashi M, Kikuchi N, Kitano H. Cell Designer 3.5: a versatile modeling tool for biochemical networks. Proc IEEE Inst Electr Electron Eng. 2008;96:1254–65.
Sorokin A, Paliy K, Selkov A, Demin OV, Dronov S, Ghazal P, et al. The Pathway Editor: a tool for managing complex biological networks. IBM Journal of Research and Development. 2006;50:561–73.
Salomonis N, Hanspers K, Zambon AC, Vranizan K, Lawlor SC, Dahlquist KD, et al. GenMAPP 2: new features and resources for pathway analysis. BMC Bioinformatics. 2007;8:217.
Sauro HM, Hucka M, Finney A, Wellock C, Bolouri H, Doyle J, et al. Next generation simulation tools: the Systems Biology Workbench and BioSPICE integration. OMICS. 2003;7:355–72.
Tokimatsu T, Sakurai N, Suzuki H, Ohta H, Nishitani K, Koyama T, et al. KaPPA-view: a web-based analysis tool for integration of transcript and metabolite data on plant metabolic pathway maps. Plant Physiol. 2005;138:1289–300.
Okuda S, Yamada T, Hamajima M, Itoh M, Katayama T, Bork P, et al. KEGG Atlas mapping for global analysis of metabolic pathways. Nucleic Acids Res. 2008;36:W423–6.
Droste P, Nöh K, Wiechert W. Omix - a visualization tool for metabolic networks with highest usability and customizability in focus. Chemie Ingenieur Technik. 2013;85:849–62.
Holford M, Li N, Nadkarni P, Zhao H. VitaPad: visualization tools for the analysis of pathway data. Bioinformatics. 2005;21:1596–602.
Chung H-J, Kim M, Park CH, Kim J, Kim JH. ArrayXPath: mapping and visualizing microarray gene-expression data with integrated biological pathway resources using Scalable Vector Graphics. Nucleic Acids Res. 2004;32:W460–4.
Weniger M, Engelmann JC, Schultz J. Genome expression pathway analysis tool--analysis and visualization of microarray gene expression data under genomic, proteomic and metabolic context. BMC Bioinformatics. 2007;8:179.
Yamada T, Letunic I, Okuda S, Kanehisa M, Bork P. iPath2.0: interactive pathway explorer. Nucleic Acids Res. 2011;39:W412–15.
Arakawa K, Kono N, Yamada Y, Mori H, Tomita M. KEGG-based pathway visualization tool for complex omics data. In Silico Biol. 2005;5:419–23.
Xia J, Wishart DS. MetPA: a web-based metabolomics tool for pathway analysis and visualization. Bioinformatics. 2010;26:2342–4.
Paley SM, Karp PD. The pathway tools cellular overview diagram and omics viewer. Nucleic Acids Res. 2006;34:3771–8.
Mlecnik B, Scheideler M, Hackl H, Hartler J, Sanchez-Cabo F, Trajanoski Z. PathwayExplorer: web service for visualizing high-throughput expression data on biological pathways. Nucleic Acids Res. 2005;33:W633–7.
Kono N, Arakawa K, Ogawa R, Kido N, Oshita K, Ikegami K, et al. Pathway projector: web-based zoomable pathway browser using KEGG atlas and Google Maps API. PloS One. 2009;4:e7710.
Pico AR, Kelder T, van Iersel MP, Hanspers K, Conklin BR, Evelo C. WikiPathways: pathway editing for the people. PLoS Biol. 2008;6:e184.
Nielsen CB, Jackman SD, Birol I, Jones SJ. ABySS-Explorer: visualizing genome sequence assemblies. IEEE Trans Vis Comput Graph. 2009;15:881–8.
Carver T, Harris SR, Otto TD, Berriman M, Parkhill J, McQuillan JA. BamView: visualizing and interpretation of next-generation sequencing read alignments. Brief Bioinform. 2013;14:203–12.
Liu C, Bonner TI, Nguyen T, Lyons JL, Christian SL, Gershon ES. DNannotator: annotation software tool kit for regional genomic sequences. Nucleic Acids Res. 2003;31:3729–35.
Yang Y, Liu J. JVM: Java Visual Mapping tool for next generation sequencing read. Adv Exp Med Biol. 2015;827:11–8.
Manske HM, Kwiatkowski DP. LookSeq: a browser-based viewer for deep sequencing data. Genome Res. 2009;19:2125–32.
Hou H, Zhao F, Zhou L, Zhu E, Teng H, Li X, et al. MagicViewer: integrated solution for next-generation sequencing data visualization and genetic variation detection and annotation. Nucleic Acids Res. 2010;38:W732–6.
Bao H, Guo H, Wang J, Zhou R, Lu X, Shi S. MapView: visualization of short reads alignment on a desktop computer. Bioinformatics. 2009;25:1554–5.
Elnitski L, Riemer C, Burhans R, Hardison R, Miller W. MultiPipMaker: comparative alignment server for multiple DNA sequences. Curr Protoc Bioinf. 2005, Chapter 10: Unit 10.14.
López-Fernández H, Glez-Peña D, Reboiro-Jato M, Gómez-López G, Pisano DG, Fdez-Riverola F. PileLineGUI: a desktop environment for handling genome position files in next-generation sequencing studies. Nucleic Acids Res. 2011;39:W562–6.
Pitt JN, Rajapakse I, Ferre-D'Amare AR. SEWAL: an open-source platform for next-generation sequence analysis and visualization. Nucleic Acids Res. 2010;38:7908–15.
Wang T, Liu J, Shen L, Tonti-Filippini J, Zhu Y, Jia H, et al. STAR: an integrated solution to management and visualization of sequencing data. Bioinformatics. 2013;29:3204–10.
Ge D, Ruzzo EK, Shianna KV, He M, Pelak K, Heinzen EL, et al. SVA: software for annotating and visualizing sequenced human genomes. Bioinformatics. 2011;27:1998–2000.
Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–92.
Zhang Z, Lin H, Ma B. ZOOM Lite: next-generation sequencing data mapping and visualization software. Nucleic Acids Res. 2010;38 Suppl 2:W743–8.
Salzberg SL, Church D, DiCuccio M, Yaschenko E, Ostell J. The genome Assembly Archive: a new public resource. PLoS Biol. 2004;2:E285.
Li P, Ji G, Dong M, Schmidt E, Lenox D, Chen L, et al. CBrowse: a SAM/BAM-based contig browser for transcriptome assembly visualization and analysis. Bioinformatics. 2012;28:2382–4.
Tang B, Wang Q, Yang M, Xie F, Zhu Y, Zhuo Y, et al. ContigScape: a Cytoscape plugin facilitating microbial genome gap closing. BMC Genomics. 2013;14:289.
Burland TG. DNASTAR's Lasergene sequence analysis software. Methods Mol Biol. 2000;132:71–91.
Huang W, Marth G. EagleView: a genome assembly viewer for next-generation sequencing technologies. Genome Res. 2008;18:1538–43.
Schatz MC, Phillippy AM, Sommer DD, Delcher AL, Puiu D, Narzisi G, et al. Hawkeye and AMOS: visualizing and assessing the quality of genome assemblies. Brief Bioinform. 2013;14:213–24.
Milne I, Bayer M, Cardle L, Shaw P, Stephen G, Wright F, et al. Tablet—next generation sequence assembly visualization. Bioinformatics. 2010;26:401–2.
Kong L, Wang J, Zhao S, Gu X, Luo J, Gao G. ABrowse--a customizable next-generation genome browser framework. BMC Bioinformatics. 2012;13:2.
Tonti-Filippini J. AnnoJ. http://www.annoj.org . Accessed: 27 July 2015.
Grant JR, Stothard P. The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acids Res. 2008;36:W181–4.
Engels R, Yu T, Burge C, Mesirov JP, DeCaprio D, Galagan JE. Combo: a whole genome comparative browser. Bioinformatics. 2006;22:1782–3.
Juan L, Liu Y, Wang Y, Teng M, Zang T, Wang Y. Family genome browser: visualizing genomes with pedigree information. Bioinformatics. 2015;31:2262–8.
Shannon PT, Reiss DJ, Bonneau R, Baliga NS. The Gaggle: an open-source software system for integrating bioinformatics software and data sources. BMC Bioinformatics. 2006;7:176.
Papanicolaou A, Heckel DG. The GMOD Drupal bioinformatic server framework. Bioinformatics. 2010;26:3119–24.
Wang H, Su Y, Mackey AJ, Kraemer ET, Kissinger JC. SynView: a GBrowse-compatible approach to visualizing comparative genome data. Bioinformatics. 2006;22:2308–9.
Sato N, Ehira S. GenoMap, a circular genome data viewer. Bioinformatics. 2003;19:1583–4.
Arakawa K, Tamaki S, Kono N, Kido N, Ikegami K, Ogawa R, et al. Genome Projector: zoomable genome map with multiple views. BMC Bioinformatics. 2009;10:31.
Abeel T, Van Parys T, Saeys Y, Galagan J, Van de Peer Y. GenomeView: a next-generation genome browser. Nucleic Acids Res. 2012;40:e12.
Lajugie J, Bouhassira EE. GenPlay, a multipurpose genome analyzer and browser. Bioinformatics. 2011;27:1889–93.
Nicol JW, Helt GA, Blanchard Jr SG, Raja A, Loraine AE. The integrated genome browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics. 2009;25:2730–1.
Skinner ME, Uzilov AV, Stein LD, Mungall CJ, Holmes IH. JBrowse: a next-generation genome browser. Genome Res. 2009;19:1630–8.
Wheeler DL, Church DM, Federhen S, Lash AE, Madden TL, Pontius JU, et al. Database resources of the National Center for Biotechnology. Nucleic Acids Res. 2003;31:28–33.
Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012;40:D1178–86.
Fiume M, Williams V, Brudno M. Savant: genome browser for high throughput sequencing data. Bioinformatics. 2010;26:1938–44.
Miller CA, Anthony J, Meyer MM, Marth G. Scribl: an HTML5 Canvas-based graphics library for visualizing genomic data over the web. Bioinformatics. 2013;29:381–3.
Axelrod N, Lin Y, Ng PC, Stockwell TB, Crabtree J, Huang J, et al. The HuRef Browser: a web resource for individual human genomics. Nucleic Acids Res. 2009;37 Suppl 1:D1018–24.
Juan L, Teng M, Zang T, Hao Y, Wang Z, Yan C, et al. The personal genome browser: visualizing functions of genetic variants. Nucleic Acids Res. 2014;42:W192–7.
Zhu J, Sanborn JZ, Benz S, Szeto C, Hsu F, Kuhn RM, et al. The UCSC cancer genomics browser. Nat Methods. 2009;6:239–40.
Goldman M, Craft B, Swatloski T, Cline M, Morozova O, Diekhans M, et al. The UCSC cancer genomics browser: update 2015. Nucleic Acids Res. 2015;43:D812–17.
Saito TL, Yoshimura J, Sasaki S, Ahsan B, Sasaki A, Kuroshu R, et al. UTGB toolkit for personalized genome browsers. Bioinformatics. 2009;25:1856–61.
Yates T, Okoniewski MJ, Miller CJ. X:Map: annotation and visualization of genome structure for Affymetrix exon array analysis. Nucleic Acids Res. 2008;36:D780–6.
Carver T, Berriman M, Tivey A, Patel C, Bohme U, Barrell BG, et al. Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics. 2008;24:2672–6.
Sinha AU, Meller J. Cinteny: flexible analysis and visualization of synteny and genome rearrangements in multiple organisms. BMC Bioinformatics. 2007;8:82.
Youens-Clark K, Faga B, Yap IV, Stein L, Ware D. CMap 1.01: a comparative mapping application for the internet. Bioinformatics. 2009;25:3040–2.
Lyons E, Pedersen B, Kane J, Alam M, Ming R, Tang H, et al. Finding and comparing syntenic regions among Arabidopsis and the outgroups papaya, poplar, and grape: CoGe with rosids. Plant Physiol. 2008;148:1772–81.
Deng X, Rayner S, Liu X, Zhang Q, Yang Y, Li N. DHPC: a new tool to express genome structural features. Genomics. 2008;91:476–83.
Carver T, Thomson N, Bleasby A, Berriman M, Parkhill J. DNAPlotter: circular and linear interactive genome visualization. Bioinformatics. 2009;25:119–20.
Zeinaly M, Soltangheis M, Shaw CD. FilooT: a visualization tool for exploring genomic data. SPIE 9017, Visualization and Data Analysis 2014. doi: 10.1117/12.2042589 .
McKay SJ, Vergara IA, Stajich JE. Using the Generic Synteny Browser (GBrowse_syn). Curr Protoc Bioinf. 2010, Chapter 9:Unit 9.12.
Yang J, Wang J, Yao ZJ, Jin Q, Shen Y, Chen R. GenomeComp: a visualization tool for microbial genome comparison. J Microbiol Methods. 2003;54:423–6.
Ohtsubo Y, Ikeda-Ohtsubo W, Nagata Y, Tsuda M. GenomeMatcher: a graphical user interface for DNA sequence comparison. BMC Bioinformatics. 2008;9:376.
Lajugie J, Fourel N, Bouhassira EE. GenPlay Multi-Genome, a tool to compare and analyze multiple human genomes in a graphical interface. Bioinformatics. 2015;31:109–11.
Yin T, Cook D, Lawrence M. ggbio: an R package for extending the grammar of graphics for genomic data. Genome Biol. 2012;13:R77.
Liang C, Jaiswal P, Hebbard C, Avraham S, Buckler ES, Casstevens T, et al. Gramene: a growing plant comparative genomics resource. Nucleic Acids Res. 2008;36:D947–53.
Ware DH, Jaiswal P, Ni J, Yap IV, Pan X, Clark KY, et al. Gramene, a tool for grass genomics. Plant Physiol. 2002;130:1606–13.
Anders S. Visualization of genomic data with the Hilbert curve. Bioinformatics. 2009;25:1231–5.
Qi J, Zhao F. inGAP-sv: a novel scheme to identify and visualize structural variation from paired end mapping data. Nucleic Acids Res. 2011;39:W567–75.
Pavlopoulos GA, Kumar P, Sifrim A, Sakai R, Lin ML, Voet T, et al. Meander: visually exploring the structural variome using space-filling curves. Nucleic Acids Res. 2013;41:e118.
Broad Institute: MEDEA: Comparative Genomic Visualization with Adobe Flash. http://www.broadinstitute.org/annotation/medea/ (2015). Accessed 27 July 2015.
Meyer M, Munzner T, Pfister H. MizBee: a multiscale synteny browser. IEEE Trans Vis Comput Graph. 2009;15:897–904.
Dees ND, Zhang Q, Kandoth C, Wendl MC, Schierding W, Koboldt DC, et al. MuSiC: identifying mutational significance in cancer genomes. Genome Res. 2012;22:1589–98.
Shen L, Shao N, Liu X, Nestler E. ngs.plot: quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics. 2014;15:284.
Dehal PS, Boore JL. A phylogenomic gene cluster resource: the Phylogenetically Inferred Groups (PhIGs) database. BMC Bioinformatics. 2006;7:201.
Fong C, Rohmer L, Radey M, Wasnick M, Brittnacher MJ. PSAT: a web tool to compare genomic neighborhoods of multiple prokaryotic genomes. BMC Bioinformatics. 2008;9:170.
Esteban-Marcos A, Darling AE, Ragan MA. Seevolution: visualizing chromosome evolution. Bioinformatics. 2009;25:960–1.
Crabtree J, Angiuoli SV, Wortman JR, White OR. Sybil: methods and software for multiple genome comparison and visualization. Methods Mol Biol. 2007;408:93–108.
Asmann YW, Middha S, Hossain A, Baheti S, Li Y, Chai HS, et al. TREAT: a bioinformatics tool for variant annotations and visualizations in targeted and exome sequencing data. Bioinformatics. 2012;28:277–8.
Miller W, Rosenbloom K, Hardison RC, Hou M, Taylor J, Raney B, et al. 28-way vertebrate alignment and conservation track in the UCSC Genome Browser. Genome Res. 2007;17:1797–808.
Huang PJ, Lee CC, Tan BC, Yeh YM, Huang KY, Gan RC, et al. Vanno: a visualization-aided variant annotation tool. Hum Mutat. 2015;36:167–74.
Ferstay JA, Nielsen CB, Munzner T. Variant view: visualizing sequence variants in their gene context. IEEE Trans Vis Comput Graph. 2013;19:2546–55.
Nordberg H, Cantor M, Dusheyko S, Hua S, Poliakov A, Shabalov I, et al. The genome portal of the Department of Energy Joint Genome Institute: 2014 updates. Nucleic Acids Res. 2014;42:D26–31.
Grigoriev IV, Nordberg H, Shabalov I, Aerts A, Cantor M, Goodstein D, et al. The genome portal of the Department of Energy Joint Genome Institute. Nucleic Acids Res. 2012;40:D26–32.
Talevich E, Invergo BM, Cock PJ, Chapman BA. Bio. Phylo: a unified toolkit for processing, analyzing and visualizing phylogenetic trees in Biopython. BMC Bioinformatics. 2012;13:209.
Huerta-Cepas J, Dopazo J, Gabaldon T. ETE: a python environment for tree exploration. BMC Bioinformatics. 2010;11:24.
Zhang H, Gao S, Lercher MJ, Hu S, Chen WH. EvolView, an online tool for visualizing, annotating and managing phylogenetic trees. Nucleic Acids Res. 2012;40:W569–72.
Smits SA, Ouverney CC. jsPhyloSVG: a javascript library for visualizing interactive and vector-based phylogenetic trees on the web. PloS One. 2010;5:e12267.
Sanderson MJ. Paloverde: an OpenGL 3D phylogeny browser. Bioinformatics. 2006;22:1004–6.
Choi JH, Jung HY, Kim HS, Cho HG. PhyloDraw: a phylogenetic tree drawing system. Bioinformatics. 2000;16:1056–8.
Ranwez V, Clairon N, Delsuc F, Pourali S, Auberval N, Diser S, et al. PhyloExplorer: a web server to validate, explore and query phylogenetic trees. BMC Evol Biol. 2009;9:108.
Jordan GE, Piel WH. PhyloWidget: web-based visualizations for the tree of life. Bioinformatics. 2008;24:1641–2.
Chevenet F, Brun C, Banuls AL, Jacq B, Christen R. TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics. 2006;7:439.
Stover BC, Muller KF. TreeGraph 2: combining and visualizing evidence from different phylogenetic analyses. BMC Bioinformatics. 2010;11:7.
Gu S, Anderson I, Kunin V, Cipriano M, Minovitsky S, Weber G, et al. TreeQ-VISTA: an interactive tree visualization tool with functional annotation query capabilities. Bioinformatics. 2007;23:764–6.
Pethica R, Barker G, Kovacs T, Gough J. TreeVector: scalable, interactive, phylogenetic trees for the web. PloS One. 2010;5:e8934.
Santamaria R, Theron R. Treevolution: visual analysis of phylogenetic trees. Bioinformatics. 2009;25:1970–1.
Boc A, Diallo AB, Makarenkov V. T-REX: a web server for inferring, validating and visualizing phylogenetic trees and networks. Nucleic Acids Res. 2012;40:W573–9.
Bremm S, von Landesberger T, Hess M, Schreck T, Weil P, Hamacherk K. Interactive visual comparison of multiple trees. 2011 IEEE Conference on Visual Analytics Science and Technology (VAST). 2011. doi: 10.1109/VAST.2011.6102439 .
Santamaria R, Theron R, Quintales L. BicOverlapper: a tool for bicluster visualization. Bioinformatics. 2008;24:1212–13.
Santamaria R, Theron R, Quintales L. BicOverlapper 2.0: visual analysis for gene expression. Bioinformatics. 2014;30:1785–6.
Goncalves JP, Madeira SC, Oliveira AL. BiGGEsTS: integrated environment for biclustering analysis of time series gene expression data. BMC research notes. 2009;2:124.
Yuan T, Huang X, Dittmar RL, Du M, Kohli M, Boardman L, et al. eRNA: a graphic user interface-based tool optimized for large data analysis from high-throughput RNA sequencing. BMC Genomics. 2014;15:176.
Kapushesky M, Kemmeren P, Culhane AC, Durinck S, Ihmels J, Korner C, et al. Expression Profiler: next generation--an online platform for analysis of microarray data. Nucleic Acids Res. 2004;32:W465–70.
Hibbs MA, Dirksen NC, Li K, Troyanskaya OG. Visualization methods for statistical analysis of microarray clusters. BMC Bioinformatics. 2005;6:115.
Floratos A, Smith K, Ji Z, Watkinson J, Califano A. geWorkbench: an open source platform for integrative genomics. Bioinformatics. 2010;26:1779–80.
Perez-Llamas C, Lopez-Bigas N. Gitools: analysis and visualisation of genomic data using interactive heat-maps. PloS One. 2011;6:e19541.
Jinwook S, Shneiderman B. Interactively exploring hierarchical clustering results [gene identification]. Computer. 2002;35:80–6.
Khomtchouk BB, Van Booven DJ, Wahlestedt C. HeatmapGenerator: high performance RNAseq and microarray visualization software suite to examine differential gene expression levels using an R and C++ hybrid computational pipeline. Source Code Biol Med. 2014;9:30.
Yachdav G, Hecht M, Pasmanik-Chor M, Yeheskel A, Rost B. HeatMapViewer: interactive display of 2D data in biology. F1000Research. 2014;3:48.
PubMed PubMed Central Google Scholar
Dietzsch J, Gehlenborg N, Nieselt K. Mayday--a microarray data analysis workbench. Bioinformatics. 2006;22:1010–12.
Weber GH, Rubel O, Huang MY, DePace AH, Fowlkes CC, Keranen SV, et al. Visual exploration of three-dimensional gene expression using physical views and linked abstract views. IEEE/ACM Trans Comput Biol Bioinform. 2009;6:296–309.
An J, Lai J, Wood DL, Sajjanhar A, Wang C, Tevz G, et al. RNASeqBrowser: a genome browser for simultaneous visualization of raw strand specific RNAseq reads and UCSC genome browser custom tracks. BMC Genomics. 2015;16:145.
Roge X, Zhang X. RNAseqViewer: visualization tool for RNA-Seq data. Bioinformatics. 2014;30:891–2.
Hochheiser H, Baehrecke EH, Mount SM, Shneiderman B. Dynamic querying for pattern identification in microarray and genomic data. International Conference on Multimedia and Expo 2003, ICME '03. 2003. doi: 10.1109/ICME.2003.1221346 .
Dietrich S, Wiegand S, Liesegang H. TraV: a genome context sensitive transcriptome browser. PloS One. 2014;9:e93677.
Download references
This work was supported by the European Commission FP7 programs INFLA-CARE (EC grant agreement number 223151), ‘Translational Potential’ (EC grant agreement number 285948).
Author information
Authors and affiliations.
Bioinformatics & Computational Biology Laboratory, Division of Basic Sciences, University of Crete, Medical School, 70013, Heraklion, Crete, Greece
Georgios A. Pavlopoulos, Nikolas Papanikolaou, Theodosis Theodosiou & Ioannis Iliopoulos
Department of Biology, University of Crete, 70013, Heraklion, Crete, Greece
Dimitris Malliarakis
EMBL - European Bioinformatics Institute, Wellcome Trust Genome Campus, Cambridge, CB10 1SD, UK
Anton J. Enright
You can also search for this author in PubMed Google Scholar
Corresponding authors
Correspondence to Georgios A. Pavlopoulos or Ioannis Iliopoulos .
Additional information
Competing interests.
The authors declare that they have no competing interests.
Authors’ contributions
GAP was the main writer of the article and the one who was inspired about the topic. DM collected all the necessary information about the tools and their citation trends. NP and TT provided fruitful feedback about the recent HCI technologies. AE provided information about the latest technological trends in the areas of genomics. II was the main supervisor of the project. All authors read and approved the final manuscript.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License ( http://creativecommons.org/licenses/by/4.0/ ), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated.
Reprints and permissions
About this article
Cite this article.
Pavlopoulos, G.A., Malliarakis, D., Papanikolaou, N. et al. Visualizing genome and systems biology: technologies, tools, implementation techniques and trends, past, present and future. GigaSci 4 , 38 (2015). https://doi.org/10.1186/s13742-015-0077-2
Download citation
Received : 26 May 2015
Accepted : 03 August 2015
Published : 25 August 2015
DOI : https://doi.org/10.1186/s13742-015-0077-2
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Biological data visualization
- Network biology
- Systems biology
- Multivariate analysis
GigaScience
ISSN: 2047-217X
- Submission enquiries: [email protected]
- General enquiries: [email protected]
- Open access
- Published: 19 July 2015
The role of visual representations in scientific practices: from conceptual understanding and knowledge generation to ‘seeing’ how science works
- Maria Evagorou 1 ,
- Sibel Erduran 2 &
- Terhi Mäntylä 3
International Journal of STEM Education volume 2 , Article number: 11 ( 2015 ) Cite this article
76k Accesses
78 Citations
13 Altmetric
Metrics details
The use of visual representations (i.e., photographs, diagrams, models) has been part of science, and their use makes it possible for scientists to interact with and represent complex phenomena, not observable in other ways. Despite a wealth of research in science education on visual representations, the emphasis of such research has mainly been on the conceptual understanding when using visual representations and less on visual representations as epistemic objects. In this paper, we argue that by positioning visual representations as epistemic objects of scientific practices, science education can bring a renewed focus on how visualization contributes to knowledge formation in science from the learners’ perspective.
This is a theoretical paper, and in order to argue about the role of visualization, we first present a case study, that of the discovery of the structure of DNA that highlights the epistemic components of visual information in science. The second case study focuses on Faraday’s use of the lines of magnetic force. Faraday is known of his exploratory, creative, and yet systemic way of experimenting, and the visual reasoning leading to theoretical development was an inherent part of the experimentation. Third, we trace a contemporary account from science focusing on the experimental practices and how reproducibility of experimental procedures can be reinforced through video data.
Conclusions
Our conclusions suggest that in teaching science, the emphasis in visualization should shift from cognitive understanding—using the products of science to understand the content—to engaging in the processes of visualization. Furthermore, we suggest that is it essential to design curriculum materials and learning environments that create a social and epistemic context and invite students to engage in the practice of visualization as evidence, reasoning, experimental procedure, or a means of communication and reflect on these practices. Implications for teacher education include the need for teacher professional development programs to problematize the use of visual representations as epistemic objects that are part of scientific practices.
During the last decades, research and reform documents in science education across the world have been calling for an emphasis not only on the content but also on the processes of science (Bybee 2014 ; Eurydice 2012 ; Duschl and Bybee 2014 ; Osborne 2014 ; Schwartz et al. 2012 ), in order to make science accessible to the students and enable them to understand the epistemic foundation of science. Scientific practices, part of the process of science, are the cognitive and discursive activities that are targeted in science education to develop epistemic understanding and appreciation of the nature of science (Duschl et al. 2008 ) and have been the emphasis of recent reform documents in science education across the world (Achieve 2013 ; Eurydice 2012 ). With the term scientific practices, we refer to the processes that take place during scientific discoveries and include among others: asking questions, developing and using models, engaging in arguments, and constructing and communicating explanations (National Research Council 2012 ). The emphasis on scientific practices aims to move the teaching of science from knowledge to the understanding of the processes and the epistemic aspects of science. Additionally, by placing an emphasis on engaging students in scientific practices, we aim to help students acquire scientific knowledge in meaningful contexts that resemble the reality of scientific discoveries.
Despite a wealth of research in science education on visual representations, the emphasis of such research has mainly been on the conceptual understanding when using visual representations and less on visual representations as epistemic objects. In this paper, we argue that by positioning visual representations as epistemic objects, science education can bring a renewed focus on how visualization contributes to knowledge formation in science from the learners’ perspective. Specifically, the use of visual representations (i.e., photographs, diagrams, tables, charts) has been part of science and over the years has evolved with the new technologies (i.e., from drawings to advanced digital images and three dimensional models). Visualization makes it possible for scientists to interact with complex phenomena (Richards 2003 ), and they might convey important evidence not observable in other ways. Visual representations as a tool to support cognitive understanding in science have been studied extensively (i.e., Gilbert 2010 ; Wu and Shah 2004 ). Studies in science education have explored the use of images in science textbooks (i.e., Dimopoulos et al. 2003 ; Bungum 2008 ), students’ representations or models when doing science (i.e., Gilbert et al. 2008 ; Dori et al. 2003 ; Lehrer and Schauble 2012 ; Schwarz et al. 2009 ), and students’ images of science and scientists (i.e., Chambers 1983 ). Therefore, studies in the field of science education have been using the term visualization as “the formation of an internal representation from an external representation” (Gilbert et al. 2008 , p. 4) or as a tool for conceptual understanding for students.
In this paper, we do not refer to visualization as mental image, model, or presentation only (Gilbert et al. 2008 ; Philips et al. 2010 ) but instead focus on visual representations or visualization as epistemic objects. Specifically, we refer to visualization as a process for knowledge production and growth in science. In this respect, modeling is an aspect of visualization, but what we are focusing on with visualization is not on the use of model as a tool for cognitive understanding (Gilbert 2010 ; Wu and Shah 2004 ) but the on the process of modeling as a scientific practice which includes the construction and use of models, the use of other representations, the communication in the groups with the use of the visual representation, and the appreciation of the difficulties that the science phase in this process. Therefore, the purpose of this paper is to present through the history of science how visualization can be considered not only as a cognitive tool in science education but also as an epistemic object that can potentially support students to understand aspects of the nature of science.
Scientific practices and science education
According to the New Generation Science Standards (Achieve 2013 ), scientific practices refer to: asking questions and defining problems; developing and using models; planning and carrying out investigations; analyzing and interpreting data; using mathematical and computational thinking; constructing explanations and designing solutions; engaging in argument from evidence; and obtaining, evaluating, and communicating information. A significant aspect of scientific practices is that science learning is more than just about learning facts, concepts, theories, and laws. A fuller appreciation of science necessitates the understanding of the science relative to its epistemological grounding and the process that are involved in the production of knowledge (Hogan and Maglienti 2001 ; Wickman 2004 ).
The New Generation Science Standards is, among other changes, shifting away from science inquiry and towards the inclusion of scientific practices (Duschl and Bybee 2014 ; Osborne 2014 ). By comparing the abilities to do scientific inquiry (National Research Council 2000 ) with the set of scientific practices, it is evident that the latter is about engaging in the processes of doing science and experiencing in that way science in a more authentic way. Engaging in scientific practices according to Osborne ( 2014 ) “presents a more authentic picture of the endeavor that is science” (p.183) and also helps the students to develop a deeper understanding of the epistemic aspects of science. Furthermore, as Bybee ( 2014 ) argues, by engaging students in scientific practices, we involve them in an understanding of the nature of science and an understanding on the nature of scientific knowledge.
Science as a practice and scientific practices as a term emerged by the philosopher of science, Kuhn (Osborne 2014 ), refers to the processes in which the scientists engage during knowledge production and communication. The work that is followed by historians, philosophers, and sociologists of science (Latour 2011 ; Longino 2002 ; Nersessian 2008 ) revealed the scientific practices in which the scientists engage in and include among others theory development and specific ways of talking, modeling, and communicating the outcomes of science.
Visualization as an epistemic object
Schematic, pictorial symbols in the design of scientific instruments and analysis of the perceptual and functional information that is being stored in those images have been areas of investigation in philosophy of scientific experimentation (Gooding et al. 1993 ). The nature of visual perception, the relationship between thought and vision, and the role of reproducibility as a norm for experimental research form a central aspect of this domain of research in philosophy of science. For instance, Rothbart ( 1997 ) has argued that visualizations are commonplace in the theoretical sciences even if every scientific theory may not be defined by visualized models.
Visual representations (i.e., photographs, diagrams, tables, charts, models) have been used in science over the years to enable scientists to interact with complex phenomena (Richards 2003 ) and might convey important evidence not observable in other ways (Barber et al. 2006 ). Some authors (e.g., Ruivenkamp and Rip 2010 ) have argued that visualization is as a core activity of some scientific communities of practice (e.g., nanotechnology) while others (e.g., Lynch and Edgerton 1988 ) have differentiated the role of particular visualization techniques (e.g., of digital image processing in astronomy). Visualization in science includes the complex process through which scientists develop or produce imagery, schemes, and graphical representation, and therefore, what is of importance in this process is not only the result but also the methodology employed by the scientists, namely, how this result was produced. Visual representations in science may refer to objects that are believed to have some kind of material or physical existence but equally might refer to purely mental, conceptual, and abstract constructs (Pauwels 2006 ). More specifically, visual representations can be found for: (a) phenomena that are not observable with the eye (i.e., microscopic or macroscopic); (b) phenomena that do not exist as visual representations but can be translated as such (i.e., sound); and (c) in experimental settings to provide visual data representations (i.e., graphs presenting velocity of moving objects). Additionally, since science is not only about replicating reality but also about making it more understandable to people (either to the public or other scientists), visual representations are not only about reproducing the nature but also about: (a) functioning in helping solving a problem, (b) filling gaps in our knowledge, and (c) facilitating knowledge building or transfer (Lynch 2006 ).
Using or developing visual representations in the scientific practice can range from a straightforward to a complicated situation. More specifically, scientists can observe a phenomenon (i.e., mitosis) and represent it visually using a picture or diagram, which is quite straightforward. But they can also use a variety of complicated techniques (i.e., crystallography in the case of DNA studies) that are either available or need to be developed or refined in order to acquire the visual information that can be used in the process of theory development (i.e., Latour and Woolgar 1979 ). Furthermore, some visual representations need decoding, and the scientists need to learn how to read these images (i.e., radiologists); therefore, using visual representations in the process of science requires learning a new language that is specific to the medium/methods that is used (i.e., understanding an X-ray picture is different from understanding an MRI scan) and then communicating that language to other scientists and the public.
There are much intent and purposes of visual representations in scientific practices, as for example to make a diagnosis, compare, describe, and preserve for future study, verify and explore new territory, generate new data (Pauwels 2006 ), or present new methodologies. According to Latour and Woolgar ( 1979 ) and Knorr Cetina ( 1999 ), visual representations can be used either as primary data (i.e., image from a microscope). or can be used to help in concept development (i.e., models of DNA used by Watson and Crick), to uncover relationships and to make the abstract more concrete (graphs of sound waves). Therefore, visual representations and visual practices, in all forms, are an important aspect of the scientific practices in developing, clarifying, and transmitting scientific knowledge (Pauwels 2006 ).
Methods and Results: Merging Visualization and scientific practices in science
In this paper, we present three case studies that embody the working practices of scientists in an effort to present visualization as a scientific practice and present our argument about how visualization is a complex process that could include among others modeling and use of representation but is not only limited to that. The first case study explores the role of visualization in the construction of knowledge about the structure of DNA, using visuals as evidence. The second case study focuses on Faraday’s use of the lines of magnetic force and the visual reasoning leading to the theoretical development that was an inherent part of the experimentation. The third case study focuses on the current practices of scientists in the context of a peer-reviewed journal called the Journal of Visualized Experiments where the methodology is communicated through videotaped procedures. The three case studies represent the research interests of the three authors of this paper and were chosen to present how visualization as a practice can be involved in all stages of doing science, from hypothesizing and evaluating evidence (case study 1) to experimenting and reasoning (case study 2) to communicating the findings and methodology with the research community (case study 3), and represent in this way the three functions of visualization as presented by Lynch ( 2006 ). Furthermore, the last case study showcases how the development of visualization technologies has contributed to the communication of findings and methodologies in science and present in that way an aspect of current scientific practices. In all three cases, our approach is guided by the observation that the visual information is an integral part of scientific practices at the least and furthermore that they are particularly central in the scientific practices of science.
Case study 1: use visual representations as evidence in the discovery of DNA
The focus of the first case study is the discovery of the structure of DNA. The DNA was first isolated in 1869 by Friedrich Miescher, and by the late 1940s, it was known that it contained phosphate, sugar, and four nitrogen-containing chemical bases. However, no one had figured the structure of the DNA until Watson and Crick presented their model of DNA in 1953. Other than the social aspects of the discovery of the DNA, another important aspect was the role of visual evidence that led to knowledge development in the area. More specifically, by studying the personal accounts of Watson ( 1968 ) and Crick ( 1988 ) about the discovery of the structure of the DNA, the following main ideas regarding the role of visual representations in the production of knowledge can be identified: (a) The use of visual representations was an important part of knowledge growth and was often dependent upon the discovery of new technologies (i.e., better microscopes or better techniques in crystallography that would provide better visual representations as evidence of the helical structure of the DNA); and (b) Models (three-dimensional) were used as a way to represent the visual images (X-ray images) and connect them to the evidence provided by other sources to see whether the theory can be supported. Therefore, the model of DNA was built based on the combination of visual evidence and experimental data.
An example showcasing the importance of visual representations in the process of knowledge production in this case is provided by Watson, in his book The Double Helix (1968):
…since the middle of the summer Rosy [Rosalind Franklin] had had evidence for a new three-dimensional form of DNA. It occurred when the DNA 2molecules were surrounded by a large amount of water. When I asked what the pattern was like, Maurice went into the adjacent room to pick up a print of the new form they called the “B” structure. The instant I saw the picture, my mouth fell open and my pulse began to race. The pattern was unbelievably simpler than those previously obtained (A form). Moreover, the black cross of reflections which dominated the picture could arise only from a helical structure. With the A form the argument for the helix was never straightforward, and considerable ambiguity existed as to exactly which type of helical symmetry was present. With the B form however, mere inspection of its X-ray picture gave several of the vital helical parameters. (p. 167-169)
As suggested by Watson’s personal account of the discovery of the DNA, the photo taken by Rosalind Franklin (Fig. 1 ) convinced him that the DNA molecule must consist of two chains arranged in a paired helix, which resembles a spiral staircase or ladder, and on March 7, 1953, Watson and Crick finished and presented their model of the structure of DNA (Watson and Berry 2004 ; Watson 1968 ) which was based on the visual information provided by the X-ray image and their knowledge of chemistry.
X-ray chrystallography of DNA
In analyzing the visualization practice in this case study, we observe the following instances that highlight how the visual information played a role:
Asking questions and defining problems: The real world in the model of science can at some points only be observed through visual representations or representations, i.e., if we are using DNA as an example, the structure of DNA was only observable through the crystallography images produced by Rosalind Franklin in the laboratory. There was no other way to observe the structure of DNA, therefore the real world.
Analyzing and interpreting data: The images that resulted from crystallography as well as their interpretations served as the data for the scientists studying the structure of DNA.
Experimenting: The data in the form of visual information were used to predict the possible structure of the DNA.
Modeling: Based on the prediction, an actual three-dimensional model was prepared by Watson and Crick. The first model did not fit with the real world (refuted by Rosalind Franklin and her research group from King’s College) and Watson and Crick had to go through the same process again to find better visual evidence (better crystallography images) and create an improved visual model.
Example excerpts from Watson’s biography provide further evidence for how visualization practices were applied in the context of the discovery of DNA (Table 1 ).
In summary, by examining the history of the discovery of DNA, we showcased how visual data is used as scientific evidence in science, identifying in that way an aspect of the nature of science that is still unexplored in the history of science and an aspect that has been ignored in the teaching of science. Visual representations are used in many ways: as images, as models, as evidence to support or rebut a model, and as interpretations of reality.
Case study 2: applying visual reasoning in knowledge production, the example of the lines of magnetic force
The focus of this case study is on Faraday’s use of the lines of magnetic force. Faraday is known of his exploratory, creative, and yet systemic way of experimenting, and the visual reasoning leading to theoretical development was an inherent part of this experimentation (Gooding 2006 ). Faraday’s articles or notebooks do not include mathematical formulations; instead, they include images and illustrations from experimental devices and setups to the recapping of his theoretical ideas (Nersessian 2008 ). According to Gooding ( 2006 ), “Faraday’s visual method was designed not to copy apparent features of the world, but to analyse and replicate them” (2006, p. 46).
The lines of force played a central role in Faraday’s research on electricity and magnetism and in the development of his “field theory” (Faraday 1852a ; Nersessian 1984 ). Before Faraday, the experiments with iron filings around magnets were known and the term “magnetic curves” was used for the iron filing patterns and also for the geometrical constructs derived from the mathematical theory of magnetism (Gooding et al. 1993 ). However, Faraday used the lines of force for explaining his experimental observations and in constructing the theory of forces in magnetism and electricity. Examples of Faraday’s different illustrations of lines of magnetic force are given in Fig. 2 . Faraday gave the following experiment-based definition for the lines of magnetic forces:
a Iron filing pattern in case of bar magnet drawn by Faraday (Faraday 1852b , Plate IX, p. 158, Fig. 1), b Faraday’s drawing of lines of magnetic force in case of cylinder magnet, where the experimental procedure, knife blade showing the direction of lines, is combined into drawing (Faraday, 1855, vol. 1, plate 1)
A line of magnetic force may be defined as that line which is described by a very small magnetic needle, when it is so moved in either direction correspondent to its length, that the needle is constantly a tangent to the line of motion; or it is that line along which, if a transverse wire be moved in either direction, there is no tendency to the formation of any current in the wire, whilst if moved in any other direction there is such a tendency; or it is that line which coincides with the direction of the magnecrystallic axis of a crystal of bismuth, which is carried in either direction along it. The direction of these lines about and amongst magnets and electric currents, is easily represented and understood, in a general manner, by the ordinary use of iron filings. (Faraday 1852a , p. 25 (3071))
The definition describes the connection between the experiments and the visual representation of the results. Initially, the lines of force were just geometric representations, but later, Faraday treated them as physical objects (Nersessian 1984 ; Pocovi and Finlay 2002 ):
I have sometimes used the term lines of force so vaguely, as to leave the reader doubtful whether I intended it as a merely representative idea of the forces, or as the description of the path along which the power was continuously exerted. … wherever the expression line of force is taken simply to represent the disposition of forces, it shall have the fullness of that meaning; but that wherever it may seem to represent the idea of the physical mode of transmission of the force, it expresses in that respect the opinion to which I incline at present. The opinion may be erroneous, and yet all that relates or refers to the disposition of the force will remain the same. (Faraday, 1852a , p. 55-56 (3075))
He also felt that the lines of force had greater explanatory power than the dominant theory of action-at-a-distance:
Now it appears to me that these lines may be employed with great advantage to represent nature, condition, direction and comparative amount of the magnetic forces; and that in many cases they have, to the physical reasoned at least, a superiority over that method which represents the forces as concentrated in centres of action… (Faraday, 1852a , p. 26 (3074))
For giving some insight to Faraday’s visual reasoning as an epistemic practice, the following examples of Faraday’s studies of the lines of magnetic force (Faraday 1852a , 1852b ) are presented:
(a) Asking questions and defining problems: The iron filing patterns formed the empirical basis for the visual model: 2D visualization of lines of magnetic force as presented in Fig. 2 . According to Faraday, these iron filing patterns were suitable for illustrating the direction and form of the magnetic lines of force (emphasis added):
It must be well understood that these forms give no indication by their appearance of the relative strength of the magnetic force at different places, inasmuch as the appearance of the lines depends greatly upon the quantity of filings and the amount of tapping; but the direction and forms of these lines are well given, and these indicate, in a considerable degree, the direction in which the forces increase and diminish . (Faraday 1852b , p.158 (3237))
Despite being static and two dimensional on paper, the lines of magnetic force were dynamical (Nersessian 1992 , 2008 ) and three dimensional for Faraday (see Fig. 2 b). For instance, Faraday described the lines of force “expanding”, “bending,” and “being cut” (Nersessian 1992 ). In Fig. 2 b, Faraday has summarized his experiment (bar magnet and knife blade) and its results (lines of force) in one picture.
(b) Analyzing and interpreting data: The model was so powerful for Faraday that he ended up thinking them as physical objects (e.g., Nersessian 1984 ), i.e., making interpretations of the way forces act. Of course, he made a lot of experiments for showing the physical existence of the lines of force, but he did not succeed in it (Nersessian 1984 ). The following quote illuminates Faraday’s use of the lines of force in different situations:
The study of these lines has, at different times, been greatly influential in leading me to various results, which I think prove their utility as well as fertility. Thus, the law of magneto-electric induction; the earth’s inductive action; the relation of magnetism and light; diamagnetic action and its law, and magnetocrystallic action, are the cases of this kind… (Faraday 1852a , p. 55 (3174))
(c) Experimenting: In Faraday's case, he used a lot of exploratory experiments; in case of lines of magnetic force, he used, e.g., iron filings, magnetic needles, or current carrying wires (see the quote above). The magnetic field is not directly observable and the representation of lines of force was a visual model, which includes the direction, form, and magnitude of field.
(d) Modeling: There is no denying that the lines of magnetic force are visual by nature. Faraday’s views of lines of force developed gradually during the years, and he applied and developed them in different contexts such as electromagnetic, electrostatic, and magnetic induction (Nersessian 1984 ). An example of Faraday’s explanation of the effect of the wire b’s position to experiment is given in Fig. 3 . In Fig. 3 , few magnetic lines of force are drawn, and in the quote below, Faraday is explaining the effect using these magnetic lines of force (emphasis added):
Picture of an experiment with different arrangements of wires ( a , b’ , b” ), magnet, and galvanometer. Note the lines of force drawn around the magnet. (Faraday 1852a , p. 34)
It will be evident by inspection of Fig. 3 , that, however the wires are carried away, the general result will, according to the assumed principles of action, be the same; for if a be the axial wire, and b’, b”, b”’ the equatorial wire, represented in three different positions, whatever magnetic lines of force pass across the latter wire in one position, will also pass it in the other, or in any other position which can be given to it. The distance of the wire at the place of intersection with the lines of force, has been shown, by the experiments (3093.), to be unimportant. (Faraday 1852a , p. 34 (3099))
In summary, by examining the history of Faraday’s use of lines of force, we showed how visual imagery and reasoning played an important part in Faraday’s construction and representation of his “field theory”. As Gooding has stated, “many of Faraday’s sketches are far more that depictions of observation, they are tools for reasoning with and about phenomena” (2006, p. 59).
Case study 3: visualizing scientific methods, the case of a journal
The focus of the third case study is the Journal of Visualized Experiments (JoVE) , a peer-reviewed publication indexed in PubMed. The journal devoted to the publication of biological, medical, chemical, and physical research in a video format. The journal describes its history as follows:
JoVE was established as a new tool in life science publication and communication, with participation of scientists from leading research institutions. JoVE takes advantage of video technology to capture and transmit the multiple facets and intricacies of life science research. Visualization greatly facilitates the understanding and efficient reproduction of both basic and complex experimental techniques, thereby addressing two of the biggest challenges faced by today's life science research community: i) low transparency and poor reproducibility of biological experiments and ii) time and labor-intensive nature of learning new experimental techniques. ( http://www.jove.com/ )
By examining the journal content, we generate a set of categories that can be considered as indicators of relevance and significance in terms of epistemic practices of science that have relevance for science education. For example, the quote above illustrates how scientists view some norms of scientific practice including the norms of “transparency” and “reproducibility” of experimental methods and results, and how the visual format of the journal facilitates the implementation of these norms. “Reproducibility” can be considered as an epistemic criterion that sits at the heart of what counts as an experimental procedure in science:
Investigating what should be reproducible and by whom leads to different types of experimental reproducibility, which can be observed to play different roles in experimental practice. A successful application of the strategy of reproducing an experiment is an achievement that may depend on certain isiosyncratic aspects of a local situation. Yet a purely local experiment that cannot be carried out by other experimenters and in other experimental contexts will, in the end be unproductive in science. (Sarkar and Pfeifer 2006 , p.270)
We now turn to an article on “Elevated Plus Maze for Mice” that is available for free on the journal website ( http://www.jove.com/video/1088/elevated-plus-maze-for-mice ). The purpose of this experiment was to investigate anxiety levels in mice through behavioral analysis. The journal article consists of a 9-min video accompanied by text. The video illustrates the handling of the mice in soundproof location with less light, worksheets with characteristics of mice, computer software, apparatus, resources, setting up the computer software, and the video recording of mouse behavior on the computer. The authors describe the apparatus that is used in the experiment and state how procedural differences exist between research groups that lead to difficulties in the interpretation of results:
The apparatus consists of open arms and closed arms, crossed in the middle perpendicularly to each other, and a center area. Mice are given access to all of the arms and are allowed to move freely between them. The number of entries into the open arms and the time spent in the open arms are used as indices of open space-induced anxiety in mice. Unfortunately, the procedural differences that exist between laboratories make it difficult to duplicate and compare results among laboratories.
The authors’ emphasis on the particularity of procedural context echoes in the observations of some philosophers of science:
It is not just the knowledge of experimental objects and phenomena but also their actual existence and occurrence that prove to be dependent on specific, productive interventions by the experimenters” (Sarkar and Pfeifer 2006 , pp. 270-271)
The inclusion of a video of the experimental procedure specifies what the apparatus looks like (Fig. 4 ) and how the behavior of the mice is captured through video recording that feeds into a computer (Fig. 5 ). Subsequently, a computer software which captures different variables such as the distance traveled, the number of entries, and the time spent on each arm of the apparatus. Here, there is visual information at different levels of representation ranging from reconfiguration of raw video data to representations that analyze the data around the variables in question (Fig. 6 ). The practice of levels of visual representations is not particular to the biological sciences. For instance, they are commonplace in nanotechnological practices:
Visual illustration of apparatus
Video processing of experimental set-up
Computer software for video input and variable recording
In the visualization processes, instruments are needed that can register the nanoscale and provide raw data, which needs to be transformed into images. Some Imaging Techniques have software incorporated already where this transformation automatically takes place, providing raw images. Raw data must be translated through the use of Graphic Software and software is also used for the further manipulation of images to highlight what is of interest to capture the (inferred) phenomena -- and to capture the reader. There are two levels of choice: Scientists have to choose which imaging technique and embedded software to use for the job at hand, and they will then have to follow the structure of the software. Within such software, there are explicit choices for the scientists, e.g. about colour coding, and ways of sharpening images. (Ruivenkamp and Rip 2010 , pp.14–15)
On the text that accompanies the video, the authors highlight the role of visualization in their experiment:
Visualization of the protocol will promote better understanding of the details of the entire experimental procedure, allowing for standardization of the protocols used in different laboratories and comparisons of the behavioral phenotypes of various strains of mutant mice assessed using this test.
The software that takes the video data and transforms it into various representations allows the researchers to collect data on mouse behavior more reliably. For instance, the distance traveled across the arms of the apparatus or the time spent on each arm would have been difficult to observe and record precisely. A further aspect to note is how the visualization of the experiment facilitates control of bias. The authors illustrate how the olfactory bias between experimental procedures carried on mice in sequence is avoided by cleaning the equipment.
Our discussion highlights the role of visualization in science, particularly with respect to presenting visualization as part of the scientific practices. We have used case studies from the history of science highlighting a scientist’s account of how visualization played a role in the discovery of DNA and the magnetic field and from a contemporary illustration of a science journal’s practices in incorporating visualization as a way to communicate new findings and methodologies. Our implicit aim in drawing from these case studies was the need to align science education with scientific practices, particularly in terms of how visual representations, stable or dynamic, can engage students in the processes of science and not only to be used as tools for cognitive development in science. Our approach was guided by the notion of “knowledge-as-practice” as advanced by Knorr Cetina ( 1999 ) who studied scientists and characterized their knowledge as practice, a characterization which shifts focus away from ideas inside scientists’ minds to practices that are cultural and deeply contextualized within fields of science. She suggests that people working together can be examined as epistemic cultures whose collective knowledge exists as practice.
It is important to stress, however, that visual representations are not used in isolation, but are supported by other types of evidence as well, or other theories (i.e., in order to understand the helical form of DNA, or the structure, chemistry knowledge was needed). More importantly, this finding can also have implications when teaching science as argument (e.g., Erduran and Jimenez-Aleixandre 2008 ), since the verbal evidence used in the science classroom to maintain an argument could be supported by visual evidence (either a model, representation, image, graph, etc.). For example, in a group of students discussing the outcomes of an introduced species in an ecosystem, pictures of the species and the ecosystem over time, and videos showing the changes in the ecosystem, and the special characteristics of the different species could serve as visual evidence to help the students support their arguments (Evagorou et al. 2012 ). Therefore, an important implication for the teaching of science is the use of visual representations as evidence in the science curriculum as part of knowledge production. Even though studies in the area of science education have focused on the use of models and modeling as a way to support students in the learning of science (Dori et al. 2003 ; Lehrer and Schauble 2012 ; Mendonça and Justi 2013 ; Papaevripidou et al. 2007 ) or on the use of images (i.e., Korfiatis et al. 2003 ), with the term using visuals as evidence, we refer to the collection of all forms of visuals and the processes involved.
Another aspect that was identified through the case studies is that of the visual reasoning (an integral part of Faraday’s investigations). Both the verbalization and visualization were part of the process of generating new knowledge (Gooding 2006 ). Even today, most of the textbooks use the lines of force (or just field lines) as a geometrical representation of field, and the number of field lines is connected to the quantity of flux. Often, the textbooks use the same kind of visual imagery than in what is used by scientists. However, when using images, only certain aspects or features of the phenomena or data are captured or highlighted, and often in tacit ways. Especially in textbooks, the process of producing the image is not presented and instead only the product—image—is left. This could easily lead to an idea of images (i.e., photos, graphs, visual model) being just representations of knowledge and, in the worse case, misinterpreted representations of knowledge as the results of Pocovi and Finlay ( 2002 ) in case of electric field lines show. In order to avoid this, the teachers should be able to explain how the images are produced (what features of phenomena or data the images captures, on what ground the features are chosen to that image, and what features are omitted); in this way, the role of visualization in knowledge production can be made “visible” to students by engaging them in the process of visualization.
The implication of these norms for science teaching and learning is numerous. The classroom contexts can model the generation, sharing and evaluation of evidence, and experimental procedures carried out by students, thereby promoting not only some contemporary cultural norms in scientific practice but also enabling the learning of criteria, standards, and heuristics that scientists use in making decisions on scientific methods. As we have demonstrated with the three case studies, visual representations are part of the process of knowledge growth and communication in science, as demonstrated with two examples from the history of science and an example from current scientific practices. Additionally, visual information, especially with the use of technology is a part of students’ everyday lives. Therefore, we suggest making use of students’ knowledge and technological skills (i.e., how to produce their own videos showing their experimental method or how to identify or provide appropriate visual evidence for a given topic), in order to teach them the aspects of the nature of science that are often neglected both in the history of science and the design of curriculum. Specifically, what we suggest in this paper is that students should actively engage in visualization processes in order to appreciate the diverse nature of doing science and engage in authentic scientific practices.
However, as a word of caution, we need to distinguish the products and processes involved in visualization practices in science:
If one considers scientific representations and the ways in which they can foster or thwart our understanding, it is clear that a mere object approach, which would devote all attention to the representation as a free-standing product of scientific labor, is inadequate. What is needed is a process approach: each visual representation should be linked with its context of production (Pauwels 2006 , p.21).
The aforementioned suggests that the emphasis in visualization should shift from cognitive understanding—using the products of science to understand the content—to engaging in the processes of visualization. Therefore, an implication for the teaching of science includes designing curriculum materials and learning environments that create a social and epistemic context and invite students to engage in the practice of visualization as evidence, reasoning, experimental procedure, or a means of communication (as presented in the three case studies) and reflect on these practices (Ryu et al. 2015 ).
Finally, a question that arises from including visualization in science education, as well as from including scientific practices in science education is whether teachers themselves are prepared to include them as part of their teaching (Bybee 2014 ). Teacher preparation programs and teacher education have been critiqued, studied, and rethought since the time they emerged (Cochran-Smith 2004 ). Despite the years of history in teacher training and teacher education, the debate about initial teacher training and its content still pertains in our community and in policy circles (Cochran-Smith 2004 ; Conway et al. 2009 ). In the last decades, the debate has shifted from a behavioral view of learning and teaching to a learning problem—focusing on that way not only on teachers’ knowledge, skills, and beliefs but also on making the connection of the aforementioned with how and if pupils learn (Cochran-Smith 2004 ). The Science Education in Europe report recommended that “Good quality teachers, with up-to-date knowledge and skills, are the foundation of any system of formal science education” (Osborne and Dillon 2008 , p.9).
However, questions such as what should be the emphasis on pre-service and in-service science teacher training, especially with the new emphasis on scientific practices, still remain unanswered. As Bybee ( 2014 ) argues, starting from the new emphasis on scientific practices in the NGSS, we should consider teacher preparation programs “that would provide undergraduates opportunities to learn the science content and practices in contexts that would be aligned with their future work as teachers” (p.218). Therefore, engaging pre- and in-service teachers in visualization as a scientific practice should be one of the purposes of teacher preparation programs.
Achieve. (2013). The next generation science standards (pp. 1–3). Retrieved from http://www.nextgenscience.org/ .
Google Scholar
Barber, J, Pearson, D, & Cervetti, G. (2006). Seeds of science/roots of reading . California: The Regents of the University of California.
Bungum, B. (2008). Images of physics: an explorative study of the changing character of visual images in Norwegian physics textbooks. NorDiNa, 4 (2), 132–141.
Bybee, RW. (2014). NGSS and the next generation of science teachers. Journal of Science Teacher Education, 25 (2), 211–221. doi: 10.1007/s10972-014-9381-4 .
Article Google Scholar
Chambers, D. (1983). Stereotypic images of the scientist: the draw-a-scientist test. Science Education, 67 (2), 255–265.
Cochran-Smith, M. (2004). The problem of teacher education. Journal of Teacher Education, 55 (4), 295–299. doi: 10.1177/0022487104268057 .
Conway, PF, Murphy, R, & Rath, A. (2009). Learning to teach and its implications for the continuum of teacher education: a nine-country cross-national study .
Crick, F. (1988). What a mad pursuit . USA: Basic Books.
Dimopoulos, K, Koulaidis, V, & Sklaveniti, S. (2003). Towards an analysis of visual images in school science textbooks and press articles about science and technology. Research in Science Education, 33 , 189–216.
Dori, YJ, Tal, RT, & Tsaushu, M. (2003). Teaching biotechnology through case studies—can we improve higher order thinking skills of nonscience majors? Science Education, 87 (6), 767–793. doi: 10.1002/sce.10081 .
Duschl, RA, & Bybee, RW. (2014). Planning and carrying out investigations: an entry to learning and to teacher professional development around NGSS science and engineering practices. International Journal of STEM Education, 1 (1), 12. doi: 10.1186/s40594-014-0012-6 .
Duschl, R., Schweingruber, H. A., & Shouse, A. (2008). Taking science to school . Washington DC: National Academies Press.
Erduran, S, & Jimenez-Aleixandre, MP (Eds.). (2008). Argumentation in science education: perspectives from classroom-based research . Dordrecht: Springer.
Eurydice. (2012). Developing key competencies at school in Europe: challenges and opportunities for policy – 2011/12 (pp. 1–72).
Evagorou, M, Jimenez-Aleixandre, MP, & Osborne, J. (2012). “Should we kill the grey squirrels?” A study exploring students’ justifications and decision-making. International Journal of Science Education, 34 (3), 401–428. doi: 10.1080/09500693.2011.619211 .
Faraday, M. (1852a). Experimental researches in electricity. – Twenty-eighth series. Philosophical Transactions of the Royal Society of London, 142 , 25–56.
Faraday, M. (1852b). Experimental researches in electricity. – Twenty-ninth series. Philosophical Transactions of the Royal Society of London, 142 , 137–159.
Gilbert, JK. (2010). The role of visual representations in the learning and teaching of science: an introduction (pp. 1–19).
Gilbert, J., Reiner, M. & Nakhleh, M. (2008). Visualization: theory and practice in science education . Dordrecht, The Netherlands: Springer.
Gooding, D. (2006). From phenomenology to field theory: Faraday’s visual reasoning. Perspectives on Science, 14 (1), 40–65.
Gooding, D, Pinch, T, & Schaffer, S (Eds.). (1993). The uses of experiment: studies in the natural sciences . Cambridge: Cambridge University Press.
Hogan, K, & Maglienti, M. (2001). Comparing the epistemological underpinnings of students’ and scientists’ reasoning about conclusions. Journal of Research in Science Teaching, 38 (6), 663–687.
Knorr Cetina, K. (1999). Epistemic cultures: how the sciences make knowledge . Cambridge: Harvard University Press.
Korfiatis, KJ, Stamou, AG, & Paraskevopoulos, S. (2003). Images of nature in Greek primary school textbooks. Science Education, 88 (1), 72–89. doi: 10.1002/sce.10133 .
Latour, B. (2011). Visualisation and cognition: drawing things together (pp. 1–32).
Latour, B, & Woolgar, S. (1979). Laboratory life: the construction of scientific facts . Princeton: Princeton University Press.
Lehrer, R, & Schauble, L. (2012). Seeding evolutionary thinking by engaging children in modeling its foundations. Science Education, 96 (4), 701–724. doi: 10.1002/sce.20475 .
Longino, H. E. (2002). The fate of knowledge . Princeton: Princeton University Press.
Lynch, M. (2006). The production of scientific images: vision and re-vision in the history, philosophy, and sociology of science. In L Pauwels (Ed.), Visual cultures of science: rethinking representational practices in knowledge building and science communication (pp. 26–40). Lebanon, NH: Darthmouth College Press.
Lynch, M. & S. Y. Edgerton Jr. (1988). ‘Aesthetic and digital image processing representational craft in contemporary astronomy’, in G. Fyfe & J. Law (eds), Picturing Power; Visual Depictions and Social Relations (London, Routledge): 184 – 220.
Mendonça, PCC, & Justi, R. (2013). An instrument for analyzing arguments produced in modeling-based chemistry lessons. Journal of Research in Science Teaching, 51 (2), 192–218. doi: 10.1002/tea.21133 .
National Research Council (2000). Inquiry and the national science education standards . Washington DC: National Academies Press.
National Research Council (2012). A framework for K-12 science education . Washington DC: National Academies Press.
Nersessian, NJ. (1984). Faraday to Einstein: constructing meaning in scientific theories . Dordrecht: Martinus Nijhoff Publishers.
Book Google Scholar
Nersessian, NJ. (1992). How do scientists think? Capturing the dynamics of conceptual change in science. In RN Giere (Ed.), Cognitive Models of Science (pp. 3–45). Minneapolis: University of Minnesota Press.
Nersessian, NJ. (2008). Creating scientific concepts . Cambridge: The MIT Press.
Osborne, J. (2014). Teaching scientific practices: meeting the challenge of change. Journal of Science Teacher Education, 25 (2), 177–196. doi: 10.1007/s10972-014-9384-1 .
Osborne, J. & Dillon, J. (2008). Science education in Europe: critical reflections . London: Nuffield Foundation.
Papaevripidou, M, Constantinou, CP, & Zacharia, ZC. (2007). Modeling complex marine ecosystems: an investigation of two teaching approaches with fifth graders. Journal of Computer Assisted Learning, 23 (2), 145–157. doi: 10.1111/j.1365-2729.2006.00217.x .
Pauwels, L. (2006). A theoretical framework for assessing visual representational practices in knowledge building and science communications. In L Pauwels (Ed.), Visual cultures of science: rethinking representational practices in knowledge building and science communication (pp. 1–25). Lebanon, NH: Darthmouth College Press.
Philips, L., Norris, S. & McNab, J. (2010). Visualization in mathematics, reading and science education . Dordrecht, The Netherlands: Springer.
Pocovi, MC, & Finlay, F. (2002). Lines of force: Faraday’s and students’ views. Science & Education, 11 , 459–474.
Richards, A. (2003). Argument and authority in the visual representations of science. Technical Communication Quarterly, 12 (2), 183–206. doi: 10.1207/s15427625tcq1202_3 .
Rothbart, D. (1997). Explaining the growth of scientific knowledge: metaphors, models and meaning . Lewiston, NY: Mellen Press.
Ruivenkamp, M, & Rip, A. (2010). Visualizing the invisible nanoscale study: visualization practices in nanotechnology community of practice. Science Studies, 23 (1), 3–36.
Ryu, S, Han, Y, & Paik, S-H. (2015). Understanding co-development of conceptual and epistemic understanding through modeling practices with mobile internet. Journal of Science Education and Technology, 24 (2-3), 330–355. doi: 10.1007/s10956-014-9545-1 .
Sarkar, S, & Pfeifer, J. (2006). The philosophy of science, chapter on experimentation (Vol. 1, A-M). New York: Taylor & Francis.
Schwartz, RS, Lederman, NG, & Abd-el-Khalick, F. (2012). A series of misrepresentations: a response to Allchin’s whole approach to assessing nature of science understandings. Science Education, 96 (4), 685–692. doi: 10.1002/sce.21013 .
Schwarz, CV, Reiser, BJ, Davis, EA, Kenyon, L, Achér, A, Fortus, D, et al. (2009). Developing a learning progression for scientific modeling: making scientific modeling accessible and meaningful for learners. Journal of Research in Science Teaching, 46 (6), 632–654. doi: 10.1002/tea.20311 .
Watson, J. (1968). The Double Helix: a personal account of the discovery of the structure of DNA . New York: Scribner.
Watson, J, & Berry, A. (2004). DNA: the secret of life . New York: Alfred A. Knopf.
Wickman, PO. (2004). The practical epistemologies of the classroom: a study of laboratory work. Science Education, 88 , 325–344.
Wu, HK, & Shah, P. (2004). Exploring visuospatial thinking in chemistry learning. Science Education, 88 (3), 465–492. doi: 10.1002/sce.10126 .
Download references
Acknowledgements
The authors would like to acknowledge all reviewers for their valuable comments that have helped us improve the manuscript.
Author information
Authors and affiliations.
University of Nicosia, 46, Makedonitissa Avenue, Egkomi, 1700, Nicosia, Cyprus
Maria Evagorou
University of Limerick, Limerick, Ireland
Sibel Erduran
University of Tampere, Tampere, Finland
Terhi Mäntylä
You can also search for this author in PubMed Google Scholar
Corresponding author
Correspondence to Maria Evagorou .
Additional information
Competing interests.
The authors declare that they have no competing interests.
Authors’ contributions
ME carried out the introductory literature review, the analysis of the first case study, and drafted the manuscript. SE carried out the analysis of the third case study and contributed towards the “Conclusions” section of the manuscript. TM carried out the second case study. All authors read and approved the final manuscript.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License ( https://creativecommons.org/licenses/by/4.0 ), which permits use, duplication, adaptation, distribution, and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
Reprints and permissions
About this article
Cite this article.
Evagorou, M., Erduran, S. & Mäntylä, T. The role of visual representations in scientific practices: from conceptual understanding and knowledge generation to ‘seeing’ how science works. IJ STEM Ed 2 , 11 (2015). https://doi.org/10.1186/s40594-015-0024-x
Download citation
Received : 29 September 2014
Accepted : 16 May 2015
Published : 19 July 2015
DOI : https://doi.org/10.1186/s40594-015-0024-x
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Visual representations
- Epistemic practices
- Science learning
- Search Menu
- Sign in through your institution
- Advance articles
- Author Guidelines
- Submission Site
- Open Access
- Why publish with this journal?
- About Bioinformatics
- Journals Career Network
- Editorial Board
- Advertising and Corporate Services
- Self-Archiving Policy
- Dispatch Dates
- Journals on Oxford Academic
- Books on Oxford Academic
Article Contents
1 introduction, 2 tool survey, 3 feature survey, 4 discussion, acknowledgement, tools for visually exploring biological networks.
Associate Editor: Jonathan Wren
- Article contents
- Figures & tables
- Supplementary Data
Matthew Suderman, Michael Hallett, Tools for visually exploring biological networks, Bioinformatics , Volume 23, Issue 20, October 2007, Pages 2651–2659, https://doi.org/10.1093/bioinformatics/btm401
- Permissions Icon Permissions
Many tools exist for visually exploring biological networks including well-known examples such as Cytoscape, VisANT, Pathway Studio and Patika. These systems play a key role in the development of integrative biology, systems biology and integrative bioinformatics. The trend in the development of these tools is to go beyond ‘static’ representations of cellular state, towards a more dynamic model of cellular processes through the incorporation of gene expression data, subcellular localization information and time-dependent behavior. We provide a comprehensive review of the relative advantages and disadvantages of existing systems with two goals in mind: to aid researchers in efficiently identifying the appropriate existing tools for data visualization; to describe the necessary and realistic goals for the next generation of visualization tools. In view of the first goal, we provide in the Supplementary Material a systematic comparison of more than 35 existing tools in terms of over 25 different features.
Contact: [email protected]
Supplementary information: Supplementary data are available at Bioinformatics online.
Networks are used ubiquitously throughout biology to represent the relationships between genes and gene products. Perhaps due to their simple discrete nature and to their amenability to visual representation, biological networks allow for the most salient properties of complex systems to be highlighted in a succinct and powerful manner. Visualization of biological networks range from small-scale descriptions of specific metabolic, regulatory or signalling pathways appearing in countless journal articles, seminars and courses, to classic efforts such as the Roche Applied Science Biochemical Pathways (Michal, 1993 ) and the Biochemical Pathway Atlas (Michal, 1998 ) that attempt to provide broad maps of cellular organization. Whereas these latter efforts required extensive manual curation to iteratively refine the maps as new information became available, the advent of high-throughput genetic-based assays that permit (near) cell-wide exploration of relationships between genes and gene products has motivated the development of software systems capable of constructing visualizations of these datasets ‘on the fly’. Assays such as genetic-based protein–protein interaction screens [e.g. (Fields and kyu Song, 1989 ; Rigaut et al. , 1999 ; Selbach and Mann, 2006 )], RNAi-based genetic interaction screens (Echeverri and Perrimon, 2006 ; Fire et al. , 1998 ) gene expression (Lockhart et al. , 1996 ) and ChIP-Chip arrays (Blat and Kleckner, 1999 ; Ren et al. , 2000 ) are readily visualized when nodes represent genes, gene products or small molecules and, links (edges) between pairs of nodes can model metabolic events, protein–protein/protein–nucleotide interactions, regulatory relationships or signaling pathways.
Such simple networks, that capture a static ‘snapshot’ of cellular state, have been shown capable of yielding insight into the underlying biology. Network analysis, e.g. has shown that many types of biological networks across a broad spectrum of organisms share important graph-theoretic properties. These properties suggest testable biological hypotheses that may in turn reveal the fundamental design principles of biological systems. For example, there is evidence that many types of biological networks are scale-free (Barabási and Oltvai, 2004), containing a few highly connected nodes (‘hubs’) and a majority of nodes linked to only a few neighbors (Barabási and Albert, 1999 ). The fundamental hypothesis here is that scale-free networks are ‘robust’ to random node removals, since a majority of the nodes are not essential to cellular function and simultaneously ‘fragile’ because targeted removal of a few hubs will disrupt cellular function. This ‘robust yet fragile’ network property is thought by some to be responsible for the ability of biological systems to survive a wide variety of attacks while being susceptible to a few specific targeted attacks (Albert et al. , 2000 ).
Network models have facilitated a shift from the study of evolutionary conservation between individual gene and gene products towards the study of conservation at the level of pathways and complexes. In particular, Kelley et al. ( 2003 ) show that the protein–protein interaction networks of two distantly related species, Saccharomyces cerevisiae and Helicobacter pylori , contain many of the same or very similar pathways. In many of these conserved pathways, the S.cerevisiae network appears to be more specialized. Resultant tools such as PathBLAST (Kelley et al. , 2004 ) are now commonly used to automate the discovery of similarities (‘alignments’) within and between protein–protein interaction networks.
These and other successes indicate that network modeling is a promising approach, however, visualization in this domain remains a difficult challenge for several reasons. First, the increasing number of interactions available for even small genomes/proteomes presents significant visualization challenges. Most available biological network tools make use of generic visualization packages (and algorithms) from the graph drawing community to address problems of scalability. However, such generic layouts often produce network drawings that resemble the infamous ‘hair ball’, since the layout optimization criteria fail to take into consideration important domain-specific knowledge such as subcellular localizations, molecular functions, protein complexes or pathways (see Fig. 1 ). Second, we lack robust query languages embedded directly within the visualization software. These querying languages allow users to filter cell-wide networks, thus restricting their attention to a core set of nodes of particular interest and to organize this information in an intuitive manner. Third, the increasing number of types of interactions raises many practical and theoretical problems related to how these datasets should be integrated into network models, how this information is best visualized and how the network can be explored. For instance, consider the visualization of a biological network containing both physical and genetic interactions. The fact that protein–protein interactions occur between molecules that are at least transiently physically co-located in the cell may suggest an approach that seeks to generate a visualization that mimics cellular or organellar location. However, such an approach may be unsuitable for genetic interactions where the interacting proteins may be implicated in two distinct, distally located pathways. Other types of biological interactions are not necessarily well modeled by simple binary, pairwise graphs (i.e. 2-hypergraphs). Consider, e.g. expressing that a certain ternary complex forms if and only if each of the three components of the complex are present. For practical reasons, including simplicity and computational tractability, most network models cannot handle such n -ary relations. Consequently, general methods of extending models such as by using n -ary relations are not practical. Extensions based on domain-specific knowledge is required. Finally, there is a general need for the biological network tools to incorporate more dynamic information within the network. A first step in this direction is the inclusion of gene expression data within several existing visualization tools. The mapping of gene expression (represented typically by color) onto network nodes can be viewed as our first glimpse of large-scale systems-wide dynamics in biological systems. However, current functionality is not capable of addressing the computational and statistical challenges that will arise as researchers begin to construct large-scale probabilistic and dynamic models of complicated, distributed biological processes, nor is it capable of addressing how dynamic models may be visually represented.

Protein–protein interaction map showing predicted subcellularlocalizations of all proteins in the secretory pathway (Scott et al. , 2005a ). The drawing was created using the Cytoscape plugin Cerebral (Barsky et al. , 2007 ). Subcellular compartments are ordered so that compartments with the most inter-compartmental links are next to one another.
The purpose of this review is 2-fold. First, it is designed to provide readers with an overview of existing software systems for biological network visualization ( Section 2 ). The existence of at least 35 such systems precludes an in-depth review of each, instead we have sought to single out systems that contain unique functionality that have broad applicability in many different studies. Furthermore, we discuss some of the most important features currently available in network visualization packages and summarize the tools that support them ( Section 3 ). Second, we discuss the future of network visualization and how current tools can be improved to satisfy future demands ( Section 4 ).
There are many network visualization tools and each meets some specific visualization need. In fact, during the preparation of this manuscript, we identified no less than 35 such tools, and the number continues to grow. A similar review (Saraiya et al. , 2005 ) published only a few years ago identifies barely half that number and, due to recent progress in this area, is silent on some of what are currently the most important visualization challenges. We focus here on six tools which together contain many of the important features necessary for network visualization in a broad range of studies. A more complete list can be found in the Supplementary Material.
Pathway Studio (Nikitin et al. , 2003 ) (formerly PathwayAssist) is a Microsoft Windows application that combines an extensive set of features with a polished graphic user interface. It includes, most notably, customizable network display styles for assigning visual attributes such as node color, size and shape, multi-user support, subcellular localization visualization and tight integration with several database including R es N et ( http://www.ariadnegenomics.com/ ), KEGG (Kanehisa, 2002 ), BIND (Alfarano et al. , 2005 ), GO (Ashburner et al. , 2000 ), DIP (Xenarios et al. , 2000 ), ERGO ( http://www.integratedgenomics.com/ ) and PathArt ( http://jubilantbiosys.com/ ). Importantly, this package also contains an SQL-like language that allows users to query the network using some simple topological contractions with node and link attributes. In comparison to other packages, this query language provides a much more flexible tool for quickly filtering nodes of interest from large networks. Pathway Studio is a commercial product although academic licenses are available.
Cytoscape (Shannon et al. , 2003 ) is a well-known network visualization tool supporting a core set of features including standard and customizable network display styles, ability to import a large variety of interaction files, and zoomable network views. Release 2.3 claims a high-performance rendering engine supporting networks with as many as 100 K nodes and edges. One of the most positive aspects of Cytoscape is the large user and developer base. In particular, since Cytoscape is a Java application whose source code is released under the Lesser General Public License (LGPL), it is straightforward for third-party developers to construct new plugins for the system. In fact, more than 40 plugins are currently available available for tasks including importing networks from various data formats, analyzing networks and generating networks from literature searches.
Osprey (Breitkreutz et al. , 2003 ) was one of the first tools specifically designed to visualize and analyze large networks. Consequently, all but one network layout in Osprey are elaborations of the circular layout (described subsequently) because they can be quickly computed. Other tools that specialize in large networks include Interviewer (Han et al. , 2004 ) and ProViz (Iragne et al. , 2005 ). Osprey was also one of the first tools to support functional comparisons between different networks. In particular, Osprey is able to superimpose one network additively on top of another in order to show similarities and differences. Osprey is a Java application and can be used free of charge.
The PATIKA Project (Demir et al. , 2002 ) provides a WWW-based visual editor, PATIKAweb, for accessing a central database containing pathway data from several sources including Reactome ( http://www.reactome.org/ ). The project is built on top of an extensive ontology supporting representation of biological objects at different levels of detail, and graph types that facilitate visualizations of molecular complexes, pathways and black-box reactions. PATIKAweb is capable of producing high-quality visualizations using the Tom Sawyer Visualization ( http://www.tomsawyer.com/ ) software. It also supports SQL-like queries on node and edge properties. It is implemented using Java Server Pages and is publicly available for non-profit use.
VisANT (Hu et al. , 2005 ) not only provides network drawing capabilities, including support for very large networks, but it is one of the first such packages to support creation, visualization and analysis of mixed networks, i.e. networks containing both directed and undirected links. The ability to use nodes to model more complex entities such as protein complexes or pathways allow for more informative visualizations. VisANT implements algorithms for analyzing node degrees, clusters, path lengths, network motifs and network randomizations. Like Cytoscape, VisANT is a Java application that can be extended using plugins and is freely available.
ProViz (Iragne et al. , 2005 ) leverages the power of the graph drawing package Tulip (David, 2001 ) for handling graphs containing millions of nodes and edges, while maintaining a guaranteed response time (i.e. ProViz will never make the user wait longer that predefined length of time). Tulip supports of a sophisticated plugin architecture allowing third-party developers to extend the system. Many plugins are currently available and shipped with the system including plugins for importing/exporting various network file formats, obtaining many different two and three-dimensional network layouts, computing various network metrics like connectivity and eccentricity, and selecting subgraphs such as spanning subtrees and paths between nodes. ProViz adds to these features the ability to import and export PSI-MI (Hermjakob et al. , 2004b ) and IntAct (Hermjakob et al. , 2004a ) data formats, and interfaces for exploring the GO (Ashburner et al. , 2000 ) and IntAct controlled vocabularies. There is functionality to define filters for large networks. Both ProViz and Tulip are implemented in C++ and can be installed in Windows, Linux or MacOSX. The source code is released under the GNU General Public License (GPL).
BiologicalNetworks/PathSys (Baitaluk et al. , 2006a ) extends Cytoscape and includes much of the functionality of Pathway Studio. In fact, Pathway Studio users will recognize the dialog box used to create pathways from a set of interactions. BiologicalNetworks is a user-interface to PathSys (Baitaluk et al. , 2006b ), a graph-based system for creating a combined database of biological pathways, gene regulatory networks and protein interaction maps. It integrates over 14 curated and publicly available data sources including BIND (Alfarano et al. , 2005 ), GO (Ashburner et al. , 2000 ) and KEGG (Kanehisa, 2002 ) for eight representative organisms. PathSys supports SQL-like queries that can explore network properties such connectivity and node degree. BiologicalNetworks improves on previous tools by better integrating expression data with network visualization and analysis. In particular, after importing expression data, users can apply sorting, normalization and clustering algorithms on the data and then create various tables, heat maps and network views of the data. It is implemented using Java Server Pages and is publicly available on the WWW.
The utility of any biological network visualization tool ultimately depends on the supported features. In this section, we consider some of the most important features including available network layout routines, supported graphical notation, integration of analysis into the visualization, variety of user input methods, integration of external biological data sources, integration of third-party software and finally availability (licensing restrictions and platform limitations).
3.1 Network layouts
Clearly, one of the most important aspects of any biological network visualization is its ability to automatically construct network drawings (or, layouts). Most tools automatically create static layouts of networks, using methods that roughly fall into one of the following categories:
3.1.1 Circular
Nearly every tool produces circular layouts. In its simplest form, each node is placed on the circumference of a circle and links are drawn as straight-line segments between them. More complicated versions attempt to order the nodes to uncover network symmetries and other versions place nodes on multiple concentric circles. Osprey (Breitkreutz et al. , 2003 ) implements a total of six different elaborations on the circular layout. The most complex, called ‘Spoked Dual Ring’, creates circular layouts of highly connected parts of the network inside a circle and places the remaining vertices on the circle circumference. The purpose of this layout is to highlight the the highly connected parts of the network and show how they relate to the remainder of the network (e.g. see Fig. S1a–c in the Supplementary Material).
3.1.2 Hierarchical
Directed edges are particularly important when visualizing regulatory networks. One approach to visualizing draws nodes on a series of horizontal lines so that edges are directed from nodes on lower horizontal lines to nodes on higher horizontal lines. This approach, invented by Sugiyama et al. ( 1981 ), is included in several systems including Pathway Studio (Nikitin et al. , 2003 ), BioPath (Schreiber, 2002 ), ROSPath (Paek et al. , 2004 ), CellDesigner (Kitano, 2003 ) and Virtual Cell ( http://www.vcell.org/ ). ProViz (Iragne et al. , 2005 ) also produces hierarchical drawings but uses a different algorithm (Messinger et al. , 1991 ). An example from CellDesigner is shown in Figure S2b.
3.1.3 Force-directed
The drawings are also known as spring embeddings (Eades, 1984 ; Frick et al. , 1994 ; Fruchterman and Reingold, 1991 ; Kamada and Kawai, 1989 ) since edges are modeled as springs that pull linked nodes together, or push unlinked nodes apart, until the layout reaches a state of equillibrium (e.g. see Figure S1a). Force-directed algorithms attempt to place nodes so that all forces are in equillibrium. This approach is quite popular because the algorithms are simple to implement, produce relatively good drawings, and are easy tweak for specific applications. In fact, nearly every network visualization tool implements the version described by Frick, Sander and Wang (Frick et al. , 1999 ).
The main drawback of force-directed algorithms is that they can require a significant amount of time before converging to equilibrium. Fortunately, these algorithms are often easy to visually animate so the user can watch the network model incrementally approach equillibrium and perhaps terminate the algorithm when a good drawing it obtained.
3.1.4 Simulated annealing
As the name suggests, simulated annealing methods model problem space as a set of states each with an associated energy so that low energy states correspond to potential solutions. Simulated annealing algorithms find solutions by traversing the space, moving from one network layout to another until it finds a layout with ‘sufficiently low energy’. GeneWays (Rzhetsky et al. , 2004 ) generalizes one such algorithm by Davidson and Harel (Davidson and Harel, 1996 ) for obtaining 3-dimensional layouts. Grid Layout (Li and Kurata, 2005 ) uses a related method and shows how, for a yeast network, their method seems to spacially cluster functionally related nodes. The main drawback of simulated annealing algorithms is that they tend to be slow (even in comparison with force-directed methods). Consequently, there are practical limits on the size of networks to which they can be applied.
All of the above mentioned layouts assume a graphical network model where interactions are between exactly two interactors. Although these simple models have been shown to yield biological insight, they are incapable of modeling more complicated biological relationships that involve more than two interactors like protein complexes, relationships that depend on external factors like cell state, or regulatory circuits. For instance, although the ‘topology’ of the ci s-regulatory network describing the set of transcription factors relevant for a particular promoter can be well described, the regulatory circuit describing the activation of the target cannot. Unfortunately, extending current visualizations to handle this added complexity is not straight-forward.
Consequently, successful visualization depends on exploiting domain-specific knowledge to reduce difficult general problems to something more manageable. There have been a few successful attempts in this direction:
3.1.5 Subcellular localization
Both Pathway Studio (Nikitin et al. , 2003 ) and Patika (Demir et al. , 2002 ) are capable of using localization to influence network visualizations, if nodes are annotated with subcellular localizations. In particular, both systems partition the drawing space into regions corresponding to the subcellular localizations and then search for layouts where nodes are forcibly constrained to their respective locations. Both systems make use of modified force-directed algorithms to achieve this. Pathway Studio uses representative cartoons as backgrounds for each region in order to improve readability. Two other tools, Cell Illustrator (Nagasaki et al. , 2003 ) and Cerebral (Barsky et al. , 2007 ), support subcellular localization in drawings where nodes are restricted to positions on a grid. For an example, see Figure 1 .
3.1.6 Composite nodes
Pathway Studio (Nikitin et al. , 2003 ), Patika (Demir et al. , 2002 ) and VisANT (Hu et al. , 2005 ) visualize composite nodes representing molecular complexes and pathways as single nodes that can be interactively expanded to show individual members or collapsed. Some improvement to this functionality is warranted. VisANT, e.g. expands composite nodes by creating a small window on top of the current network view and then draws the composite node members inside the window. Unfortunately, these windows cover nearby neighbors making it difficult to see how the composite node members relate to the greater network. When the user attempts to expand a node in Patika (e.g. see Fig. S4), there must be space for the node to be expanded in the drawing, otherwise Patika will not expand the node.
3.1.7 Hierarchical clusters
Hierarchical clusters of the nodes or edges can be very useful for obtaining simplified visualizations of large, complex networks. Schwikowski et al. ( 2000 ) show three views of a yeast protein–protein interaction network: either display all nodes and edges without any graph drawing constraints, collapse nodes of the same function into composite nodes, or collapse proteins of the same subcellular localization into composite nodes. Similar to quotient graphs (e.g. Bourqui et al. , 2006 ), an edge between two composite nodes in the last two views indicates the existence of a threshhold number of edges between composite node members. More general hierarchical clusters of proteins exist, including, e.g. the Structural Classification of Proteins (SCOP) (Murzin et al. , 1995 ) and Gene Ontology (Ashburner et al. , 2000 ). The SCOP hierarchy contains several levels of protein domain similarity differentiated by increasing specificity: class, fold, superfamily, family, protein and species. Lappe et al. ( 2001 ) show that a large yeast protein-protein interaction network at the SCOP level of superfamily has a very simple drawing. Edge thickness between superfamilies indicates the number of links between proteins in each superfamily.
A recent Cytoscape plugin, GenePro (Vlasblom et al. , 2006 ), partially supports interactive exploration of hierarchical network clusters (e.g. see Fig. S5). Given a protein interaction network and a predefined clustering on its nodes, GenePro initially presents the user with the most abstract view and then allows the user to expand clusters in a new window to see cluster members. GenePro can also render nodes as pie charts showing the fractions of proteins sharing a common feature.
3.1.8 Time series
Most tools do not attempt to visualize time series data. Those that do, e.g. BioTapestry (Longabaugh et al. 2005 ), simply highlight links and nodes that are active during a given time period but otherwise present a static picture. Unfortunately, this approach fails to take advantage of the reduced network size at each time point and the small number of changes from one time point to the next.
3.1.9 Neighbor expansion
Rather than show the entire network in one display, many tools including VisANT (Hu et al. , 2005 ), Osprey (Breitkreutz et al. , 2003 ) and MINT Viewer (Zanzoni et al. , 2002 ; Chatr-aryamontri et al. , 2007 ) initially show a small subnetwork and then allow the user to click on a node in order to add all of its neighbors to the current view (e.g. see Fig. S3). Later, the user may click the node again to hide those neighbors. The best tools provide animations from one network view to another so that the user can easily maintain a mental mapping from the previous to the current view.
3.1.10 Three dimensions
Currently, only a few tools including GeneWays (Rzhetsky et al. , 2004 ) and ProViz (Iragne et al. , 2005 ) offer three-dimensional visualizations, and these are all very rudimentary. Making use of higher dimensions is difficult because users are generally viewing the network on a two-dimensional screen. Consequently, dynamic navigation is a necessity and is often awkward without specialized equipment. In addition, graphical complexity increases because network entities must not only represent data but also simulate distance from the user.
3.1.11 Matrix representations
Although networks are typically visualized using the so-called ‘ball-and-stick’ representation, a complementary representation uses adjacency matrices and is often useful for uncovering topological patterns in, e.g. dense networks. In Bioinformatics, these are more commonly called clustergrams, heatmaps with dendrograms, or cluster maps. Here, each node corresponds to exactly one row and one column in the matrix and the intersection of a row and column is colored to represent the existence or strength of the link between the corresponding nodes. Drawing this matrix with different row and column orderings can uncover monochromatic patches indicating clusters of nodes with similar ‘behavior’ (e.g. Fig. 1 of Collins et al. , 2007 ). To create a matrix representation, researchers generally use one or more methods for ‘optimally’ ordering rows and columns provided by their preferred statistical software package. Unfortunately, general-purpose orderings are rarely sufficient for specific biological applications so there has been some recent investigation of support for computing user-assisted orderings (e.g. Henry and Fekete, 2006 ).
3.2 Graphical notation
Until recently, most tools relied heavily on node/edge shape (e.g. to represent whether the node corresponds to a protein, gene or small molecule) and colors (e.g. gene expression, subcellular localization, molecular function) to visualize network properties. Unfortunately, the lack of standards describing typical biological objects results in the need for each visualization to come equipped with a legend describing symbols and colors. In some cases, the use of such node and edge attributes actually detracts from readability. This has motivated the development of informal standards that have evolved via imitation and the popularity of some tools. For example, elements of the Pathway Studio layout style has migrated into the BiologicalNetworks package. However, concern about a lack of formal standards has been increasing (Klipp et al. , 2007 ), and there now exist several efforts towards a fully standardized graphics vocabulary (Cook et al. , 2001 ; Kitano, 2003 ; Kohn, 1999 , 2001 ; Kohn et al. , 2006 ; Kurata et al. , 2005 ; Pirson et al. , 2000 ). Cook et al. ( 2001 ) outline a carefully reasoned approach that can describe complex biological systems unambiguously. The authors hope that such a standard would be amenable to various automations including simulation and resolution of queries like ‘Find models where a potassium channel blocker affects cell-cycle progression’. Unfortunately, according to Kohn et al. ( 2006 ), this proposal is too cumbersome for biologists to actually use so they propose their own molecular interaction map (MIM) notation for regulatory networks that deliberately permits some forms of ambiguity. For example, in the interest of visual compactness, MIM diagrams do not specify an order for steps in a reaction. Kohn et al. ( 2006 ) downplay concerns for automation claiming that the process of manually creating MIM diagrams is in itself and important tool for exposing gaps in our knowledge of biological processes. Others however (Kurata et al. , 2003 , 2005 ) have implemented a tool called CADLIVE that supports a slight modification of MIM. Kitano ( 2003 ) defines an alternative graphical notation called Systems Biology Graphical Notation (SBGN) that uses state-transition diagrams to model regulatory networks. In constrast to MIM diagrams, SBGN diagrams do enforce an event sequence and are consequently less compact because they may contain multiple nodes for a single molecule. In addition, they have implemented a tool called CellDesigner that generates SBGN diagrams (Funahashi et al. , 2003 ) as well as created a website ( http://sbgn.org/ ) in order to promote wider community use and development of SBGN. Figure S2 shows an example from CellDesigner.
3.3 Integration of analysis
There are currently few network tools designed for both the visualization of biological networks and the analysis of these networks. Consequently, users are forced to switch between tools, resulting in the need to continually import/export and reformat data. Clearly, a more optimal choice would be a network tool that supports both visualization and analysis, with a seamless integration between these two procedures. BiologicalNetworks is an early leader here allowing pathway visualizations from queries that combine gene expression data analysis with simple topological patterns. Several specialized tools exist for the integration of very specific types of visualization and data. Wilmascope (Dwyer et al. , 2004 ) shows time series data using ‘2.5 dimensional layouts’ (networks are stacked one on top of the other by time points in order to highlight changes in the network). MAVisto (Schreiber and Schwöbbermeyer, 2005 ) searches for over-represented network motifs and highlights these in drawings of the network. GridLayout (Li and Kurata, 2005 ) highlights functionally related nodes by placing them in roughly the same regions of the drawing. A large number of tools (Dahlquist et al. , 2002 ; Grosu et al. , 2002 ; Karp et al. , 2002 ; Khatri et al. , 2005 ; Mlecnik et al. , 2005 ; Pan et al. , 2003 , Thimm et al. , 2004 ) are designed mainly for mapping gene or protein expression profiles onto existing network diagrams.
3.4 User input and customization
Most of the major tools support a graphic user interface where mouse clicks/movements, simple dialog boxes and data imports allow most functionality to be accessed. However, in many cases the functionality offered by the biological network tool is insufficient or cumbersome for the task at hand. Several tools offer various means for third parties to develop new functionality and integrate it directly within the tool.
Plugins are an important way for advanced users to customize and extend an application. Of the major tools, Cytoscape, VisANT and ProViz support plugins. In fact, all three of these tools are based on a somewhat generic software design in order to stimulate a community of third party develops capable of expanding their system to address a greater range of biological applications. Plugins require however, a significant amount of effort and expertise to create, and thus do not represent a feasible avenue for tailoring systems to meet the needs of a particular scientific application for most labs.
Scripting and query languages can provide a convenient trade-off between the power and flexibility of plugins whilst conserving the convenience of features available through graphic user interfaces. Currently, Pathway Studio offers a wizard interface for creating very simple network and data queries and only BiologicalNetworks provides a language interface for expressing such queries. A recent tool, GUESS (Adar, 2006 ), provides the user with a powerful scripting language based on Python ( http://www.python.org/ ) that includes a convenient notation for networks. The GUESS user interface consists of a component for network visualization and a component for entering commands in the scripting language. The language allows for the analysis and modification of network layouts, however, it is sufficiently simple that users do not require previous programming experience. In addition, it is relatively easy to extend its functionality by creating interfaces to other libraries such as the R statistical library. Currently, GUESS is more widely used in the social network community but has been used for biological networks.
To the best of our knowledge, only Pathway Studio supports a multi-user environment that allows users to set sharing permissions on data and computed results.
3.5 Incorporation of external data sources
Most of the biological network tools are distributed with pre-formated versions of popular interaction databases [e.g. BIND (Alfarano et al. , 2005 ) and DIP (Xenarios et al. , 2000 )], metabolic pathway databases [e.g. KEGG (Kanehisa, 2002 )], gene ontologies [GO (Ashburner et al. , 2000 )] and molecular sequence databases [e.g. UniProt (Bairoch et al. , 2005 ), Entrez Gene http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=gene ]. There are of course many additional bioinformatics public repositories and datasets produced by individual labs that may be relevant to the user's current study. The translation of such datasets into the format required by the specific network tool is in many cases non-trivial, especially for laboratories without significant bioinformatics expertise. Partial solutions to this ubiquitous problem can be found in several biological network tools.
In VisANT, Pathway Studio and BiologicalNetworks, all data are ported to a central repository and represented in an application-specific format. This forces the end-user to build parsers for each new data resource and reformat the data to the precise format expected by the tool. This also requires that the end-users rely on developers to maintain up-to-date versions of each imported database.
Fortunately, a small number of data format standards are becoming wide spread. When both the dataset is expressed in a standardized format and the network visualizaton tool supports the importation of this format, the incorporation of these datasets requires neither programming expertise nor an understanding of the format itself. The three most popular examples, SBML, PSI-MI and BioPAX, are open, XML-based ( http://www.w3.org/XML/ ) standards that use a leveled approach , meaning that each standard is described at various levels of complexity and specificity. Users may then choose the simplest level sufficient to represent all of the necessary information in their dataset. The SBML (Systems Biology Markup Language) (Hucka and Finney, 2003 ; Hucka et al. , 2003 ) is supported by over 100 software systems, including Cytoscape, PATIKA and BiologicalNetworks. SBML is designed for modeling biochemical reaction networks at a level that admits automated simulation. Currently, SBML levels 1.0 and 2.0 are defined but a third, more detailed level is planned that would support the composition of models from component submodels, rule-based generation of states and interactions, and descriptions of cell geometries (Hucka and Finney, 2003 ).
Unlike SBML, PSI-MI (Proteomics Standards Initiative-Molecular Interactions) (Hermjakob et al. , 2004b ) has the more pragmatic mandate of describing molecular interactions rather than complete cellular models. Several visualization tools including Cytoscape and VisANT can import and export PSI-MI formated data and several public databases accept submissions in PSI-MI format.
A third standard, BioPAX http://www.biopax.org/ ), has released two levels. Level 1 is used for representing metabolic pathways, while level 2 is for representing molecular interactions. PATIKA can export data in BioPAX format whereas VisANT and Cytoscape (using the BioPAX plugin) can import BioPAX files. In a comparative study of PSI-MI, SBML and BioPAX, Strömbäck and Lambrix ( 2005 ) claim that BioPAX is the most general and expressive of the three while PSI-MI is most appropriate for describing molecular interactions and SBML is best suited for simulation models of molecular pathways.
3.6 Integration of third-party software
Like external datasources, interfacing with external software tools can be very difficult or even impossible. However, overcoming this hurdle is key to contributing to the trend toward integrated visualization and analysis. There have been a few early attempts. For example, GUESS (described earlier) contains a nearly transparent interface to the R statistical package ( http://www.r-project.org/ ) in its scripting language. Use of the CytoTalk plugin for Cytoscape is a quick way to add network visualization facilities to analysis software. The plugin simply allows external processes (including, e.g. R, Python and Perl) to remotely manipulate networks displayed in Cytoscape.
High-throughput experimental biology has already managed to create nearly cell-wide maps of protein–protein, protein–nucleotide and genetic interactions. As the cost, efficiency and accuracy of these assays improve, we can expect datasets several orders of magnitude larger than we currently have at our disposal, over a greater number of organisms. Furthermore, the variety of data types will also increase to include, e.g. a greater emphasis on protein-small molecule interactions, more comprehensive time-series gene and protein expression data and more comprehensive genetic interaction studies. There is a natural trend towards combining this information within probabilistic or dynamical models, capable of capturing the most salient aspects of complex biological processes. In order to extract as much information as we can from these meta-datasets, current biological network tools must evolve in functionality to address issues of scalability, integration and visualization.
With respect to scalability, visualization tools will need to employ novel approaches to providing access to massive datasets while respecting end-user limitations. More than ever before, users will depend on sophisticated network analysis algorithms to uncover interesting biological stories that they used to find ‘by eye’ in much smaller networks. There are several positive examples in this direction. Given a pathway in some organism, PathBLAST is able to identify evolutionarily conserved pathways in a second organism, by solving a restricted version of the subgraph isomorphism problem. Given a list of distinguished genes or gene products corresponding to, e.g. differentially expressed genes from a microarray experiment, the Steiner approach of Scott et al. ( 2005b ) searches large interaction networks for the minimal connecting set of nodes that ‘hook up’ the members of the input list. The resultant subnetwork is typically of a size that is more amenable to visualization and analysis.
Unfortunately, the size of cell-wide interaction networks renders many network analysis problems intractable. For instance, MAVisto (Schreiber and Schwöbbermeyer, 2005 ) determines whether or not certain motifs are over-represented in a network. Since determining the presence of a motif is equivalent to solving the subgraph isomorphism problem, it is very unlikely that any computer will ever be able to search for motifs of more than a few nodes in any cell-wide interaction network. Fortunately, the computational intractability of a problem in general does not imply intractability in a specific biological context. Creatively rephrasing the question can lead to tractable solutions, e.g. by restricting attention to the Steiner tree rather than the original network, or to a single cell state or location.
With respect to integration, visualization tools will need to go beyond simplistic graphical models and mere compliance with accepted standards in order to truly integrate new data types. Whereas interaction network models assume a static list of interacting pairs, there are many examples of proteins whose function and therefore interaction types and partners differ depending on cell state or subcellular location. For example, in the absence of glucocorticoids, the glucocorticoid receptor (GR) is bound to the cytosolic chaperone Hsp90 in the cytosol. The introduction of glucocorticoids causes the release of GR from Hsp90 and its subsequent retrotranslocation into the nucleus, where it functions either as a transcription factor (protein–nucleotide interactions) or an adapter protein within large transcriptional complexes (protein–protein interactions). We are not yet, however, close to obtaining complete information about cell state so models will need to strike a delicate balance between being immediately useful and attempting to model true biological networks. Standards like SBML (Hucka and Finney, 2003 ; Hucka et al. , 2003 ] that aim at completeness can be used to measure progress.
With respect to visualization, single network views will provide little more than brief glimpses of the large datasets. Visualization tools will then need to support many different types of views, each network view at a different level of detail. Dynamic navigation from one view to another will be a key to showing the connection between different views. Navigating from one time series point to another, for instance, could involve a view showing only the differences between the two time points. If the time points are consecutive, the number of differences will tend to be quite small. A similar approach could be applied to subcellular localization information as well.
To adequately address each of these issues, active cooperation will be required between a variety research fields including graph drawing , information visualization , network analysis and of course biology . It is unfortunate to witness the number of new tools designed for biology that have earlier analogs in other research areas. Pajek (Batagelj and Mrvar, 2001 ), for instance, is a general visualization and analysis tool that has been used extensively to study social networks but has seen limited use with biological networks [Ho et al. , 2002 ; Ludemann et al. , 2004 ; Tong et al. , 2001 ]. This waste of resources is caused by a simple lack of communication between fields. For instance, graph drawing researchers ignorant of emerging biological models tend to tackle layout problems of little use in biology or, at best, express solutions to relevant problems in a way that is inaccessible to biologists. A biologically informed graph drawing community, however, would be capable of generating clever ideas for optimization criteria that produce layouts more easily interpreted by biologists. This would have tremendous benefits by better facilitating the evaluation of correctness of existing interaction networks. Consider, e.g. a visualization of protein-protein interactions where proteins are placed in the layout according to subcellular localizations. Edges that span any membrane would be identified as potential mistakes, either because they are false positives identified by the assay, or because the subcellular location may be improperly or incompletely described in the dataset.
The key to success of this endeavor is the involvement of all these communities towards standards, open access software, and distributed development. Tools developed in this environment will attract large user communities, avoid duplication of effort and ultimately lead the way towards the goals of systems biology.
Funding was provided by NSERC (Postdoctoral Fellowship to M.S.; Discovery to M.H.).
Conflict of Interest : none declared.
Google Scholar
Google Preview
Author notes
Supplementary data.
| Month: | Total Views: |
|---|---|
| November 2016 | 8 |
| December 2016 | 3 |
| January 2017 | 20 |
| February 2017 | 38 |
| March 2017 | 29 |
| April 2017 | 24 |
| May 2017 | 33 |
| June 2017 | 40 |
| July 2017 | 17 |
| August 2017 | 19 |
| September 2017 | 19 |
| October 2017 | 25 |
| November 2017 | 26 |
| December 2017 | 71 |
| January 2018 | 88 |
| February 2018 | 71 |
| March 2018 | 113 |
| April 2018 | 222 |
| May 2018 | 270 |
| June 2018 | 219 |
| July 2018 | 212 |
| August 2018 | 194 |
| September 2018 | 190 |
| October 2018 | 123 |
| November 2018 | 122 |
| December 2018 | 56 |
| January 2019 | 54 |
| February 2019 | 67 |
| March 2019 | 92 |
| April 2019 | 82 |
| May 2019 | 80 |
| June 2019 | 37 |
| July 2019 | 78 |
| August 2019 | 40 |
| September 2019 | 47 |
| October 2019 | 64 |
| November 2019 | 63 |
| December 2019 | 54 |
| January 2020 | 54 |
| February 2020 | 32 |
| March 2020 | 70 |
| April 2020 | 52 |
| May 2020 | 30 |
| June 2020 | 51 |
| July 2020 | 34 |
| August 2020 | 46 |
| September 2020 | 67 |
| October 2020 | 64 |
| November 2020 | 59 |
| December 2020 | 42 |
| January 2021 | 58 |
| February 2021 | 106 |
| March 2021 | 69 |
| April 2021 | 52 |
| May 2021 | 67 |
| June 2021 | 69 |
| July 2021 | 100 |
| August 2021 | 57 |
| September 2021 | 53 |
| October 2021 | 50 |
| November 2021 | 60 |
| December 2021 | 71 |
| January 2022 | 56 |
| February 2022 | 67 |
| March 2022 | 74 |
| April 2022 | 144 |
| May 2022 | 91 |
| June 2022 | 75 |
| July 2022 | 95 |
| August 2022 | 49 |
| September 2022 | 72 |
| October 2022 | 102 |
| November 2022 | 83 |
| December 2022 | 66 |
| January 2023 | 86 |
| February 2023 | 70 |
| March 2023 | 80 |
| April 2023 | 51 |
| May 2023 | 71 |
| June 2023 | 42 |
| July 2023 | 48 |
| August 2023 | 85 |
| September 2023 | 78 |
| October 2023 | 68 |
| November 2023 | 73 |
| December 2023 | 75 |
| January 2024 | 78 |
| February 2024 | 52 |
| March 2024 | 98 |
| April 2024 | 81 |
| May 2024 | 99 |
| June 2024 | 107 |
| July 2024 | 49 |
| August 2024 | 31 |
Email alerts
Citing articles via, looking for your next opportunity.
- Recommend to your Library
Affiliations
- Online ISSN 1367-4811
- Copyright © 2024 Oxford University Press
- About Oxford Academic
- Publish journals with us
- University press partners
- What we publish
- New features
- Open access
- Institutional account management
- Rights and permissions
- Get help with access
- Accessibility
- Advertising
- Media enquiries
- Oxford University Press
- Oxford Languages
- University of Oxford
Oxford University Press is a department of the University of Oxford. It furthers the University's objective of excellence in research, scholarship, and education by publishing worldwide
- Copyright © 2024 Oxford University Press
- Cookie settings
- Cookie policy
- Privacy policy
- Legal notice
This Feature Is Available To Subscribers Only
Sign In or Create an Account
This PDF is available to Subscribers Only
For full access to this pdf, sign in to an existing account, or purchase an annual subscription.
- Docs »
- 4. Network Visualization »
- 4.1. Visual representation of biological entities and interactions
- Edit on GitHub
4.1. Visual representation of biological entities and interactions ¶
Graphical representations of biological networks includes how to intuitively visualize a set of connected nodes (or vertice) corresponding to biological entities, including genes, gene products (protein, transcript factor, miRNA, etc.), small molecules (compound, metabolite etc.), protein family and complex, and their links (or edges), such as physical or genetic interactions, regulatory events (transcriptional and translational activation or inhibition, phosphorylation, etc.), co-expression, shared protein domain, complex formation, trans-location and other biochemical reactions.
Fig. 4.1 An example of visualization of biological networks ¶
MONGKIE provides the sophisticated data models for visualization of biological networks with advanced graph drawing techniques, and therefore can represent different types of biological entities and interactions between them with out-of-the-box visual styles, shown in Fig. 4.1 . Both nodes and edges differ in their style according to their biological meaning. The style of nodes - e.g. label, font, shape, color, size and icon image - shows the type and state of biological components, and edges linking a relation participant with the information about the role also differ in their style - e.g. shape or thickness or color of lines, shape or color of arrows as well as label and font.
Classification Systems of Visual Representations Included in Biology Textbooks
- Conference: International Conference The Future of Education
- At: Firenze, Italy

- Ionian University

- National and Kapodistrian University of Athens

Discover the world's research
- 25+ million members
- 160+ million publication pages
- 2.3+ billion citations

- Chi-Yan Tsui

- Jennifer Wiley

- Thomas D. Griffin

- Gunther Kress

- Gary J. Anglin

- Scott Slough

- Susan K. Jennings

- Patricia A. Carpenter

- Recruit researchers
- Join for free
- Login Email Tip: Most researchers use their institutional email address as their ResearchGate login Password Forgot password? Keep me logged in Log in or Continue with Google Welcome back! Please log in. Email · Hint Tip: Most researchers use their institutional email address as their ResearchGate login Password Forgot password? Keep me logged in Log in or Continue with Google No account? Sign up
Loading metrics
Open Access
Peer-reviewed
Research Article
A High-Throughput Screening Approach to Discovering Good Forms of Biologically Inspired Visual Representation
* E-mail: [email protected] (NP); [email protected] (DDC)
Affiliations McGovern Institute for Brain Research, Massachusetts Institute of Technology, Cambridge, Massachussetts, United States of America, Department of Brain and Cognitive Sciences, Massachusetts Institute of Technology, Cambridge, Massachussetts, United States of America
Affiliations McGovern Institute for Brain Research, Massachusetts Institute of Technology, Cambridge, Massachussetts, United States of America, Department of Brain and Cognitive Sciences, Massachusetts Institute of Technology, Cambridge, Massachussetts, United States of America, The Rowland Institute at Harvard, Harvard University, Cambridge, Massachusetts, United States of America
- Nicolas Pinto,
- David Doukhan,
- James J. DiCarlo,
- David D. Cox

- Published: November 26, 2009
- https://doi.org/10.1371/journal.pcbi.1000579
- Reader Comments
While many models of biological object recognition share a common set of “broad-stroke” properties, the performance of any one model depends strongly on the choice of parameters in a particular instantiation of that model—e.g., the number of units per layer, the size of pooling kernels, exponents in normalization operations, etc. Since the number of such parameters (explicit or implicit) is typically large and the computational cost of evaluating one particular parameter set is high, the space of possible model instantiations goes largely unexplored. Thus, when a model fails to approach the abilities of biological visual systems, we are left uncertain whether this failure is because we are missing a fundamental idea or because the correct “parts” have not been tuned correctly, assembled at sufficient scale, or provided with enough training. Here, we present a high-throughput approach to the exploration of such parameter sets, leveraging recent advances in stream processing hardware (high-end NVIDIA graphic cards and the PlayStation 3's IBM Cell Processor). In analogy to high-throughput screening approaches in molecular biology and genetics, we explored thousands of potential network architectures and parameter instantiations, screening those that show promising object recognition performance for further analysis. We show that this approach can yield significant, reproducible gains in performance across an array of basic object recognition tasks, consistently outperforming a variety of state-of-the-art purpose-built vision systems from the literature. As the scale of available computational power continues to expand, we argue that this approach has the potential to greatly accelerate progress in both artificial vision and our understanding of the computational underpinning of biological vision.
Author Summary
One of the primary obstacles to understanding the computational underpinnings of biological vision is its sheer scale—the visual system is a massively parallel computer, comprised of billions of elements. While this scale has historically been beyond the reach of even the fastest super-computing systems, recent advances in commodity graphics processors (such as those found in the PlayStation 3 and high-end NVIDIA graphics cards) have made unprecedented computational resources broadly available. Here, we describe a high-throughput approach that harnesses the power of modern graphics hardware to search a vast space of large-scale, biologically inspired candidate models of the visual system. The best of these models, drawn from thousands of candidates, outperformed a variety of state-of-the-art vision systems across a range of object and face recognition tasks. We argue that these experiments point a new way forward, both in the creation of machine vision systems and in providing insights into the computational underpinnings of biological vision.
Citation: Pinto N, Doukhan D, DiCarlo JJ, Cox DD (2009) A High-Throughput Screening Approach to Discovering Good Forms of Biologically Inspired Visual Representation. PLoS Comput Biol 5(11): e1000579. https://doi.org/10.1371/journal.pcbi.1000579
Editor: Karl J. Friston, University College London, United Kingdom
Received: June 11, 2009; Accepted: October 26, 2009; Published: November 26, 2009
Copyright: © 2009 Pinto et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Funding: This study was funded in part by The National Institutes of Health (NEI R01EY014970), The McKnight Endowment for Neuroscience, Dr. Gerald Burnett and Marjorie Burnett, and The Rowland Institute of Harvard. Hardware support was generously provided by the NVIDIA Corporation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Competing interests: The authors have declared that no competing interests exist.
Introduction
The study of biological vision and the creation of artificial vision systems are naturally intertwined—exploration of the neuronal substrates of visual processing provides clues and inspiration for artificial systems, and artificial systems, in turn, serve as important generators of new ideas and working hypotheses. The results of this synergy have been powerful: in addition to providing important theoretical frameworks for empirical investigations (e.g. [1] – [6] ), biologically-inspired models are routinely among the highest-performing artificial vision systems in practical tests of object and face recognition [7] – [12] .
However, while neuroscience has provided inspiration for some of the “broad-stroke” properties of the visual system, much is still unknown. Even for those qualitative properties that most biologically-inspired models share, experimental data currently provide little constraint on their key parameters. As a result, even the most faithfully biomimetic vision models necessarily represent just one of many possible realizations of a collection of computational ideas.
Truly evaluating the set of biologically-inspired computational ideas is difficult, since the performance of a model depends strongly on its particular instantiation–the size of the pooling kernels, the number of units per layer, exponents in normalization operations, etc. Because the number of such parameters (explicit or implicit) is typically large, and the computational cost of evaluating one particular model is high, it is difficult to adequately explore the space of possible model instantiations. At the same time, there is no guarantee that even the “correct” set of principles will work when instantiated on a small scale (in terms of dimensionality, amount of training, etc.). Thus, when a model fails to approach the abilities of biological visual systems, we cannot tell if this is because the ideas are wrong, or they are simply not put together correctly or on a large enough scale.
As a result of these factors, the availability of computational resources plays a critical role in shaping what kinds of computational investigations are possible. Traditionally, this bound has grown according to Moore's Law [13] , however, recently, advances in highly-parallel graphics processing hardware (such as high-end NVIDIA graphics cards, and the PlayStation 3's IBM Cell processor) have disrupted this status quo for some classes of computational problems. In particular, this new class of modern graphics processing hardware has enabled over hundred-fold speed-ups in some of the key computations that most biologically-inspired visual models share in common. As is already occurring in other scientific fields [14] , [15] , the large quantitative performance improvements offered by this new class of hardware hold the potential to effect qualitative changes in how science is done.
In the present work, we take advantage of these recent advances in graphics processing hardware [16] , [17] to more expansively explore the range of biologically-inspired models–including models of larger, more realistic scale. In analogy to high-throughput screening approaches in molecular biology and genetics, we generated and trained thousands of potential network architectures and parameter instantiations, and we “screened” the visual representations produced by these models using tasks that engage the core problem of object recognition–tolerance to image variation [10] – [12] , [18] , [19] . From these candidate models, the most promising were selected for further analysis.
We show that this large-scale screening approach can yield significant, reproducible gains in performance in a variety of basic object recognitions tasks and that it holds the promise of offering insight into which computational ideas are most important for achieving this performance. Critically, such insights can then be fed back into the design of candidate models (constraining the search space and suggesting additional model features), further guiding evolutionary progress. As the scale of available computational power continues to expand, high-throughput exploration of ideas in computational vision holds great potential both for accelerating progress in artificial vision, and for generating new, experimentally-testable hypotheses for the study of biological vision.
A Family of Candidate Models
In order to generate a large number of candidate model instantiations, it is necessary to parameterize the family of all possible models that will be considered. A schematic of the overall architecture of this model family, and some of its parameters, is shown in Figure 2 . The parameterization of this family of models was designed to be as inclusive as possible–that is, the set of model operations and parameters was chosen so that the family of possible models would encompass (as special cases) many of the biologically-inspired models already described in the extant literature (e.g. [1] – [4] , [7] , [9] ). For instance, the full model includes an optional “trace” term, which allows learning behavior akin to that described in previous work (e.g. [4] , [20] – [22] ). While some of the variation within this family of possible models might best be described as variation in parameter tuning within a fixed model architecture, many parameters produce significant architectural changes in the model (e.g. number of filters in each layer). The primary purpose of this report is to present an overarching approach to high-throughput screening. While precise choices of parameters and parameter ranges are clearly important, one could change which parameters were explored, and over what ranges, without disrupting the integrity of the overarching approach. An exhaustive description of specific model parameters used here is included in the Supplemental Text S1 , and is briefly described next.
- PPT PowerPoint slide
- PNG larger image
- TIFF original image
Our implemented performance speed-ups for a key filtering operation in our biologically-inspired model implementation. Performance and price are shown across a collection of different GPUs, relative to a commonly used MATLAB CPU-based implementation (using a single CPU core with the filter2 function, which is coded in C++). We contrast this standard implementation with a multi-core MATLAB version, a highly-optimized C/SSE2 multi-core implementation on the same CPU, and highly-optimized GPU implementations. We have implemented speedups of over thousands of times with GPUs, resulting in qualitative changes in what kinds of model investigations are possible. More technical details and a throughout discussion of the computational framework enabling these speedups can be found in Supplemental Figure S1 and Supplemental Text S2 . * These costs are based on multi-GPU systems containing four GPUs in addition to the quad-core CPU (Q9450). ** These costs include both the hardware and MATLAB yearly licenses (based on an academic discount pricing, for one year).
https://doi.org/10.1371/journal.pcbi.1000579.g001
Model parameters were organized into four basic groups. The first group of parameters controlled structural properties of the system, such as the number of filters in each layer and their sizes. The second group of parameters controlled the properties of nonlinearities within each layer, such as divisive normalization coeffients and activation functions. The third group of parameters controlled how the models learned filter weights in response to video inputs during an Unsupervised Learning Phase (this class includes parameters such as learning rate, trace factors, etc.; see Phase 2: Unsupervised Learning below). A final set of parameters controlled details of how the resulting representation vectors are classified during screening and validation (e.g. parameters of dimensionality reduction, classification parameters, etc.). For the purposes of the work presented here, this class of classification-related parameters was held constant for all analyses below. Briefly, the output values of the final model layer corresponding to each test example image were “unrolled” into a vector, their dimensionality was reduced using Principal Component Analysis (PCA) keeping as many dimensions as there were data points in the training set, and labeled examples were used to train a linear Support Vector Machine (SVM).
Each model consisted of three layers, with each layer consisting of a “stack” of between 16 and 256 linear filters that were applied at each position to a region of the layer below. At each stage, the output of each unit was normalized by the activity of its neighbors within a parametrically-defined radius. Unit outputs were also subject to parameterized threshold and saturation functions, and the output of a given layer could be spatially resampled before being given to the next layer as input. Filter kernels within each stack within each layer were initialized to random starting values, and learned their weights during the Unsupervised Learning Phase (see below, see Supplemental Text S1 ). Briefly, during this phase, under parametric control, a “winning” filter or filters were selected for each input patch, and the kernel of these filters was adapted to more closely resemble that patch, achieving a form of online non-parametric density estimation. Building upon recent findings from visual neuroscience [18] , [23] , [24] , unsupervised learning could also be biased by temporal factors, such that filters that “won” in previous frames were biased to win again (see Supplemental Text S1 for details).
It should be noted that while the parameter set describing the model family is large, it is not without constraints. While our model family includes a wide variety of feed-forward architectures with local intrinsic processing (normalization), we have not yet included long-range feedback mechanisms (e.g. layer to layer). While such mechanisms may very well turn out to be critically important for achieving the performance of natural visual systems, the intent of the current work is to present a framework to approach the problem. Other parameters and mechanisms could be added to this framework, without loss of generality. Indeed, the addition of new mechanisms and refinement of existing ones is a major area for future research (see Discussion ).
Parallel Computing Using Commodity Graphics Hardware
While details of the implementation of our model class are not essential to the theoretical implications of our approach, attention must nonetheless be paid to speed in order to ensure the practical tractability, since the models used here are large (i.e. they have many units), and because the space of possible models is enormous. Fortunately, the computations underlying our particular family of candidate models are intrinsically parallel at a number of levels. In addition to coarse-grain parallelism at the level of individual model instantiations (e.g. multiple models can be evaluated at the same time) and video frames (e.g. feedforward processing can be done in parallel on multiple frames at once), there is a high degree of fine-grained parallelism in the processing of each individual frame. For instance, when a filter kernel is applied to an image, the same filter is applied to many regions of the image, and many filters are applied to each region of the image, and these operations are largely independent. The large number of arithmetic operations per region of image also results in high arithmetic intensity (numbers of arithmetic operations per memory fetch), which is desirable for high-performance computing, since memory accesses are typically several orders of magnitude less efficient than arithmetic operations (when arithmetic intensity is high, caching of fetched results leads to better utilization of a processor's compute resources). These considerations are especially important for making use of modern graphics hardware (such as the Cell processor and GPUs) where many processors are available. Highly-optimized implementations of core operations (e.g. linear filtering, local normalization) were created for both the IBM Cell Processor (PlayStation 3), and for NVIDIA graphics processing units (GPUs) using the Tesla Architecture and the CUDA programming model [25] . These implementations achieve highly significant speed-ups relative to conventional CPU-based implementations (see Figure 1 and Supplemental Figure S1 ). High-level “outer loop” coordination of these highly optimized operations was accomplished using the Python programming language (e.g. using PyCUDA [26] ), allowing for a favorable balance between ease of programming and raw speed (see Supplemental Text S2 ). In principle, all of the analyses presented here could have been performed using traditional computational hardware; however, the cost (in terms of time and/or money) of doing so with current CPU hardware is prohibitive.
The system consists of three feedforward filtering layers, with the filters in each layer being applied across the previous layer. Red colored labels indicate a selection of configurable parameters (only a subset of parameters are shown).
https://doi.org/10.1371/journal.pcbi.1000579.g002
Figure 1 shows the relative speedup and performance/cost of each implementation (IBM Cell on Sony's PlayStation 3 and several NVIDIA GPUs) relative to traditional MATLAB and multi-threaded C code for the linear filtering operation (more details such as the raw floating point performance can be found in the Supplemental Figure S1 ). This operation is not only a key component of the candidate model family (see below) but it's also the most computationally demanding, reaching up to 94% of the total processing time (for the PlayStation 3 implementation), depending on model parameters (average fraction is 28%). The use of commodity graphics hardware affords orders-of-magnitude increases in performance. In particular, it should be noted that the data presented in this work took approximately one week to generate using our PlayStation 3-based implementation (222x speedup with one system) on a cluster of 23 machines. We estimate that producing the same results at the same cost using a conventional MATLAB implementation would have taken more than two years (see Figure S1 ).
Screening for Good Forms of Representation
Our approach is to sample a large number of model instantiations, using a well-chosen “screening” task to find promising architectures and parameter ranges within the model family. Our approach to this search was divided into four phases (see Figure 3 ): Candidate Model Generation, Unsupervised Learning, Screening, and Validation/Analysis of high-performing models.
The experiments described here consist of five phases. (A) First, a large collection of model instantiations are generated with randomly selected parameter values. (B) Each of these models then undergoes an unsupervised learning period, during which its filter kernels are adapted to spatio-temporal statistics of the video inputs, using a learning algorithm that is influenced by the particular parameter instantiation of that model. After the Unsupervised Learning Phase is complete, filter kernels are fixed, and (C) each model is subjected to a screening object recognition test, where labeled images are represented using each model instantiation, and these re-represented images are used to train an SVM to perform a simple two-class discrimination task. Performance of each candidate model is assessed using a standard cross-validation procedure. (D) From all of the model instantiations, the best are selected for further analysis. (E) Finally, these models are tested on other object recognition tasks.
https://doi.org/10.1371/journal.pcbi.1000579.g003
Phase 1: candidate model generation.
Candidate model parameter sets were randomly sampled with a uniform distribution from the full space of possible models in the family considered here (see Figure 2 and Figure S2 for a schematic diagram of the models, and Supplemental Materials for an exhaustive description of model parameters and value ranges that were explored; Supplemental Text S1 ).
Phase 2: unsupervised learning.
All models were subjected to a period of unsupervised learning, during which filter kernels were adapted to spatiotemporal statistics of a stream of input images. Since the family of models considered here includes features designed to take advantage of the temporal statistics of natural inputs (see Supplementary Methods), models were learned using video data. In the current version of our family of models, learning influenced the form of the linear kernels of units at each layer of the hierarchy, but did not influence any other parameters of the model.
We used three video sets for unsupervised learning: “Cars and Planes”, “Boats”, and “Law and Order”. The “Law and Order” video set consisted of clips from the television program of the same name (Copyright NBC Universal), taken from DVDs, with clips selected to avoid the inclusion of text subtitles. These clips included a variety of objects moving through the frame, including characters' bodies and faces.
The “Cars and Planes” and “Boats” video sets consisted of 3D ray-traced cars, planes and boats undergoing 6-degree-of-freedom view transformations (roughly speaking, “tumbling” through space). These same 3D models were also used in a previous study [11] . Video clips were generated where an object would appear for approximately 300 frames, performing a random walk in position (3 degrees of freedom) and rotation (3 degrees of freedom) for a total of 15,000 frames. Examples are shown in Figures 4A and 4B .
(A) Sequences of a rendered car undergoing a random walk through the possible range of rigid body movements. (B) A similar random walk with a rendered boat.
https://doi.org/10.1371/journal.pcbi.1000579.g004
For the sake of convenience, we refer to each unsupervised learning video set as a “petri dish,” carrying forward the analogy to high-throughput screening from biology. In the results presented here, 2,500 model instantiations were independently generated in each “petri dish” by randomly drawing parameter values from a uniform distribution (a total of 7,500 models were trained). Examples of filter kernels resulting from this unsupervised learning procedure are shown in Supplemental Figures S3 , S4 , S5 and S6 .
After the end of the Unsupervised Learning Phase , the linear filter kernels were not modified further, and the resulting model was treated as a fixed transformation (e.g. a static image is entered as input, and a vector of responses from the units of the final layer is outputted).
Phase 3: screening.
Following the Unsupervised Learning Phase , each “petri dish” was subjected to a Screening Phase to determine which model instantiations produced image representations that are well-suited for performing invariant object recognition tasks.
During the Screening Phase , individual static images were supplied as input to each model, and the vector of responses from the units of its final layer were taken as that model's “representation” of the image. The labeled, “re-represented” images were then reduced in dimensionality by PCA and taken as inputs (training examples) for a classifier (in our case, a linear SVM).
We used a simple “Cars vs. Planes” synthetic object recognition test as a screening task (see [11] for details). In this task, 3D models from two categories (cars and planes), were rendered across a wide range of variation in position, scale, view, and background. The rendered grayscale images (200 by 200 pixels) were provided as input to each model, and a classifier was trained to distinguish car images from plane images (150 training images per category). Performance of each model was then tested on a new set of unlabeled re-represented car and plane images (150 testing images per category). This recognition test has the benefit of being relatively quick to evaluate (because it only contains two classes), while at the same time having previous empirical grounding as a challenging object recognition test due to the large amount of position, scale, view, and background variation [11] (see Figure 5A ).
(A) A new set of rendered cars and planes composited onto random natural backgrounds. (B) Rendered boats and animals. (C) Rendered female and male faces. (D) A subset of the MultiPIE face test set [27] with the faces manually removed from the background, and composited onto random image backgrounds, with additional variation in position, scale, and planar rotation added.
https://doi.org/10.1371/journal.pcbi.1000579.g005
Phase 4: validation.
The best models selected during the Screening Phase were submitted to validation tests using other image sets, to determine if the representations generated by the models were useful beyond the immediate screening task. For the present work, four validation sets were used: 1) a new set of rendered cars and planes (generated by the same random process that generated the screening set, but with different specific examplars), 2) a set of rendered boats and animals 3) a set of rendered images of two synthetic faces (one male, one female, [10] , [12] ), and 4) a modified subset of the standard MultiPIE face recognition test set ( [27] ; here dubbed the “MultiPIE Hybrid” set). In the case of the rendered sets (sets 1–3), as with the screening set, the objects were rendered across a wide range of views, positions, and scales.
For the “MultiPIE hybrid” set, 50 images each of two individuals from the standard MultiPIE set were randomly selected from the full range of camera angles, lighting, expressions, and sessions included in the MultiPIE set. These faces were manually removed from their backgrounds and were further transformed in scale, position, planar rotation and were composited onto random natural backgrounds. Examples of the resulting images are shown in Figure 5 .
For all sets (as with the screening set) classifiers were trained with labeled examples to perform a two-choice task (i.e. Cars vs. Planes, Boats vs. Animals, Face 1 vs. Face 2), and were subsequently tested with images not included in the training set.
While a number of standardized “natural” object and face recognition test sets exist [28] – [34] , we made a deliberate choice not to use these sets. Previous investigations [10] – [12] , [35] , [36] have raised concerns with many of these sets, calling into question whether they appropriately capture the problem of interest. As a result, we chose to focus here on image sets that include substantial image variation by design, be they synthetic (as in our rendered set) or natural (as in the MultiPIE Hybrid set) in origin.
Performance Comparison with Other Algorithms
“v1-like” baseline..
Since object recognition performance measures are impossible to interpret in a vacuum, we used a simple V1-like model to serve as one baseline against which model performance can be compared. This V1-like model was taken, without modification, from Pinto et al. [11] , and was shown previously to match or exceed the performance of a variety of purpose-built vision systems on the popular (but, we argue, flawed as a test of invariant object recognition) Caltech101 object recognition set and a wide variety of standard face recognition sets (ORL, Yale, CVL, AR, and Labeled Faces in the Wild [10] , [12] ). Importantly, this model is based on only a first-order description of the first stage of visual processing in the brain, and it contains no mechanisms that should allow it to tolerate the substantial image variation that makes object recognition hard in the first place [11] , [19] . Here, this model serves as a lower bound on the amount of trivial regularity that exists in the test set. To be considered promising object recognition systems, models should at least exceed the performance of the V1-like model.
Comparison with state-of-the-art algorithms.
To facilitate comparison with other models in the literature, we obtained code for, or re-implemented five “state of the art” object recognition algorithms from the extant literature: “Pyramid Histogram of Oriented Gradients” (PHOG) [37] , “Pyramid Histogram of Words” (PHOW) (also known as the Spatial Pyramid [38] ), the “Geometric Blur” shape descriptors [39] , the descriptors from the “Scale Invariant Feature Transformation” (SIFT) [40] , and the “Sparse Localized Features” (SLF) features of Mutch and Lowe [8] (a sparse extension of the C2 features from the Serre et al. HMAX model [7] ). In all cases, we were able to reproduce or exceed the authors' reported performance for each system on the Caltech101 test set, which served as a sanity check that we had correctly implemented and used each algorithm as intended by its creators.
Each algorithm was applied using an identical testing protocol to our validation sets. In cases where an algorithm from the literature dictated that filters be optimized relative to each training set (e.g. [38] and [8] ), we remained faithful to the authors' published descriptions and allowed this optimization, resulting in a different individually tailored model for each validation set. This was done even though our own high-throughput-derived models were not allowed such per-set optimizations (i.e. the same representation was used for all validation sets), and could therefore theoretically be “handicapped” relative to the state-of-the-art models.
Object Recognition Performance
As a first exploration of our high-throughput approach, we generated 7,500 model instantiations, in three groups of 2,500, with each group corresponding to a different class of unsupervised learning videos (“petri dishes”; see Methods ). During the Screening Phase , we used the “Cars vs. Planes” object discrimination task [11] to assess the performance of each model, and the most promising five models from each set of 2,500 models was submitted to further analysis. The raw computation required to generate, train and screen these 7,500 models was completed in approximately one week, using 23 PlayStation 3 systems [41] . Results for models trained with the “Law and Order” petri dish during the Unsupervised Learning Phase are shown in Figure 6A . As expected, the population of randomly-generated models exhibited a broad distribution of performance on the screening task, ranging from chance performance (50%) to better than 80% correct. Figure 6B shows the performance of the best five models drawn from the pool of 2,500 models in the “Law and Order” petri dish. These models consistently outperformed the V1-like model baseline ( Figure 7 ), and this performance was roughly maintained even when the model was retrained with a different video set (e.g. a different clip from Law and Order), or with a different random initialization of the filter kernel weights ( Figure 6C ).
(A) Histogram of the performance of 2,500 models on the “Cars vs. Planes” screening task (averaged over 10 random splits; error bars represent standard error of the mean). The top five performing models were selected for further analysis. (B) Performance of the top five models (1–5), and the performance achieved by averaging the five SVM kernels (red bar labelled “blend”) (C) Performance of the top five models (1–5) when trained with a different random initialization of filter weights (top) or with a different set of video clips taken from the “Law and Order” television program (bottom).
https://doi.org/10.1371/journal.pcbi.1000579.g006
Performance of the top five models from the Screening Phase on a variety of other object recognition challenges. Example images from each object recognition test are shown in Figure 5 . For each validation set, the performance (averaged over 10 random splits; error bars represent standard error of the mean) is first plotted for V1-like and V1-like+ baseline models (see [10] – [12] for a detailed description of these two variants) (gray bars), and for five state-of-the-art vision systems (green bars): Scale Invariant Feature Transform (SIFT, [40] ), Geometric Blur Descriptor (GB, [39] ), Pyramidal Histogram of Gradients (PHOG, [37] ), Pyramidal Histogram of Words (PHOW, [38] ), and a biologically-inspired hierarchical model (“Sparse Localized Features” SLF, [8] ). Finally, performance of the five best models derived from the high-throughput screening approach presented in this paper (black bars), and the performance achieved by averaging the five SVM kernels (red bar labelled “blend”). In general, high-throughput-derived models outperformed the V1-like baseline models, and tended to outperform a variety of state-of-the-art systems from the literature. Model instantiation 3281 and the blend of all five top models uniformly produced the best results across all test sets considered here.
https://doi.org/10.1371/journal.pcbi.1000579.g007
Since these top models were selected for their high performance on the screening task, it is perhaps not surprising that they all show a high level of performance on that task. To determine whether the performance of these models generalized to other test sets, a series of Validation tests were performed. Specifically, we tested the best five models from each Unsupervised Learning petri dish on four test sets: two rendered object sets, one rendered face set, and a modified subset of the MultiPIE face recognition image set (see Validation Phase in Methods ). Performance across each of these validation sets is shown in Figure 7 (black bars). While the exact ordering of model performance varied somewhat from validation set to validation set, the models selected during the Screening Phase performed well across the range of validation tasks.
The top five models found by our high-throughput screening procedure generally outperformed state-of-the-art models from the literature (see Methods ) across all sets, with the best model found by the high-throughput search uniformly yielding the highest performance across all validation sets. Even greater performance was achieved by a simple summing of the SVM kernels from the top five models (red bar, Figure 7 ). Of note, the nearest contender from the set of state-of-the-art models is another biologically-inspired model [7] , [8] .
Interestingly, a large performance advantage between our high-throughput-derived models and state-of-the-art models was observed for the MultiPIE hybrid set, even though this is arguably the most different from the task used for screening, since it is composed from natural images (photographs), rather than synthetic (rendered) ones. It should be noted that several of the state-of-the-art models, including the sparse C2 features (“SLF” in Figure 7 ), which was consistently the nearest competitor to our models, used filters that were individually tailored to each validation test–i.e. the representation used for “Boats vs. Planes” was optimized for that set, and was different from the representation used for the MultiPIE Hybrid set. This is in contrast to our models, which learned their filters from a completely unrelated video data set (Law and Order) and were screened using an unrelated task (“Cars vs. Planes”, see Methods ). While even better performance could no doubt be obtained by screening with a subset taken from each individual validation test, the generalizability of performance across a range of different tasks argues that our approach may be uncovering features and representations that are broadly useful. Such generality is in keeping with the models' biological inspiration, since biological visual representations must be flexible enough to represent a massive diversity of objects in order to be useful.
Results for the 2,500 models in each of the other two “petri dishes” (i.e. models trained with alternate video sets during unsupervised learning) were appreciably similar, and are shown in Supplemental Figures S7 and S8 , using the same display conventions set forth in Figures 6 and 7 .
We have demonstrated a high-throughput framework, within which a massive number of candidate vision models can be generated, screened, and analyzed. Models found in this way were found to consistently outperform an experimentally-motivated baseline model (a V1-like model; [10] – [12] ), and the representations of visual space instantiated by these models were found to be useful generally across a variety of object recognition tasks. The best of these models and the blend of the five best models were both found to consistently outperform a variety of state-of-the-art machine vision systems for all of the test sets explored here, even without any additional optimization.
This work builds on a long tradition of machine vision systems inspired by biology (e.g. [1] – [4] , [7] , [9] ). However, while this past work has generated impressive progress towards building artificial visual systems, it has explored only a few examples drawn from the larger space of biologically-inspired models. While the task of exploring the full space of possible model instantiations remains daunting (even within the relatively restricted “first-order” class of models explored here), our results suggest that even a relatively simple, brute-force high-throughput search strategy is effective in identifying promising models for further study. In the parameter space used here, we found that a handful of model instantiations performed substantially better than the rest, with these “good” models occurring at a rate of approximately one in five-hundred. The relative rarity of these models underscores the importance of performing large-scale experiments with many model instantiations, since these models would be easy to miss in a “one-off” mode of exploration. Importantly, these rare, high-performing models performed well across a range of object recognition tasks, indicating that our approach does not simply optimize for a given task, but can uncover visual representations of general utility.
Though not conceptually critical to our approach, modern graphics hardware played an essential role in making our experiments possible. In approximately one week, we were able to test 7,500 model instantiations, which would have taken approximately two years using a conventional (e.g. MATLAB-based) approach. While it is certainly possible to use better-optimized CPU-based implementations, GPU hardware provides large increases in attainable computational power (see Figure 1 and Supplemental Figure S1 ).
An important theme in this work is the use of parametrically controlled objects as a way of guiding progress. While we are ultimately interested in building systems that tolerate image variation in real-world settings, such sets are difficult to create, and many popular currently-available “natural” object sets have been shown to lack realistic amounts of variation [10] – [12] . Our results show that it is possible to design a small synthetic set to screen and select models that generalize well across various visual classification tasks, suggesting that parametric sets can capture the essence of the invariant object recognition problem. Another critical advantage of the parametric screening approach presented here is that task difficulty can be increased on demand–that is, as models are found that succeed for a given level of image variation, the level of variation (and therefore the level of task difficulty), can be “ratcheted up” as well, maintaining evolutionary “pressure” towards better and better models.
While we have used a variety of synthetic (rendered) object image sets, images need not be synthetic to meet the requirements of our approach. The modified subset of the MultiPIE set used here (“MultiPIE Hybrid”, Figure 5 ) is an example of how parametric variation can also be achieved using carefully controlled photography.
Future Directions
While our approach has yielded a first crop of promising biologically-inspired visual representations, it is another, larger task to understand how these models work, and why they are better than other alternatives. While such insights are beyond the scope of the present paper, our framework provides a number of promising avenues for further understanding.
One obvious direction is to directly analyze the parameter values of the best models in order to understand which parameters are critical for performance. Figure 8 shows distributions of parameter values for four arbitrarily chosen parameters. While in no way conclusive, there are hints that some particular parameter values may be more important for performance than others (for quantitative analysis of the relationship between model parameters and performance, see Supplemental Text S3 , Figures S9 and S10 ). The speed with which large collections of models can be evaluated opens up the possibility of running large-scale experiments where given parameters are held fixed, or varied systematically. Insights derived from such experiments can then be fed back into the next round of high-throughput search, either by adjusting the parameter search space or by fundamentally adjusting the algorithm itself. Such iterative refinement is an active area of research in our group.
See Supplemental Text S1 for an exhaustive description of the meaning of each parameter. The top five best performing models are plotted in red, with the other models overplotted in semi-transparent blue. The parameters considered in (A) and (B) show hints of a relationship between parameter value and inclusion in the top five. In (A) all of the five best models had the same value of the parameter, and in (B) best models were clustered in lower ranges of parameter value. (C) and (D) show parameters where the best models were distributed across a range of parameter values. Such examinations of parameter values are in no way conclusive, but can provide hints as to which parameters might be important for performance. See also Supplemental Text S3 , Figures S9 and S10 .
https://doi.org/10.1371/journal.pcbi.1000579.g008
The search procedure presented here has already uncovered promising visual representations, however, it represents just the simplest first step one might take in conducting a large-scale search. For the sake of minimizing conceptual complexity, and maximizing the diversity of models analyzed, we chose to use random, brute-force search strategy. However, a rich set of search algorithms exist for potentially increasingly the efficiency with which this search is done (e.g. genetic algorithms [42] , simulated annealing [43] , and particle swarm techniques [44] ). Interestingly, our brute-force search found strong models with relatively high probability, suggesting that, while these models would be hard to find by “manual” trial-and-error, they are not especially rare in the context of our high-throughput search.
While better search algorithms will no doubt find better instances from the model class used here, an important future direction is to refine the parameter-ranges searched and to refine the algorithms themselves. While the model class described here is large, the class of all models that would count as “biologically-inspired” is even larger. A critical component of future work will be to adjust existing mechanisms to achieve better performance, and to add new mechanisms (including more complex features such as long-range feedback projections). Importantly, the high-throughput search framework presented here provides a coherent means to find and compare models and algorithms, without being unduly led astray by weak sampling of the potential parameter space.
Another area of future work is the application of high-throughput screening to new problem domains. While we have here searched for visual representations that are good for object recognition, our approach could also be applied to a variety of other related problems, such as object tracking, texture recognition, gesture recognition, feature-based stereo-matching, etc. Indeed, to the extent that natural visual representations are flexibly able to solve all of these tasks, we might likewise hope to mine artificial representations that are useful in a wide range of tasks.
Finally, as the scale of available computational resources steadily increases, our approach naturally scales as well, allowing more numerous, larger, and more complex models to be examined. This will give us both the ability to generate more powerful machine vision systems, and to generate models that better match the scale of natural systems, providing more direct footing for comparison and hypothesis generation. Such scaling holds great potential to accelerate both artificial vision research, as well as our understanding of the computational underpinnings of biological vision.
Supporting Information
Processing Performance of the Linear Filtering Operation. The theoretical and observed processing performance in GFLOPS (billions of floating point operations per second) is plotted for a key filtering operation in our biologically-inspired model implementation. Theoretical performance numbers were taken from manufacturer marketing materials and are generally not achievable in real-world conditions, as they consider multiple floating operations per clock cycle, without regard to memory communication latencies (which typically are the key determinant of real-world performance). Observed processing performance for the filtering operation varied across candidate models in the search space, as input and filter sizes varied. Note that the choice of search space can be adjusted to take maximum advantage of the underlying hardware at hand. We plot the “max” observed performance for a range of CPU and GPU implementations, as well as the “mean” and “min” performance of our PlayStation 3 implementation observed while running the 7,500 models presented in this study. The relative speedup denotes the peak performance ratio of our optimized implementations over a reference MATLAB code on one of the Intel QX9450's core (e.g. using filter2, which is itself coded in C++), whereas the relative GFLOPS per dollar indicates the peak performance per dollar ratio. Costs of typical hardware for each approach and cost per FLOPS are shown at the bottom. * These ranges indicate the performance and cost of a single system containing from one (left) to four (right) GPUs. ** These costs include both the hardware and MATLAB yearly licenses (based on an academic discount pricing, for one year).
https://doi.org/10.1371/journal.pcbi.1000579.s001
(1.19 MB TIF)
A schematic of the flow of transformations performed in our family of biologically-inspired models. Blue-labeled boxes indicate the cascade of operations performed in each of the three layers in the canonical model. Gray-labeled boxes to the right indicate filter weight update steps that take place during the Unsupervised Learning Phase after the processing of each input video frame. The top gray-labeled box shows processing steps undertaken during the Screening and Validation Phases to evaluate the performance achievable with each model instantiation.
https://doi.org/10.1371/journal.pcbi.1000579.s002
(0.95 MB TIF)
Examples of Layer 1 filters taken from different models. A random assortment of linear filter kernels taken from the first layers of the top five (A) and fifteen randomly chosen other model instantiations (B) taken from the “Law and Order” petri dish. Each square represents a single two-dimensional filter kernel, with the values of each filter element represented in gray scale (the gray-scale is assigned on a per-filter basis, such that black is the smallest value found in the kernel, and white is the largest). For purposes of comparison, a fixed number of filters were taken from each model's Layer 1, even though different models have differing number of filters in each layer. Filter kernels are initialized with random values and learn their structure during the Unsupervised Learning Phase of model generation. Interestingly, oriented structures are common in filter from both the top five models and from non-top-five models.
https://doi.org/10.1371/journal.pcbi.1000579.s003
(3.71 MB TIF)
Examples of Layer 2 filters taken from different models. Following the same basic convention as in Supplemental Figure S3 , a random assortment of portions of filter kernels from Layer 2 of the top five (A) and fifteen other randomly-chosen model instantiations (B) are shown in gray-scale to provide a qualitative sense of what the linear filters (produced as a result of the Unsupervised Learning Phase) look like. Note that since each Layer 1 is itself a stack of k l = 1 two-dimensional planes (or “feature maps”) resulting from filtering with a stack of k l = 1 filters (see Supplemental Text S1 and Supplemental Figure S6 , each Layer 2 filter is actually a f s l = 2 × f s l = 2 × k l = 1 kernel For the sake of visual clarity, we here present just one randomly-chosen f s l = 2 × f s l = 2 “slice” from each of the randomly-chosen filters. As in Supplemental Figure S3 , there are signs of “structure” in the filters of both the top five and non-top-five models.
https://doi.org/10.1371/journal.pcbi.1000579.s004
(3.76 MB TIF)
Examples of Layer 3 filters taken from different models. Following the same basic convention as in Supplemental Figures S3 and S4 , a random assortment of portions of filter kernels from Layer 3 of the top five (A) and fifteen other randomly-chosen model instantiations (B) are shown in gray-scale to provide a qualitative sense of what the linear filters (produced as a result of the Unsupervised Learning Phase) look like. Note that since each Layer 2 is itself a stack of k l = 2 two-dimensional planes (or “feature maps”) resulting from filtering with a stack of k l = 2 filters (see Supplemental Text S1 and Supplemental Figure S6 ), each Layer 3 filter is actually a f s l = 3 × f s l = 3 × k l = 2 kernel. For the sake of visual clarity, we here present just one randomly-chosen f s l = 3 × f s l = 3 “slice” from each of the randomly-chosen filters. As in Supplemental Figures S3 and S4 , there are signs of “structure” in the filters of both the top five and non-top-five models.
https://doi.org/10.1371/journal.pcbi.1000579.s005
(3.72 MB TIF)
Example filterbanks from the best model instantiation in the “Law and Order” Petri Dish. Filter kernels were learned during the Unsupervised Learning Phase, after which filter weights were fixed. Colors indicate filter weights, and were individually normalized to make filter structure clearer (black-body color scale with black indicating the smallest filter weight, white representing the largest filter weight). The filter stack for each layer consists of k l filters, with size f s . Because the Layer 1 filterbank for this model includes 16 filters, the Layer 1 output will have a feature “depth” of 16, and thus each Layer 2 filter is a stack of 16 f s × f s kernels. One filter (filter 61) is shown expanded for illustration purposes. Similarly, since the Layer 2 filterbank in this example model includes 64 filters, the output of Layer 2 will have a depth of 64, and thus each filter in Layer 3 filterbank must also be 64-deep.
https://doi.org/10.1371/journal.pcbi.1000579.s006
(1.65 MB TIF)
High-throughput screening in the “Cars and Planes” Petri Dish. Data are shown according to the same display convention set forth in the main paper. (A) Histogram of the performance of 2,500 models on the “Cars vs. Planes” screening task. The top five performing models were selected for further analysis. (B) Performance of the top five models (1–5). (C) Performance of the top five models when trained with a different random initialization of filter weights (top) or with a different set of video clips (bottom). (D) Performance of the top five models from the Screening Phase on a variety of other object recognition challenges.
https://doi.org/10.1371/journal.pcbi.1000579.s007
(0.21 MB TIF)
High-throughput screening and validation in the “Boats'” Petri Dish. Data are shown according to the same display convention set forth in the main paper. (A) Histogram of the performance of 2,500 models on the “Cars vs. Planes” screening task. The top five performing models were selected for further analysis. (B) Performance of the top five models (1–5). (C) Performance of the top five models when trained with a different random initialization of filter weights (top) or with a different set of video clips (bottom). (D) Performance of the top five models from the Screening Phase on a variety of other object recognition challenges.
https://doi.org/10.1371/journal.pcbi.1000579.s008
(0.22 MB TIF)
Linear regression analysis of relationship between parameter values and model performance. As a first-order analysis of the relationship between model parameters and model performance, we performed a linear regression analysis in which the values of each of the 52 parameters were included as predictors in a multiple linear regression analysis. Next, p-values were computed for the t statistic on each beta weight in the regression. A histogram of the negative natural log of the p-values is shown here, with the bin including significant p-values highlighted in orange (each count corresponds to one model parameter). For reference, the histogram is divided into three ranges (low-nonsignificant, medium-nonsignificant, and significant) and a listing of parameters included each significance range is printed below the histogram. Each parameter listing includes a 1) verbal description of the parameter, 2) its symbol according to the terminology in the Supplemental Methods, 3) the section number where it is referenced, and 4) whether it was positively (“+”) or negatively (“−”) correlated with performance. In addition, the parameters were divided into three rough conceptual groups and were color-coded accordingly: Filtering (green), Normalization/Activation/Pooling (red), and Learning (blue). Beneath the bin corresponding to significantly predictive parameters, a bar plot showing the fraction of each group found in the set of significant parameters. The expected fraction, if the parameters were distributed randomly, is shown as a dotted line. Activation/Normalization/Pooling parameters were slightly over-represented in the set of significantly-predictive parameters, but no group was found to be significantly over- or under-represented (p = 0.338; Fischer's exact test).
https://doi.org/10.1371/journal.pcbi.1000579.s009
(2.28 MB TIF)
Figure S10.
How similar are the top models? (A) Model similarity on the basis of parameter values (L0 or Hamming Distance). Each model is specified by a vector of 52 parameter values. As a first attempt at comparing models, we generated an expanded binary parameter vector in which every possible parameter/value combination was represented as a separate variable (e.g. a parameter ω that can take on values 3, 5, and 7 would be included in the expanded vector as three binary values [ω = 3], [ω = 5], and [ω = 7]). The Hamming distance distance between any two vectors can then serve as a metric of the similarity between any two models. In order to determine if the top five models taken from the “Law and Order” petri dish were more similar to each than would be expected of five randomly selected models, we computed the median pairwise Hamming distance between the top five models, and between a random sampling of 100,000 sets of five models taken from the remaining (non-top-five) models. The distribution of randomly selected model pairs is shown in (A), and the observed median distance amongst the top five models is indicated by an arrow. The top-five models tended to be more similar to one another than to a random selection of models from the full population, but this effect was not significant (p = 0.136; permutation test). (B) Model similarity on the basis of output (“Representation” similarity). As another way to compare model similarity, for each model we computed model output vectors for a selection of 600 images taken from the Screening task image sets. We then computed the L2 (Euclidean) distance matrix between these “re-represented” image vectors as a proxy for the structure of the output space of each model. A distance metric between any two models was then defined as the L2 distance between the unrolled upper-diagonal portion of the two models' similarity matrices (this distance metric is similar to the Frobenius norm). Finally, as in (A), the median distances between the top five models and between a collection of 10,000 randomly drawn sets of five models were computed. The histogram in (B) shows the distribution of median distances from randomly drawn sets of five models, and the arrow indicates the median distance observed in the top-five set. As in (A), the top-five models tended to be more similar to one another (lower distance), but this effect was not significant (p = 0.082; permutation test).
https://doi.org/10.1371/journal.pcbi.1000579.s010
(6.31 MB TIF)
Search Space of Candidate Models.
https://doi.org/10.1371/journal.pcbi.1000579.s011
(0.14 MB PDF)
Technical Details of the Computational Framework.
https://doi.org/10.1371/journal.pcbi.1000579.s012
(0.08 MB PDF)
First-Order Analyses of Model Parameters and Behavior.
https://doi.org/10.1371/journal.pcbi.1000579.s013
(0.05 MB PDF)
Acknowledgments
We would like to thank Tomaso Poggio and Thomas Serre for helpful discussions; Roman Stanchak, Youssef Barhomi and Jennie Deutsch for technical assistance, and Andreas Klöckner for supporting PyCUDA. Hardware support was generously provided by the NVIDIA Corporation.
Author Contributions
Conceived and designed the experiments: NP JJD DDC. Performed the experiments: NP. Analyzed the data: NP. Contributed reagents/materials/analysis tools: NP DD. Wrote the paper: NP JJD DDC.
- View Article
- Google Scholar
- 5. Rolls ET, Deco G (2002) Computational neuroscience of vision. Oxford University Press New York.
- 6. Haykin S (1998) Neural Networks: A Comprehensive Foundation. Prentice Hall PTR Upper Saddle River, NJ, USA.
- 10. Pinto N, DiCarlo JJ, Cox DD (2008) Establishing Good Benchmarks and Baselines for Face Recognition. European Conference on Computer Vision (ECCV).
- 12. Pinto N, DiCarlo JJ, Cox DD (2009) How far can you get with a modern face recognition test set using only simple features? Computer Vision and Pattern Recognition Conference (CVPR).
- 26. PyCUDA http://mathema.tician.de/software/pycuda .
- 27. Gross R, Matthews I, Cohn J, Kanade T, Baker S (2007) The CMU multi-pose, illumination, and expression (Multi-PIE) face database. Technical report, Tech. rep., Robotics Institute, Carnegie Mellon University, 2007. TR-07-08.
- 29. Griffin G, Holub A, Perona P (2007) The caltech-256 object category dataset. Technical Report 7694, California Institute of Technology. http://authors.library.caltech.edu/7694 .
- 30. Huang GB, Ramesh M, Berg T, Learned-Miller E (October 2007) Labeled Faces in the Wild: A Database for Studying Face Recognition in Unconstrained Environments. Technical report, 07–49, University of Massachusetts, Amherst.
- 31. ORL Face Set http://www.cl.cam.ac.uk/Research/DTG/attarchive/facedatabase.html .
- 32. Yale Face Set http://cvc.yale.edu .
- 33. CVL Face Set http://www.lrv.fri.uni-lj.si/facedb.html .
- 34. AR Face Set http://cobweb.ecn.purdue.edu/aleix/ar.html .
- 42. Deb K (2001) Multi-Objective Optimization Using Evolutionary Algorithms. Wiley.
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- Proc Natl Acad Sci U S A
- v.111(13); 2014 Apr 1

How biological vision succeeds in the physical world
Dale purves.
a Neuroscience and Behavioral Disorders Program, Duke-NUS Graduate Medical School, Republic of Singapore, 169857;
b Department of Neurobiology, Duke University Medical Center, Durham, NC, 27710; and
c Duke Institute for Brain Sciences, Duke University, Durham, NC, 27708
Brian B. Monson
Janani sundararajan, william t. wojtach.
Author contributions: D.P., B.B.M., J.S., and W.T.W. analyzed data and wrote the paper.
Biological visual systems cannot measure the properties that define the physical world. Nonetheless, visually guided behaviors of humans and other animals are routinely successful. The purpose of this article is to consider how this feat is accomplished. Most concepts of vision propose, explicitly or implicitly, that visual behavior depends on recovering the sources of stimulus features either directly or by a process of statistical inference. Here we argue that, given the inability of the visual system to access the properties of the world, these conceptual frameworks cannot account for the behavioral success of biological vision. The alternative we present is that the visual system links the frequency of occurrence of biologically determined stimuli to useful perceptual and behavioral responses without recovering real-world properties. The evidence for this interpretation of vision is that the frequency of occurrence of stimulus patterns predicts many basic aspects of what we actually see. This strategy provides a different way of conceiving the relationship between objective reality and subjective experience, and offers a way to understand the operating principles of visual circuitry without invoking feature detection, representation, or probabilistic inference.
In the 1960s and for the following few decades, it seemed all but certain that the rapidly growing body of information about the electrophysiological and anatomical properties of neurons in the primary visual pathway of experimental animals would reveal how the brain uses retinal stimuli to generate perceptions and appropriate visually guided behaviors ( 1 ). However, despite the passage of 50 years, this expectation has not been met. In retrospect, the missing piece is understanding how stimuli that cannot specify the properties of physical sources can nevertheless give rise to generally successful perceptions and behaviors.
The problematic relationship between visual stimuli and the physical world was recognized by Ptolemy in the 2nd century, Alhazen in the 11th century, Berkeley in the 18th century, Helmholtz in the 19th century, and many others since ( 2 – 12 ). To explain how accurate perceptions and behaviors could arise from stimuli that cannot specify their sources, Helmholtz, arguably the most influential figure over this history, proposed that observers augmented the information in retinal stimuli by making “unconscious inferences” about the world based on past experience. The idea of vision as inference has been revived in the last two decades using Bayesian decision theory, which posits that the uncertain provenance of retinal images illustrated in Fig. 1 is resolved by making use of the probabilistic relationship between image features and their possible physical sources ( 13 – 16 ).

The uncertain provenance of retinal stimuli. Images formed on the retina cannot specify physical properties such as illumination, surface reflectance, atmospheric transmittance, and the many other factors that determine the luminance values in visual stimuli. The same conflation of physical information holds for geometrical, spectral (color), and sequential (motion) stimulus properties. Thus, the behavioral significance of any visual stimulus is uncertain. Understanding how the image formation process might be inverted to recover properties of the environment under these circumstances is referred to as the inverse optics problem.
The different concept of vision we consider here is based on a more radical reading of the challenge of responding to stimuli that cannot specify the metrics of the environment ( 17 – 20 ). The central point is that because there is no biologically feasible way to solve this problem by mapping retinal image features onto real-world properties, visual systems like ours circumvent it by generating perceptions and behaviors that depend on the frequency of occurrence of biologically determined stimuli that are tied to reproductive success. In what follows, we describe how this strategy of vision operates, how it explains the anomalous way we experience the physical world, and what it implies about visual system circuitry.
Vision in Empirical Terms
Although it is often assumed that the purpose of the evolved properties of the eye and early-level visual processing is to present stimulus features to the brain so that neural computations can recreate a representation of the environment, there is overwhelming evidence that we do not see the physical world for what it is ( 17 , 18 , 20 – 24 ). Whatever else this evidence may suggest, it indicates that to be useful, perceptions need not accord with measured reality. Indeed, generating veridical perceptions seems impossible given the uncertain significance of information conveyed by retinal stimuli ( Fig. 1 ), even when the constraints of physics that define the world are taken into account ( 10 – 12 ).
In terms of neo-Darwinian evolution, however, a visual strategy that can circumvent the inverse optics problem and explain why perceptions differ from the measured properties of the world is straightforward. Random changes in the structure and function of visual systems in ancestral forms would be favored by natural selection according to how well the ensuing percepts guided behaviors that promoted reproductive success. Any configuration of an eye and/or neural circuitry that strengthened the empirical link between visual stimuli and useful behavior would tend to increase in the population, whereas less beneficial ocular properties and circuit configurations would not. As a result, both perceptions and, ultimately, behaviors would depend on previously instantiated neural circuitry that promoted reproductive success; consequently, the recovery or representation of the actual properties of the world would be unnecessary.
Stimulus Biogenesis
The key to understanding how and why this general strategy explains the anomalous way we perceive the world when the properties of objects cannot be directly determined is recognizing that visual stimuli are not the passive result of physics or the statistics of physical properties in the environment, but are actively created according to their influence on reproductive success.
In contrast to the intuition that vision begins with a retinal image that is then processed and eventually represented in the visual brain according to a series of more-or-less logical steps, in the present argument the retinal image is just one of a series of stages in the biological transformation of disordered photon energy that begins at the corneal surface and continues in the processing carried out by the retina, thalamus, and cortex. In this framework, the “visual stimulus” is defined by the transformation of information by a recurrent network of ascending and descending connections, where the instrumental goal of generating perceptions and behaviors that work is met despite the absence of information about the actual properties of the world in which the animal must survive. Thus, although visual stimuli are usually taken to be images determined by the physical environment, they are better understood as determined by the biological properties of the eye and the rest of the visual system.
Many of these properties are already well known. For a visual stimulus to exist, photons must first be transformed into a topographical array ordered by the evolved properties of the eye. The evolved preneural properties that accomplish this are the dimensions of the eye, the shape and refractive index of the cornea, the dynamic characteristics of the lens, and the properties of ocular media, all of which serve to filter and focus photons impinging on a small region of the corneal surface. This process is continued by an arrangement of photoreceptors that restricts transduction to a limited range of photon energies, and the chain of early-level neural receptive field properties that continue to transform the biologically crafted input at the level of the retina. Although the nature of neural processing is less clear as one ascends in the primary visual system, enough is known about the organization of early-level receptive fields to provide a general idea of how they contribute to this overall strategy of relying on the frequency of occurrence of visual stimuli to generate successful perceptions, as described in the following section. The major role of the physical world in this understanding of vision is simply to provide empirical feedback regarding which perceptions and behaviors promoted reproductive success, and which did not.
An Example: The Perception of Lightness
To illustrate how this concept of vision works, consider the biological transformation of radiant energy into stimuli at an early stage where the preneural and neural events are best understood. Because increasing the luminance of any region of a retinal image increases the number of photons captured by the relevant photoreceptors, common sense suggests that physical measurements of light intensity and its perceived lightness should be proportional, and that two regions returning the same amount of light should appear to be equally light or dark. Perceptions of lightness, however, do not meet these expectations: In psychophysical experiments, the apparent lightness elicited by the luminance values at any particular region of a retinal image is clearly nonlinear and depends heavily on the surrounding luminance values ( 20 , 21 , 24 ).
To understand the significance of these discrepancies, take a typical luminance pattern on the retina arising from photons that are ordered by the evolved properties of the eye. For all intents and purposes, an image such as the example in Fig. 2 A will have occurred only once; it is highly unlikely that the retina of an observer would ever again be activated by exactly the same pattern of luminance values falling on the same topographical array of millions of receptors. Because patterns like this are effectively unique, even a large catalog of such images would be of little or no help in promoting useful visual behavior on an empirical (trial and error) basis. However, smaller regions of the image, such as those sampled by the templates in Fig. 2 A , would have occurred more than once, some many times, as shown by the distributions in Fig. 2 B .

Accumulated human experience with luminance patterns. ( A ) To evaluate the concept that perception arises as a function of accumulated experience over evolutionary time, calibrated digital photographs can be sampled with templates about the size of visual receptive fields to measure how often different patterns of luminance occur in visual stimuli. ( B ) By repeated sampling, the frequency of occurrence of the luminance of any target region in a pattern of luminance values (indicated by a question mark) can be represented as a frequency distribution. The frequency of occurrence of the central region’s luminance is different in the two surrounds, as would be true for any other pattern of luminance values assessed in this way. (The background image in A is from ref. 50 ; the data in B are after ref. 27 ).
There is, of course, a lower limit to the size of samples that would be useful. If, for example, the size of the sample were reduced to a single point, the frequency of occurrence of the “pattern” would be maximal, but the resulting perceptions and behaviors would be based on a minimum of information. The greatest biological success would presumably arise from frequently occurring samples that comprised relatively small patterns in which the responses of the relevant neurons used information supplied by both the luminance value at any point and a tractable number of surrounding luminance values. This arrangement corresponds to the way retinal images are in fact processed by the receptive fields of early-level visual neurons, which, in the central vision of rhesus macaques (and presumably humans), are on the order of a degree or less of visual arc ( 25 , 26 )—roughly the size of the templates used in Fig. 2 A .
To explore the merits of this concept of vision, templates like those in Fig. 2 A can be used to sample the patterns that are routinely processed at the early stages of the visual pathway (the information extracted at other stages would, in principle, work as well). If perceptions of lightness indeed depend on the frequency of occurrence of small patterns of luminance values, then these data should predict what we see. One way of representing the frequency of occurrence of such stimuli is by transforming the distributions in Fig. 2 B into cumulative distribution functions, thereby allowing the target luminance values in different surrounds to be ranked relative to one another ( Fig. 3 ). In this way, the lightness values that would be elicited by the luminance value of any region of a pattern in the context of surrounding luminance values can be specified. In the present concept of vision, the differences in these ranks account for the perceived differences in lightness of the identical target luminance values in Fig. 3 .

Predicting lightness percepts based on the frequency of occurrence of stimulus patterns. The frequency distributions from Fig. 2 B are here transformed to distribution functions that indicate the cumulative frequency of occurrence of the central target luminance given the luminance of the ( Inset ) surround. The dashed lines show the percentile rank of a specific central luminance value ( T ) in each distribution. As Insets show, central squares with identical photometric values elicit different lightness percepts (called “simultaneous lightness contrast”) predicted by their relative rankings (relative frequencies of occurrence).
Similar analyses have been used to explain not only the perception of simple luminance patterns like those in Figs. 2 and and3 3 but also perceptions elicited by a variety of complex luminance patterns ( 20 , 27 ), geometrical patterns ( 18 ), spectral patterns ( 28 ), and moving stimuli ( 29 , 30 ). In addition, artificial neural networks that evolve on the basis of ranking the frequency of luminance patterns can rationalize major aspects of early-level receptive field properties in experimental animals ( 31 , 32 ).
Why Stimulus Frequency Predicts Perception and Behavior
Missing from this account, however, is why the frequencies of occurrence of visual stimuli sampled in this way predict perception. The reason, we maintain, is that the relative number of times biologically generated patterns are transduced and processed in accumulated experience tracks reproductive success. In Fig. 3 , for example, the frequencies of occurrence of the patterns at the stage of photoreception have caused the central luminance value to occur more often when in the lower luminance surround than in the higher one, resulting in a steeper slope at that point on the cumulative distribution function. If the relative ranking along this function corresponds to the perception of lightness, then the higher the rank of a target luminance (T) in a given surround relative to another target luminance with the same surround, the lighter the target should appear. Therefore, because the target luminance in a darker surround ( Fig. 3 , Left ) has a higher rank than the same target luminance in a lighter surround ( Fig. 3 , Right ), the former should be seen as lighter than the latter, as it is. Because the frequency of occurrence of patterns is an evolved property—and because these relative rankings along the function correspond to perception—the visually guided behaviors that result will in varying degrees have contributed to reproductive success. Thus, by aligning the frequencies of occurrence of light patterns over evolutionary time with perceptions of light and dark and the behaviors they elicit, this strategy can explain vision without solving the inverse optics problem.
Visual Perception on This Basis
Despite the inclination to do so, it would be misleading to imagine that the perceptions predicted by the relative ranking of luminance or other patterns depend on information about the “statistics of the environment.” It is, of course, true that because physical objects tend to be uniform in their local composition, nearby luminance values in evolved retinal image patterns tend to be similar; indeed, the work of Brünswik ( 4 ) and, later, Gibson ( 33 ), which focused on how constraints of the environment might be conveyed in the structure of images, relied on this and other statistical information to explain vision. However, as illustrated in Fig. 1 , the relationship between properties of the physical world and retinal images conflates such information, undermining strategies that rely on statistical features of the environment to explain perception.
Although circumventing the inverse problem empirically gives the subjective impression that we perceive the actual properties of objects and conditions in the world, this is not the case. Nor does responding to luminance values (or other image attributes) according to the frequency of occurrence of local patterns reveal reality or bring subjective values “closer” to objective ones. It therefore follows that these discrepancies between lightness and luminance—or any other visual qualities and their physical correlates—are not “illusions” ( 22 , 23 ) but simply signatures of the strategy we and, presumably, other visual animals have evolved to promote useful behaviors despite the inability of biological visual systems to measure physical parameters.
In sum, successful perceptions and behavior arise not because the actual properties of the world are recovered from images, but because the perceptual values assigned by the frequency of occurrence of visual stimuli accord with the reproductive success of the species and individual. As a result, the visual qualities that we see are better understood as signifying perceptions and behaviors that led to reproductive success in the past rather than encoding information, statistical or otherwise, about the world in the present.

Other Interpretations of Vision
What, then, can be said about other concepts of vision, and how they compare with the strategy of vision presented here? Three current frameworks are considered: vision as detecting and representing image features, vision as probabilistic inference, and vision as efficient coding.
Vision as Feature Detection and Representation.
An early and still widely accepted idea is that visual (and other) sensory systems operate analytically, detecting behaviorally important features in retinal images that are then used to construct neural representations of the world at the level of the visual cortex. This interpretation of visual processing accords with electrophysiological evidence that demonstrates the selectivity of neuronal receptive fields, as well as with the compelling impression that what we see is external reality. Although attractive on these grounds, this interpretation of vision is ruled out by the inability of the visual system to measure the physical parameters of the world ( Fig. 1 ), as well as its inability to explain a host of phenomena in luminance, color, form, distance, depth, and motion psychophysics on this basis ( 20 ).
Vision as Probabilistic Inference.
More difficult to assess is the idea that vision is based on a strategy of probabilistic inference. Helmholtz introduced the idea of unconscious inference in the 19th century to explain how vision might improve responses to retinal images that he took to be inherently inadequate stimuli ( 3 ). In the first half of the 20th century, visual inferences were conceived in terms of gestalt laws or other heuristics. More recently, many mathematical psychologists and computer scientists have endorsed the idea of vision as statistical inference by proposing that images map back onto the properties of objects and conditions in the world as Bayesian probabilities ( 13 , 15 , 16 , 34 – 37 ).
Bayes’ theorem ( 38 ) states that the probability of a conditional inference about A given B being true (the posterior probability) is determined by the probability of B given A (the likelihood function) multiplied by the ratio of the independent probabilities of A (the prior probability) and B. This way of making rational predictions in the face of uncertainty is widely and successfully used in applications ranging from weather forecasting and medical diagnosis to poker and sports betting.
The value of Bayes’ theorem as a tool to understand vision, however, is another matter. To be biologically useful, the posterior probability would have to indicate the probability of a property of the world (e.g., surface reflectance or illumination values) underlying a given visual stimulus. This, in turn, would depend on the probability of the visual stimulus given the physical property (the likelihood) and the prior probability of that state of the world. Although this approach is logical, information about the likelihood and prior probabilities is simply not available to the visual system given the inverse problem, thereby negating the biological feasibility of this explanation. In contrast, the empirical concept of vision described here avoids these problems by pursuing a different goal: fomenting reproductive success despite an inability to recover properties of the physical world in which behavior must take place. Although the frequency of occurrence of stimuli is often used to infer the probability of an underlying property of the physical world given an image, no such inferences are being made in this empirical strategy. Nor does the approach rely on a probabilistic solution: The biologically determined frequency of occurrence of visual stimuli simply generates useful perceptions and behaviors according to reproductive success.
These reservations add to other criticisms of Bayesian decision theory applied to cognitive issues, and to neuroscience generally ( 39 , 40 ).
Vision as Efficient Coding.
Another popular framework for understanding vision and its underlying circuitry is efficient coding ( 5 , 41 – 45 ). A code is a rule for converting information from one form to another. In vision, coding is understood as the conversion of retinal stimulus patterns into the electrochemical signals (receptor, synaptic and action potentials) used for communication with the rest of the brain; this information is then taken to be decoded by further computational processes to achieve perceptual and behavioral effects. Given the nature of sensory transduction and the distribution of peripheral sensory effects to distant sites by action potentials, coding for the purpose of neural computation seems an especially apt metaphor, and has been widely accepted ( 44 , 46 , 47 ).
Such approaches variously interpret visual circuits as carrying out optimal coding procedures based on minimizing energy use ( 5 , 42 , 43 , 48 – 50 ), making accurate predictions ( 51 – 53 ), eliminating redundancy ( 54 ), or normalizing information ( 55 , 56 ). The common theme of these overlapping ideas is that optimizing information transfer by minimizing redundancy, lowering wiring costs, and/or maximizing the entropy of sensory outputs will all have been advantageous to visual animals ( 57 ).
The importance of efficiency (whether in coding or otherwise) is clearly a factor in any evolutionary process, and the importance of these several ways of achieving it is not in doubt. However, generating perceptions by means of circuitry that contends with a world whose physical parameters cannot be measured by biological vision is a different goal, in much the same way that the goals of any organ system differ from the concurrent need to achieve them as efficiently as possible. Thus, these efforts are not explanations of visual perception, which no more depends on efficiency than the meaning of a verbal message depends on how efficiently it is transmitted.
Implications for Future Research
Given the central role it has played in modern neuroscience, the way scientists conceive vision is broadly relevant to the future direction of brain research, its potential benefits, and its economic value. An issue much debated at present is the intention to invest heavily over the coming decade in a complete analysis of human brain connectivity at both macroscopic and microscopic levels ( 58 – 60 ) (also http://blogs.nature.com/news/2013/04/obama-launches-ambitious-brain-map-project-with-100-million.html , accessed February 24, 2014). The impetus for this initiative is largely based on the success of the human genome project in scientific, health, technical, and financial terms. To underscore this parallel, the goal of the project is referred to as obtaining the “brain connectome.”
Although neuroscientists rightly applaud this investment in better understanding brain connectivity, the related technology and possible health benefits, a weakness in the comparison with the human genome project (and with genetics in general) is that the basic functional and structural principles of genes were already well established at the outset. In contrast, the principles underlying the structure and function of the human brain and its component circuits remain unknown. Indeed, the stated aim of the brain connectome project is the hope that additional anatomical information will help establish these principles.
Given this goal, the operation of the visual system—the brain region about which most is now known—is especially relevant. If the function of visual circuitry, a presumptive bellwether for operations in the rest of the brain, has been determined by evolutionary and individual history rather than by logical “design” principles, then understanding function by examining brain connectivity may be far more challenging than imagined. Perhaps the most daunting obstacle is that reproductive success—the driver of any evolved strategy of vision—is influenced by a very large number of factors, many of which will be difficult to discern, let alone quantify. Thus, the relation between accumulated experience and reproductive success may never be specified in more than qualitative or semiquantitative terms.
In light of these obstacles, it may be that the best way to understand the principles underlying neural connectivity is to evolve increasingly complex networks in progressively more realistic environments. Until relatively recently, pursuing this goal would have been fanciful. However, the advent of genetic and other computer algorithms has made evolving artificial neural networks in simple environments relatively easy ( 31 , 32 ). This approach should eventually be able to link evolved visual functions and their operating principles with the wealth of detail already known from physiological and anatomical studies over the last 50 y.
A central challenge in understanding vision is that biological visual systems cannot measure or otherwise access the properties of the physical world. We have argued that vision like ours addresses this challenge by evolving the ability to form and transduce small, biologically determined image patterns whose frequencies of occurrence directly link perceptions and behaviors with reproductive success. In this way, perceptions and behaviors come to work in the physical world without sensory measurements of the environment, and without inferences or the complex computations that are often imagined. As a result, however, vision does not accord with reality but with perceptions and behaviors that succeed in a world whose actual properties are not revealed. This framework for vision, supported by evidence from human psychophysics and predictions of perceptions based on accumulated experience (i.e., the frequency of occurrence of biogenic stimuli), implies that Gustav Fechner’s goal of understanding the relationship between objective (physical) and subjective (psychological) domains ( 61 ) can be met if pursued in these biological terms rather than in the statistical, logical, and computational terms that are more appropriate to physics, mathematics, and algorithm-based computer science. Although it may not be easy to relate this understanding of vision to higher-order tasks such as object recognition, if the argument here is correct, then all further uses of visual information must be built up from the way we see these foundational qualities.
Acknowledgments
We are grateful for helpful criticism from Dan Bowling, Jeff Lichtman, Yaniv Morgenstern, and Cherlyn Ng.
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
arXiv's Accessibility Forum starts next month!
Help | Advanced Search
Computer Science > Machine Learning
Title: time to augment self-supervised visual representation learning.
Abstract: Biological vision systems are unparalleled in their ability to learn visual representations without supervision. In machine learning, self-supervised learning (SSL) has led to major advances in forming object representations in an unsupervised fashion. Such systems learn representations invariant to augmentation operations over images, like cropping or flipping. In contrast, biological vision systems exploit the temporal structure of the visual experience during natural interactions with objects. This gives access to "augmentations" not commonly used in SSL, like watching the same object from multiple viewpoints or against different backgrounds. Here, we systematically investigate and compare the potential benefits of such time-based augmentations during natural interactions for learning object categories. Our results show that time-based augmentations achieve large performance gains over state-of-the-art image augmentations. Specifically, our analyses reveal that: 1) 3-D object manipulations drastically improve the learning of object categories; 2) viewing objects against changing backgrounds is important for learning to discard background-related information from the latent representation. Overall, we conclude that time-based augmentations during natural interactions with objects can substantially improve self-supervised learning, narrowing the gap between artificial and biological vision systems.
| Comments: | 20 pages |
| Subjects: | Machine Learning (cs.LG); Computer Vision and Pattern Recognition (cs.CV) |
| Cite as: | [cs.LG] |
| (or [cs.LG] for this version) | |
| Focus to learn more arXiv-issued DOI via DataCite |
Submission history
Access paper:.
- Other Formats
References & Citations
- Google Scholar
- Semantic Scholar
BibTeX formatted citation
Bibliographic and Citation Tools
Code, data and media associated with this article, recommenders and search tools.
- Institution
arXivLabs: experimental projects with community collaborators
arXivLabs is a framework that allows collaborators to develop and share new arXiv features directly on our website.
Both individuals and organizations that work with arXivLabs have embraced and accepted our values of openness, community, excellence, and user data privacy. arXiv is committed to these values and only works with partners that adhere to them.
Have an idea for a project that will add value for arXiv's community? Learn more about arXivLabs .

- Research Article
- Neuroscience
Factorized visual representations in the primate visual system and deep neural networks
- Jack W Lindsey
- Zuckerman Mind Brain Behavior Institute, Columbia University, United States ;
- Department of Neuroscience, Columbia University, United States ;
- Open access
- Copyright information
Share this article
Cite this article.
- Elias B Issa
- Copy to clipboard
- Download BibTeX
- Download .RIS
eLife assessment
Elife digest, introduction, data availability, article and author information.
Object classification has been proposed as a principal objective of the primate ventral visual stream and has been used as an optimization target for deep neural network models (DNNs) of the visual system. However, visual brain areas represent many different types of information, and optimizing for classification of object identity alone does not constrain how other information may be encoded in visual representations. Information about different scene parameters may be discarded altogether (‘invariance’), represented in non-interfering subspaces of population activity (‘factorization’) or encoded in an entangled fashion. In this work, we provide evidence that factorization is a normative principle of biological visual representations. In the monkey ventral visual hierarchy, we found that factorization of object pose and background information from object identity increased in higher-level regions and strongly contributed to improving object identity decoding performance. We then conducted a large-scale analysis of factorization of individual scene parameters – lighting, background, camera viewpoint, and object pose – in a diverse library of DNN models of the visual system. Models which best matched neural, fMRI, and behavioral data from both monkeys and humans across 12 datasets tended to be those which factorized scene parameters most strongly. Notably, invariance to these parameters was not as consistently associated with matches to neural and behavioral data, suggesting that maintaining non-class information in factorized activity subspaces is often preferred to dropping it altogether. Thus, we propose that factorization of visual scene information is a widely used strategy in brains and DNN models thereof.
The study makes a valuable empirical contribution to our understanding of visual processing in primates and deep neural networks, with a specific focus on the concept of factorization. The analyses provide convincing evidence that high factorization scores are correlated with neural predictivity. This work will be of interest to systems neuroscientists studying vision and could inspire further research that ultimately may lead to better models of or a better understanding of the brain.
- Read the peer reviews
- About eLife assessments
When looking at a picture, we can quickly identify a recognizable object, such as an apple, applying a single word label to it. Although extensive neuroscience research has focused on how human and monkey brains achieve this recognition, our understanding of how the brain and brain-like computer models interpret other complex aspects of a visual scene – such as object position and environmental context – remains incomplete.
In particular, it was not clear to what extent object recognition comes at the expense of other important scene details. For example, various aspects of the scene might be processed simultaneously. On the other hand, general object recognition may interfere with processing of such details.
To investigate this, Lindsey and Issa analyzed 12 monkey and human brain datasets, as well as numerous computer models, to explore how different aspects of a scene are encoded in neurons and how these aspects are represented by computational models. The analysis revealed that preventing effective separation and retention of information about object pose and environmental context worsened object identification in monkey cortex neurons. In addition, the computer models that were the most brain-like could independently preserve the other scene details without interfering with object identification.
The findings suggest that human and monkey high level ventral visual processing systems are capable of representing the environment in a more complex way than previously appreciated. In the future, studying more brain activity data could help to identify how rich the encoded information is and how it might support other functions like spatial navigation. This knowledge could help to build computational models that process the information in the same way, potentially improving their understanding of real-world scenes.
Artificial deep neural networks (DNNs) are the most predictive models of neural responses to images in the primate high-level visual cortex ( Cadieu et al., 2014 ; Schrimpf et al., 2020 ). Many studies have reported that DNNs trained to perform image classification produce internal feature representations broadly similar to those in areas V4 and IT of the primate cortex, and that this similarity tends to be greater in models with better classification performance ( Yamins et al., 2014 ). However, it remains opaque what aspects of the representations of these more performant models drive them to better match neural data. Moreover, beyond a certain threshold level of object classification performance, further improvement fails to produce a concomitant improvement in predicting primate neural responses ( Schrimpf et al., 2020 ; Nonaka et al., 2021 ; Linsley, 2023 ). This weakening trend motivates finding new normative principles, besides object classification ability, that push models to better match primate visual representations.
One strategy for achieving high object classification performance is to form neural representations that discard some (are tolerant to) or all (are invariant to) information besides object class. Invariance in neural representations is in some sense a zero-sum strategy: building invariance to some parameters improves the ability to decode others. We also note that our use of ‘invariance’ in this context refers to invariance in neural representations, rather than behavioral or perceptual invariance ( DiCarlo and Cox, 2007 ). However, high-level cortical neurons in the primate ventral visual stream are known to simultaneously encode many forms of information about visual input besides object identity, such as object pose ( Freiwald and Tsao, 2010 ; Hong et al., 2016 ; Kravitz et al., 2013 ; Peters and Kriegeskorte, 2021 ). In this work, we seek to characterize how the brain simultaneously represents different forms of information.
In particular, we introduce methods to quantify the relationships between different types of visual information in a population code (e.g., object pose vs. camera viewpoint), and specifically the degree to which different forms of information are ‘factorized’. Intuitively, if the variance driven by one parameter is encoded along orthogonal dimensions of population activity space compared to the variance driven by other scene parameters, we say that this representation is factorized. We note that our definition of factorization is closely related to the existing concept of manifold disentanglement ( DiCarlo and Cox, 2007 ; Chung et al., 2018 ) and can be seen as a generalization of disentanglement to high-dimensional visual scene parameters like object pose. Factorization can enable simultaneous decoding of many parameters at once, supporting diverse visually guided behaviors (e.g., spatial navigation, object manipulation, or object classification) ( Johnston and Fusi, 2023 ).
Using existing neural datasets, we found that both factorization of and invariance to object category and position information increase across the macaque ventral visual cortical hierarchy. Next, we leveraged the flexibility afforded by in silico models of visual representations to probe different forms of factorization and invariance in more detail, focusing on several scene parameters of interest: background content, lighting conditions, object pose, and camera viewpoint. Across a broad library of DNN models that varied in their architecture and training objectives, we found that factorization of all of the above scene parameters in DNN feature representations was positively correlated with models’ matches to neural and behavioral data. Interestingly, while neural invariance to some scene parameters (background scene and lighting conditions) predicted neural fits, invariance to others (object pose and camera viewpoint) did not. Our results generalized across both monkey and human datasets using different measures (neural spiking, fMRI, and behavior; 12 datasets total) and could not be accounted for by models’ classification performance. Thus, we suggest that factorized encoding of multiple behaviorally relevant scene variables is an important consideration, alongside other desiderata such as classification performance, in building more brain-like models of vision.
Disentangling object identity manifolds in neural population responses can be achieved by qualitatively different strategies. These include building invariance of responses to non-identity scene parameters (or, more realistically, partial invariance; DiCarlo and Cox, 2007 ) and/or factorizing non-identity-driven response variance into isolated (factorized) subspaces ( Figure 1A , left vs. center panels, cylindrical/spherical-shaded regions represent object manifolds). Both strategies maintain an ‘identity subspace’ in which object manifolds are linearly separable. In a non-invariant, non-factorized representation, other variables like camera viewpoint also drive variance within the identity subspace, ‘entangling’ the representations of the two variables ( Figure 1A , right; viewpoint-driven variance is mainly in identity subspace, orange flat-shaded region).
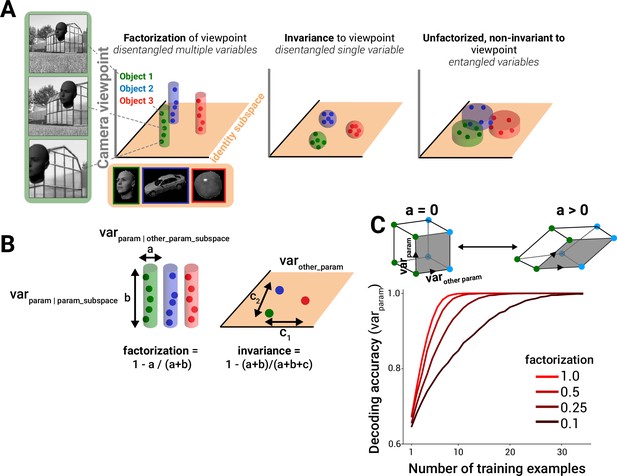
Framework for quantifying factorization in neural and model representations.
( A ) A subspace for encoding a variable, for example, object identity, in a linearly separable manner can be achieved by becoming invariant to non-class variables (compact spheres, middle column, where the volume of the sphere corresponds to the degree of neural invariance, or tolerance, for non-class variables; colored dots represent example images within each class) and/or by encoding variance induced by non-identity variables in orthogonal neural axes to the identity subspace (extended cylinders, left column). Only the factorization strategy simultaneously represents multiple variables in a disentangled fashion. A code that is sensitive to non-identity parameters within the identity subspace corrupts the ability to decode identity (right column) (identity subspace denoted by orange plane). ( B ) Variance across images within a class can be measured in two different linear subspaces: that containing the majority of variance for all other parameters ( a, ‘other_param_subspace’ ) and that containing the majority of the variance for that parameter ( b, ‘param_subspace’ ). Factorization is defined as the fraction of parameter-induced variance that avoids the other-parameter subspace (left). By contrast, invariance to the parameter of interest is computed by comparing the overall parameter-induced variance to the variance in response to other parameters ( c, ‘var_other_param’ ) (right). ( C ) In a simulation of coding strategies for two binary variables out of 10 total dimensions that are varying (see ‘Methods ’ ), a decrease in orthogonality of the relationship between the encoding of the two variables (alignment a > 0, or going from a square to a parallelogram geometry), despite maintaining linear separability of variables, results in poor classifier performance in the few training-samples regime when i.i.d. Gaussian noise is present in the data samples (only 3 of 10 dimensions used in simulation are shown).
To formalize these different representational strategies, we introduced measures of factorization and invariance to scene parameters in neural population responses ( Figure 1B ; see Equations 2–4 in ‘Methods’). Concretely, invariance to a scene variable (e.g., object motion) is computed by measuring the degree to which varying that parameter alone changes neural responses, relative to the changes induced by varying other parameters (lower relative influence on neural activity corresponds to higher invariance, or tolerance, to that parameter). Factorization is computed by identifying the axes in neural population activity space that are influenced by varying the parameter of interest and assessing how much it overlaps the axes influenced by other parameters (‘ a’ in Figure 1B and C ; lower overlap corresponds to higher factorization). We quantified this overlap in two different ways (‘principal components analysis (PCA)-based’ and ‘covariance-based’ factorization, corresponding to Equations 2 and 4 in ‘Methods’), which produced similar results when compared in subsequent analyses (unless otherwise noted, factorization scores will generally refer to the PCA-based method, and the covariance method is shown in Figures 5–7 for comparison). Intuitively, a neural population in which one neural subpopulation encodes object identity and another separate subpopulation encodes object position exhibits a high degree of factorization of those two parameters (however, note that factorization may also be achieved by neural populations with mixed selectivity in which the ‘subpopulations’ correspond to subspaces, or independent orthogonal linear projections, of neural activity space rather than physical subpopulations). Though the example presented in Figure 1 focused on factorization of and invariance to object identity versus non-identity variables, we stress that our definitions can be applied to any scene variables of interest. Furthermore, we presented a simplified visual depiction of the geometry within each scene variable subspace in Figure 1 . We emphasize that our factorization metric does not require a particular geometry within a variable’s subspace, whether parallel linearly ordered coding of viewpoint as in the cylindrical class manifolds shown in Figure 1A and B , or a more complex geometry where there is a lack of parallelism and/or a more nonlinear layout.
While factorization and invariance are not mutually exclusive representational strategies, they are qualitatively different. Factorization, unlike invariance, has the potential to enable the simultaneous representation of multiple scene parameters in a decodable fashion. Intuitively, factorization increases with higher dimensionality as this decreases overlap, all other things being equal (in the limit, the angle between points will approach 90 o or a fully orthogonal code in high dimensions), and for a given finite, fixed dimension, factorization is mainly driven by the angle between this dimension and the other variable subspaces which measures the degree of contamination ( Figure 1C ; square vs. parallelogram). In a simulation, we found that the extent to which the variables of interest were represented in a factorized way (i.e., along orthogonal axes, rather than correlated axes) influenced the ability of a linear discriminator to successfully decode both variables in a generalizable fashion from a few training samples ( Figure 1C ).
Given the theoretically desirable properties of factorized representations, we next asked whether such representations are observed in neural data, and how much factorization contributes empirically to downstream decoding performance in real data. Specifically, we took advantage of an existing dataset in which the tested images independently varied object identity versus object pose plus background context ( Majaj et al., 2015 ; https://github.com/brain-score/vision/blob/master/examples/data_metrics_benchmarks.ipynb ). We found that both V4 and IT responses exhibited more significant factorization of object identity information from non-identity information than a shuffle control (which accounts for effects on factorization due to dimensionality of these regions) ( Figure 2—figure supplement 1 ; see ’Methods’). Furthermore, the degree of factorization increased from V4 to IT ( Figure 2A ). Consistent with prior studies, we also found that invariance to non-identity information increased from V4 to IT in our analysis ( Figure 2A , right, solid lines; Rust and DiCarlo, 2010 ). Invariance to non-identity information was even more pronounced when measured in the subspace of population activity capturing the bulk (90%) of identity-driven variance as a consequence of increased factorization of identity from non-identity information ( Figure 2A , right, dashed lines).
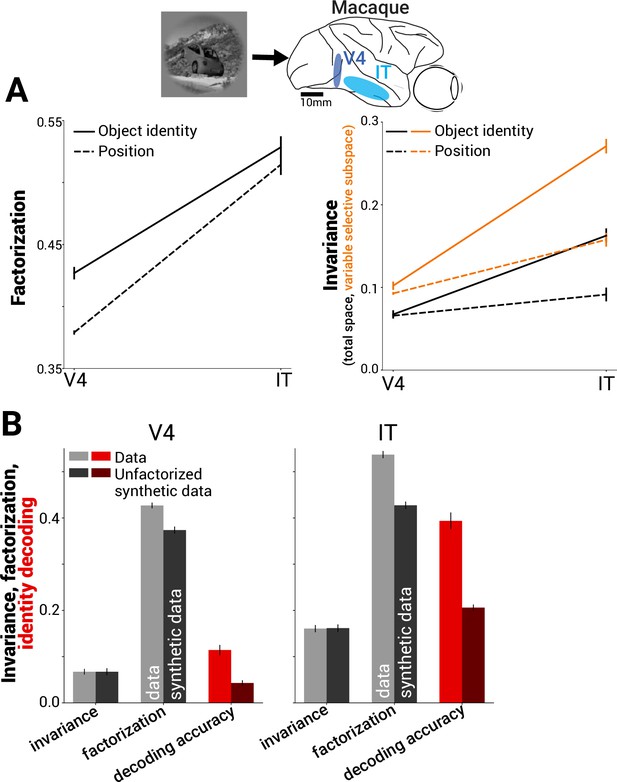
Benefit of factorization to neural decoding in macaque V4 and IT.
( A ) Factorization of object identity and position increased from macaque V4 to IT (PCA-based factorization, see ‘Methods’; dataset E1 – multiunit activity in macaque visual cortex) (left). Like factorization, invariance also increased from V4 to IT (note, ‘identity’ refers to invariance to all non-identity position factors, solid black line) (right). Combined with increased factorization of the remaining variance, this led to higher invariance within the variable’s subspace (orange lines), representing a neural subspace for identity information with invariance to nuisance parameters which decoders can target for read-out. ( B ) An experiment to test the importance of factorization for supporting object class decoding performance in neural responses. We applied a transformation to the neural data (linear basis rotation) that rotated the relative positions of mean responses to object classes without changing the relative proportion of within- vs. between-class variance (Equation 1 in ’Methods’). This transformation preserved invariance to non-class factors (leftmost pair of bars in each plot), while decreasing factorization of class information from non-class factors (center pair of bars in each plot). Concurrently, it had the effect of significantly reducing object class decoding performance (light vs. dark red bars in each plot, chance = 1/64; n = 128 multi-unit sites in V4 and 128 in IT).
To illustrate the beneficial effect of factorization on decoding performance, we performed a statistical lesion experiment that precisely targeted this aspect of representational geometry. Specifically, we analyzed a transformed neural representation obtained by rotating the population data so that inter-class variance more strongly overlapped with the principal components (PCs) of the intra-class variance in the data (see Equation 1 in ’Methods’). Note that this transformation, designed to decrease factorization, acts on the angle between latent variable subspaces. The applied linear basis rotation leaves all other activity statistics completely intact (such as mean neural firing rates, covariance structure of the population, and its invariance to non-class variables) yet has the effect of strongly reducing object identity decoding performance in both V4 and IT ( Figure 2B ). Our analysis shows that maintaining invariance alone in the neural population code was insufficient to account for a large fraction of decoding performance in high-level visual cortex; factorization of non-identity variables is key to the decoding performance achieved by V4 and IT representations.
We next asked whether factorization is found in DNN model representations and whether this novel, heretofore unconsidered metric, is a strong indicator of more brainlike models. When working with computational models, we have the liberty to test an arbitrary number of stimuli; therefore, we could independently vary multiple scene parameters at sufficient scale to enable computing factorization and invariance for each, and we explored factorization in DNN model representations in more depth than previously measured in existing neural experiments. To gain insight back into neural representations, we also assessed the ability of each model to predict separately collected neural and behavioral data. In this fashion, we may indirectly assess the relative significance of geometric properties like factorization and invariance to biological visual representations – if, for instance, models with more factorized representations consistently match neural data more closely, we may infer that those neural representations likely exhibit factorization themselves ( Figure 3 ). To measure factorization, invariance, and decoding properties of DNN models, we generated an augmented image set, based on the images used in the previous dataset ( Figure 2 ), in which we independently varied the foreground object identity, foreground object pose, background identity, scene lighting, and 2D scene viewpoint. Specifically for each base image from the original dataset, we generated sets of images that varied exactly one of the above scene parameters while keeping the others constant, allowing us to measure the variance induced by each parameter relative to the variance across all scene parameters ( Figure 3 , top left; 100 base scenes and 10 transformed images for each source of variation). We presented this large image dataset to models (4000 images total) to assess the relative degree of representational factorization of and invariance to each scene parameter. We conducted this analysis across a broad range of DNNs varying in architecture and objective as well as other implementational choices to obtain the widest possible range of DNN representations for testing our hypothesis. These included models using supervised training for object classification ( Krizhevsky et al., 2012 ; He et al., 2016 ), contrastive self-supervised training ( He et al., 2020 ; Chen et al., 2020 ), and self-supervised models trained using auxiliary objective functions ( Tian et al., 2019 ; Doersch et al., 2015 ; He et al., 2017 ; Donahue and Simonyan, 2019 ; see ’Methods’ and Supplementary file 1b ).
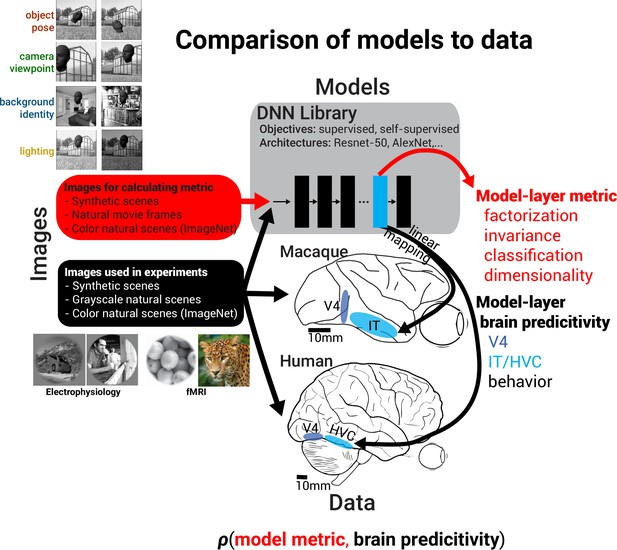
Measurement of factorization in deep neural network (DNN) models and comparison to brain data.
Schematic showing how meta-analysis on models and brain data was conducted by first computing various representational metrics on models and then measuring a model’s predictive power across a variety of datasets. For computing the representational metrics of factorization of and invariance to a scene parameter, variance in model responses was induced by individually varying each of four scene parameters (n = 10 parameter levels) for each base scene (n = 100 base scenes) (see images on the top left). The combination of model-layer metric and model-layer dataset predictivity for a choice of model, layer, metric, and dataset specifies the coordinates of a single dot on the scatter plots in Figures 4 and 7 , and the across-model correlation coefficient between a particular representational metric and neural predictivity for a dataset summarizes the potential importance of the metric in producing more brainlike models (see Figures 5 and 6 ).
First, we asked whether, in the course of training, DNN models develop factorized representations at all. We found that the final layers of trained networks exhibited consistent increases in factorization of all tested scene parameters relative to a randomly initialized (untrained) baseline with the same architecture ( Figure 4A , top row, rightward shift relative to black cross, a randomly initialized ResNet-50). By contrast, training DNNs produced mixed effects on invariance, typically increasing it for background and lighting but reducing it for object pose and camera viewpoint ( Figure 4A , bottom row, leftward shift relative to black cross for left two panels). Moreover, we found that the degree of factorization in models correlated with the degree to which they predicted neural activity for single-unit IT data ( Figure 4A , top row), which can be seen as correlative evidence that neural representations in IT exhibit factorization of all scene variables tested. Interestingly, we saw a different pattern for representational invariance to a scene parameter. Invariance showed mixed correlations with neural predictivity ( Figure 4A , bottom row), suggesting that IT neural representations build invariance to some scene information (background and lighting) but not to others (object pose and observer viewpoint). Similar effects were observed when we assessed correlations between these metrics and fits to human behavioral data rather than macaque neural data ( Figure 4B ).
Neural and behavioral predictivity of models versus their factorization and invariance properties.
( A ) Scatter plots, for example, neural dataset (IT single units, macaque E2 dataset) showing the correlation between a model’s predictive power as an encoding model for IT neural data versus a model’s ability to factorize or become invariant to different scene parameters (each dot is a different model, using each model’s penultimate layer). Note that factorization (PCA-based, see ‘Methods’) in trained models is consistently higher than that for an untrained, randomly initialized Resnet-50 DNN architecture (rightward shift relative to black cross). Invariance to background and lighting but not to object pose and viewpoint increased in trained models relative to the untrained control (rightward versus leftward shift relative to black cross). ( B ) Same as ( A ) except for human behavior performance patterns across images (human I2 dataset). Increasing scene parameter factorization in models generally correlated with better neural predictivity (top row). A noticeable drop in neural predictivity was seen for high levels of invariance to object pose (bottom row, second panel).
To assess the robustness of these findings to choice of images and brain regions used in an experiment, we conducted the same analyses across a large and diverse set of previously collected neural and behavioral datasets, from different primate species and visual regions (six macaque datasets [ Majaj et al., 2015 ; Rust and DiCarlo, 2012 ; Rajalingham et al., 2018 ]: two V4, two ITC (inferior temporal cortex), and two behavior; six human datasets [ Rajalingham et al., 2018 ; Kay et al., 2008 ; Shen et al., 2019 ]: two V4, two HVC (higher visual cortex), and two behavior; Supplementary file 1a ). Consistently, increased factorization of scene parameters in model representations correlated with models being more predictive of neural spiking responses, voxel BOLD signal, and behavioral responses to images ( Figure 5A , black bars; see Figure 4—figure supplements 1 – 3 for scatter plots across all datasets). Although invariance to appearance factors (background identity and scene lighting) correlated with more brainlike models, invariance for spatial transforms (object pose and camera viewpoint) consistently did not (zero or negative correlation values; Figure 5C , red and green open circles). Our results were preserved when we re-ran the analyses using only the subset of models with the identical ResNet-50 architecture ( Figure 5—figure supplement 1 ) or when we evaluated model predictivity using representational dissimilarity matrices of the population (RDMs) instead of linear regression (encoding) fits of individual neurons or voxels ( Figure 5—figure supplement 2 ). Furthermore, the main finding of a positive correlation between factorization and neural predictivity was robust to the particular choice of PCA threshold we used to quantify factorization ( Figure 5—figure supplement 3 ). We found similar results using a covariance-based method for computing factorization that does not have any free parameters ( Figure 5C , faded filled circles; see Equations 4 in ‘Methods’).
Scene parameter factorization correlates with more brainlike deep neural network (DNN) models.
( A ) Factorization of scene parameters in model representations computed using the PCA-based method consistently correlated with a model being more brainlike across multiple independent datasets measuring monkey neurons, human fMRI voxels, or behavioral performance in both macaques and humans (left vs. right column) (black bars). By contrast, increased invariance to camera viewpoint or object pose was not indicative of brainlike models (gray bars). In all cases, model representational metric and neural predictivity score were computed by averaging scores across the last 5 model layers. ( B ) Instead of computing factorization scores using our synthetic images ( Figure 3 , top left), recomputing camera viewpoint or object pose factorization from natural movie datasets that primarily contained camera or object motion, respectively, gave similar results for predicting which model representations would be more brainlike (right: example movie frames; also see ’Methods’). Error bars in ( A and B ) are standard deviations over bootstrapped resampling of the models. ( C ) Summary of the results from ( A ) across datasets (x-axis) for invariance (open symbols) versus factorization (closed symbols) (for reference, ‘ x ’ symbols indicate predictive power when using model classification performance). Results using a comparable, alternative method for computing factorization (covariance-based, Equation 4 in ’Methods’; light closed symbols) are shown adjacent to the original factorization metric (PCA-based, Equation 2 in ‘Methods’; dark closed symbols).
Finally, we tested whether our results generalized across the particular image set used for computing the model factorization scores in the first place. Here, instead of relying on our synthetically generated images, where each scene parameter was directly controlled, we re-computed factorization from two types of relatively unconstrained natural movies, one where the observer moves in an urban environment (approximates camera viewpoint changes) ( Lee et al., 2012 ) and another where objects move in front of a fairly stationary observer (approximates object pose changes) ( Monfort, 2019 ). Similar to the result found for factorization measured using augmentations of synthetic images, factorization of frame-by-frame variance (local in time, presumably dominated by either observer or camera motion; see ‘Methods’) from other sources of variance across natural movies (non-local in time) was correlated with improved neural predictivity in both macaque and human data while invariance to local frame-by-frame differences was not ( Figure 5B ; black versus gray bars). Thus, we have shown that a main finding – the importance of object pose and camera viewpoint factorization for achieving brainlike representations – holds across types of brain signal (spiking vs. BOLD), species (monkey vs. human), cortical brain areas (V4 vs. IT), images for testing in experiments (synthetic, grayscale vs. natural, color), and image sets for computing the metric (synthetic images vs. natural movies).
Our analysis of DNN models provides strong evidence that greater factorization of a variety of scene variables is consistently associated with a stronger match to neural and behavioral data. Prior work had identified a similar correlation between object classification performance (measured fitting a decoder for object class using model representations) and fidelity to neural data ( Yamins et al., 2014 ). A priori, it is possible that the correlations we have demonstrated between scene parameter factorization and neural fit can be entirely captured by the known correlation between classification performance and neural fits ( Schrimpf et al., 2020 ; Yamins et al., 2014 ) as factorization and classification may themselves be correlated. However, we found that factorization scores significantly boosted cross-validated predictive power of neural/behavioral fit performance compared to simply using object classification alone, and factorization boosted predictive power as much if not slightly more when using RDMs instead of linear regression fits to quantify the match to the brain/behavior ( Figure 6 ). Thus, considering factorization in addition to object classification performance improves upon our prior understanding of the properties of more brainlike models ( Figure 7 ).
Scene parameter factorization combined with object identity classification improves correlations with neural predictivity.
Average across datasets of brain predictivity of classification (faded black bar), dimensionality (faded pink bar), and factorization (remaining faded colored bars) in a model representation. Linearly combining factorization with classification in a regression model (unfaded bars at right) produced significant improvements in predicting the most brainlike models (performance cross-validated across models and averaged across datasets, n = 4 datasets for each of V4, IT/HVC and behavior). The boost from factorization in predicting the most brainlike models was not observed for neural and fMRI data when combining classification with a model’s overall dimensionality (solid pink bars; compared to black dashed line for brain predictivity when using classification alone). Results are shown for both the PCA-based and covariance-based factorization metric (top versus bottom row). Error bars are standard deviations over bootstrapped resampling of the models.
Combining classification performance with object pose factorization improves predictions of the most brainlike models on IT/HVC data.
Example scatter plots for neural and fMRI datasets (macaque E1 and E2, IT multi units and single units; human F1 and F2, fMRI voxels) showing a saturating and sometimes reversing trend in neural (voxel) predictivity for models that are increasingly good at classification (top row). This saturating/reversing trend is no longer present when adding object pose factorization to classification as a combined, predictive metric for brainlikeness of a model (middle and bottom rows). The x-axis of each plot indicates the predicted encoding fit or representational dissimilarity matrix (RDM) correlation after fitting a linear regression model with the indicated metrics as input (either classification or classification + factorization).
Object classification, which has been proposed as a normative principle for the function of the ventral visual stream, can be supported by qualitatively different representational geometries ( Yamins et al., 2014 ; Nayebi, 2021 ). These include representations that are completely invariant to non-class information ( Caron et al., 2019b ; Caron, 2019a ) and representations that retain a high-dimensional but factorized encoding of non-class information, which disentangles the representation of multiple variables ( Figure 1A ). Here, we presented evidence that factorization of non-class information is an important strategy used, alongside invariance, by the high-level visual cortex ( Figure 2 ) and by DNNs that are predictive of primate neural and behavioral data ( Figures 4 and 5 ).
Prior work has indicated that building representations that support object classification performance and representations that preserve high-dimensional information about natural images are both important principles of the primate visual system ( Cadieu et al., 2014 ; Elmoznino and Bonner, 2022 ; though see Conwell et al., 2022 ). Critically, our results cannot be accounted for by classification performance or dimensionality alone ( Figure 6 , gray and pink bars); that is, the relationship between factorization and matches to neural data was not entirely mediated by classification or dimensionality. That said, we do not regard factorization and dimensionality, or factorization and object classification performance, as mutually exclusive hypotheses for useful principles of visual representations. Indeed, high-dimensional representations could be regarded as a means to facilitate factorization, and likewise factorized representations can better support classification ( Figure 1C ).
Our notion of factorization is related to, but distinct from, several other concepts in the literature. Many prior studies in machine learning have considered the notion of disentanglement, often defined as the problem of inferring independent factors responsible for generating the observed data ( Kim and Mnih, 2018 ; Eastwood and Williams, 2018 ; Higgins, 2018 ). One prior study notably found that machine learning models designed to infer disentangled representations of visual data displayed single-unit responses that resembled those of individual neurons in macaque IT ( Higgins et al., 2021 ). Our definition of factorization is more flexible, requiring only that independent factors be encoded in orthogonal subspaces, rather than by distinct individual neurons. Moreover, our definition applies to generative factors, such as camera viewpoint or object pose, that are multidimensional and context dependent. Factorization is also related to a measure of ‘abstraction’ in representational geometry introduced in a recent line of work ( Bernardi et al., 2020 ; Boyle et al., 2024 ), which is observed to emerge in trained neural networks ( Johnston and Fusi, 2023 ; Alleman et al., 2024 ). In these studies, an abstract representation is defined as one in which variables are encoded and can be decoded in a consistent fashion regardless of the values of other variables. A fully factorized representation should be highly abstract according to this definition, though factorization emphasizes the geometric properties of the population representation while these studies emphasize the consequences for decoding performance in training downstream linear read-outs. Relatedly, another recent study found that orthogonal encoding of class and non-class information is one of several factors that determines few-shot classification performance ( Sorscher et al., 2022 ). Our work can be seen as complementary to work on representational straightening of natural movie trajectories in the population space ( Hénaff et al., 2021 ). This work suggested that visual representations maintain a locally linear code of latent variables like camera viewpoint, while our work focused on the global arrangement of the linear subspaces affected by different variables (e.g., overall coding of camera viewpoint-driven variance versus sources of variance from other scene variables in a movie). Local straightening of natural movies was found to be important for early visual cortex neural responses but not necessarily for high-level visual cortex ( Toosi and Issa, 2022 ), where the present work suggests factorization may play a role.
Our work has several limitations. First, our analysis is primarily correlative. Going forward, we suggest that factorization could prove to be a useful objective function for optimizing neural network models that better resemble primate visual systems, or that factorization of latent variables should at least be a by-product of other objectives that lead to more brain-like models. An important direction for future work is finding ways to directly incentivize factorization in model objective functions so as to test its causal impact on the fidelity of learned representations to neural data. Second, our choice of scene variables to analyze in this study was heuristic and somewhat arbitrary. Future work could consider unsupervised methods (in the vein of independent components analysis) for uncovering the latent sources of variance that generate visual data, and assessing to what extent these latent factors are encoded in factorized form. Third, in our work we do not specify the details of how a particular scene parameter is encoded within its factorized subspace, including whether the code is linear (‘straightened’) or nonlinear ( Hénaff et al., 2021 ; Hénaff et al., 2019 ). Neural codes could adopt different strategies, resulting in similar factorization scores at the population level, each with some support in visual cortex literature: (1) each neuron encodes a single latent variable ( Field, 1994 ; Chang and Tsao, 2017 ), (2) separate brain subregions encode qualitatively different latent variables but using distributed representations within each region ( Tsao et al., 2006 ; Lafer-Sousa and Conway, 2013 ; Vaziri et al., 2014 ), and (3) each neuron encodes multiple variables in a distributed population code, such that the factorization of different variables is only apparent as independent directions when assessed in high-dimensional population activity space ( Field, 1994 ; Rigotti et al., 2013 ). Future work can disambiguate among these possibilities by systematically examining ventral visual stream subregions ( Kravitz et al., 2013 ; Vaziri et al., 2014 ; Kravitz et al., 2011 ) and the single neuron tuning curves within them ( Leopold et al., 2006 ; Freiwald et al., 2009 ).
Monkey datasets
Macaque monkey datasets were of single-unit neural recordings ( Rust and DiCarlo, 2012 ), multi-unit neural recordings ( Majaj et al., 2015 ), and object recognition behavior ( Rajalingham et al., 2018 ). Single-unit spiking responses to natural images were measured in V4 and anterior ventral IT ( Rust and DiCarlo, 2012 ). The advantages of this dataset are that it contains well-isolated single neurons, the gold standard for electrophysiology. Furthermore, the IT recordings were obtained from penetrating electrodes targeting the anterior ventral portion of IT near the base of skull, reflecting the highest level of the IT hierarchy. On the other hand, the multi-unit dataset was obtained from across IT with a bias toward where multi-unit arrays are more easily placed such as CIT and PIT ( Majaj et al., 2015 ), complementing the recording locations of the single-unit dataset. An advantage of the multi-unit dataset using chronic recording arrays is that an order of magnitude more images were tested per recording site (see dataset comparisons in Supplementary file 1a ). Finally, the monkey behavioral dataset came from a third study examining the image-by-image object classification performance of macaques and humans ( Rajalingham et al., 2018 ).
Human datasets
Three datasets from humans were used, two fMRI datasets and one object recognition behavior dataset ( Nonaka et al., 2021 ; Rajalingham et al., 2018 ; Kay et al., 2008 ). The fMRI datasets used different images (color versus grayscale) but otherwise used a fairly similar number of images and voxel resolution in MR imaging. Human fMRI studies have found that different DNN layers tend to map to V4 and HVC human fMRI voxels ( Nonaka et al., 2021 ). The human behavioral dataset measured image-by-image classification performance and was collected in the same study as the monkey behavioral signatures ( Rajalingham et al., 2018 ).
Computational models
In recent years, a variety of approaches to training DNN vision models have been developed that learn representations that can be used for downstream classification (and other) tasks. Models differ in a variety of implementational choices including in their architecture, objective function, and training dataset. In the models we sampled, objectives included supervised learning of object classification (AlexNet, ResNet), self-supervised contrastive learning (MoCo, SimCLR), and other unsupervised learning algorithms based on auxiliary tasks (e.g., reconstruction or colorization). A majority of the models that we considered relied on the widely used, performant ResNet-50 architecture, though some in our library utilized different architectures. The randomly initialized network control utilized ResNet-50 (see Figure 4A and B ). The set of models we used is listed in Supplementary file 1b .
Simulation of factorized versus non-factorized representational geometries
For the simulation in Figure 1C , we generated data in the following way. First, we randomly sampled the values of N = 10 binary features. Feature values corresponded to positions in an N-dimensional vector space as follows: each feature was assigned an axis in N-dimensional space, and the value of each feature (+1 or –1) was treated as a coefficient indicating the position along that axis. All but two of the feature axes were orthogonal to the rest. The last two features, which served as targets for the trained linear decoders, were assigned axes whose alignment ranged from 0 (orthogonal) to 1 (identical). In the noiseless case, factorization of these two variables with respect to one another is given by subtracting the square of the cosine of the angle between the axes from 1. We added Gaussian noise to the positions of each data point and randomly sampled K positive and negative examples for each variable of interest to use as training data for the linear classifier (a support vector machine).
Macaque neural data analyses
For the shuffle control used as a null model for factorization, we shuffled the object identity labels of the images ( Figure 2—figure supplement 1 ). For the transformation used in Figure 2B , we computed the PCs of the mean neural activity response to each object class (‘class centers,’ x c ), referred to as the inter-class PCs, v 1 inter , v 2 inter , …, v N inter . We also computed the PCs of the data with corresponding class centers subtracted (i.e., x - x c ) from each activity pattern, referred to as the intra-class PCs v 1 intra , v 2 intra , …, v N intra . We transformed the data by applying to the class centers a change of basis matrix W inter→intra that rotated each inter-class PC into the corresponding intra-class PC: W inter→intra = v 1 intra ( v 1 inter ) T + …1 v N intra ( v N inter ) T . That is, the class centers were transformed by this matrix, but the relative positions of activity patterns within a given class were fixed. For an activation vector x belonging to a class c for which the average activity vector over all images of class c is x c , the transformed vector was
This transformation has the effect of preserving intra-class variance statistics exactly from the original data and preserving everything about the statistics of inter-class variance except its orientation relative to intra-class variance. That is, the transformation is designed to affect (specifically decrease) factorization while controlling for all other statistics of the activity data that may be relevant to object classification performance (considering the simulation in Figure 1C of two binary variables, this basis change of the neural data in Figure 2B is equivalent to turning a square into the maximally flat parallelogram, the degenerate one where all the points are collinear).
Scene parameter variation
Our generated scenes consisted of foreground objects imposed upon natural backgrounds. To measure variance associated with a particular parameter like the background identity, we randomly sampled 10 different backgrounds while holding the other variables (e.g., foreground object identity and pose constant). To measure variance associated with foreground object pose, we randomly varied object angle from [–90, 90] along all three axes independently, object position on the two in-plane axes, horizontal [–30%, 30%] and vertical [–60%, 60%], and object size [×1/1.6, ×1.6]. To measure variance associated with camera position, we took crops of the image with scale uniformly varying from 20 to 100% of the image size, and position uniformly distributed across the image. To measure variance associated with lighting conditions, we applied random jitters to the brightness, contrast, saturation, and hue of an image, with jitter value bounds of [–0.4, 0.4] for brightness, contrast, and saturation and [–0.1, 0.1] for hue. These parameter choices follow standard data augmentation practices for self-supervised neural network training, as used, for example, in the SimCLR and MoCo models tested here ( He et al., 2020 ; Chen et al., 2020 ).
Factorization and invariance metrics
Factorization and invariance were measured according to the following equations:
Variance induced by a parameter ( var param ) is computed by measuring the variance (summed across all dimensions of neural activity space) of neural responses to the 10 augmented versions of a base image where the augmentations are those obtained by varying the parameter of interest. This quantity is then averaged across the 100 base images. The variance induced by all parameters is simply the sum of the variances across all images and augmentations. To define the ‘other-parameter subspace,’ we averaged neural responses for a given base image over all augmentations using the parameter of interest, and ran PCA on the resulting set of averaged responses. The subspace was defined as the space spanned by top PCA components containing 90% of the variance of these responses. Intuitively, this space captures the bulk of the variance driven by all parameters other than the parameter of interest (due to the averaging step). The variance of the parameter of interest within this ‘other-parameter subspace,’ var param|other_param_subspace , was computed the same way as var param but using the projections of neural activity responses onto the other-parameter subspace. In the main text, we refer to this method of computing factorization as PCA-based factorization.
We also considered an alternative definition of factorization referred to as covariance-based factorization. In this alternative definition, we measured the covariance matrices cov param and cov other_param induced by varying (in the same fashion as above) the parameter of interest, and all other parameters. Factorization was measured by the following equation:
This is equal to 1 minus the dot product between the normalized, flattened covariance matrices, and thus covariance-based factorization is a measure of the discrepancy of the covariance structure induced by the parameter of interest and other parameters. The main findings were unaffected by our choice of method for computing the factorization metric, whether PCA or covariance based ( Figures 5 — 7 ). An advantage of the PCA-based method is that as an intermediate one recovers the linear subspaces containing parameter variance, but in so doing requires an arbitrary choice of the explained variance threshold used to choose the number of PCs. By contrast, the covariance-based method is more straightforward to compute and has no free parameters. Thus, these two metrics are complementary and somewhat analogous in methodology to two metrics commonly used for measuring dimensionality (the number of components needed to explain a certain fraction of the variance, analogous to our original PCA-based definition, and the participation ratio, analogous to our covariance-based definition) ( Ding and Glanzman, 2010 ; Litwin-Kumar et al., 2017 ).
Natural movie factorization metrics
For natural movies, variance is not induced by explicit control of a parameter as in our synthetic scenes but implicitly, by considering contiguous frames (separated by 200 ms in real time) as reflective of changes in one of two motion parameters (object versus observer motion) depending on how stationary the observer is (MIT Moments in Time movie set: stationary observer; UT-Austin Egocentric movie set: nonstationary) ( Lee et al., 2012 ; Monfort, 2019 ). Here, the all parameters condition is simply the variance across all movie frames, which in the case of MIT Moments in Time dataset includes variance across thousands of video clips taken in many different settings and in the case of the UT-Austin Egocentric movie dataset includes variance across only four movies but over long durations of time during which an observer translates extensively in an environment (3–5 hr). Thus, movie clips in the MIT Moments in Time movie set contained new scenes with different object identities, backgrounds, and lightings and thus effectively captured variance induced by these non-spatial parameters ( Monfort, 2019 ). In the UT-Austin Egocentric movie set, new objects and backgrounds are encountered as the subject navigates around the urban landscape ( Lee et al., 2012 ).
Model neural encoding fits
Linear mappings between model features and neuron (or voxel) responses were computed using ridge regression (with regularization coefficient selected by cross-validation) on a low-dimensional linear projection of model features (top 300 PCA components computed using images in each dataset). We also tested an alternative approach to measuring representational similarity between models and experimental data based on representational similarity analysis ( Kriegeskorte and Kievit, 2013 ), computing dot product similarities of the representations of all pairs of images and measuring the Spearman correlation coefficient between these pairwise similarity matrices obtained from a given model and neural dataset, respectively.
Model behavioral signatures
We followed the approach of Rajalingham et al., 2018 . We took human and macaque behavioral data from the object classification task and used it to create signatures of image-level difficulty (the ‘I1’ vector) and image-by-distractor-object confusion rates (the ‘I2’ matrix). We did the same for the DNN models, extracting model ‘behavior’ by training logistic regression classifiers to classify object identity in the same image dataset used in the experiments of Rajalingham et al., 2018 , using model layer activations as inputs. Model behavioral accuracy rates on image by distractor object pairs were assessed using the classification probabilities output by the logistic regression model, and these were used to compute I1 and I2 metrics as was done for the true behavioral data. Behavioral similarity between models and data was assessed by measuring the correlation between the entries of the I1 vectors and I2 matrices (both I1 and I2 results are reported).
Model layer choices
The scatter plots in Figure 4A and B and Figure 4—figure supplements 1 – 3 use metrics (factorization, invariance, and goodness of neural fit) taken from the final representational layer of the network (the layer prior to the logits layer used for classification in supervised network, prior to the embedding head in contrastive learning models, or prior to any auxiliary task-specific layers in unsupervised models trained using auxiliary tasks). However, representational geometries of model activations, and their match to neural activity and behavior, vary across layers. This variability arises because different model layers correspond to different stages of processing in the model (convolutional layers in some cases, and pooling operations in others), and may even have different dimensionalities. To ensure that our results do not depend on idiosyncrasies of representations in one particular model layer and the particular network operations that precede it, summary correlation statistics in all other figures ( Figures 5 — 7 , Figure 5—figure supplements 1 – 3 ) show the results of the analysis in question averaged over the five final representational layers of the model. That is, the metrics of interest (factorization, invariance, neural encoding fits, RDM correlation, behavioral similarity scores) were computed independently for each of the five final representational layers of each model, and these five values were averaged prior to computing correlations between different metrics.
Correlation of model predictions and experimental data
A Spearman linear correlation coefficient was calculated for each model layer by biological dataset combination (six monkey datasets and six human datasets). Here, we do not correct for noise in the biological data when computing the correlation coefficient as this would require trial repeats (for computing intertrial variability) that were limited or not available in the fMRI data used. In any event, normalizing by the data noise ceiling applies a uniform scaling to all model prediction scores and does not affect model comparison, which only depends on ranking models as being relatively better or worse in predicting brain data. Finally, we estimated the effectiveness of model factorization, invariance, or dimensionality in combination with model object classification performance for predicting model neural and behavioral fit by performing a linear regression on the particular dual metric combination (e.g., classification plus object pose factorization) and reporting the Spearman correlation coefficient of the linearly weighted metric combination. The correlation was assessed on held-out models (80% used for training, 20% for testing), and the results were averaged over 100 randomly sampled train/test splits.
The current manuscript is a computational study, so no data have been generated for this manuscript. Publicly available datasets and models were used. Analysis code is available at https://github.com/issalab/Lindsey-Issa-Factorization , (copy archived at Issa, 2024 ).
- Naselaris T
- Google Scholar
- Siegelbaum SA
- Bojanowski P
- Kornblith S
- Sompolinsky H
- Williams CKI
- Elmoznino E
- Freiwald WA
- Livingstone MS
- Simoncelli EP
- Charlton JA
- Summerfield C
- Botvinick M
- Johnston WJ
- Ungerleider LG
- Kriegeskorte N
- Krizhevsky A
- Sutskever I
- Lafer-Sousa R
- Litwin-Kumar A
- Rajalingham R
- Prescott-Roy J
- Tootell RBH
Author details
- Zuckerman Mind Brain Behavior Institute, Columbia University, New York, United States
- Department of Neuroscience, Columbia University, New York, United States
Contribution
Competing interests.
For correspondence
Doe csgf (de-sc0020347), klingenstein-simons foundation (fellowship in neuroscience), sloan foundation (fellowship), grossman-kavli center at columbia (scholar award).
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Acknowledgements
This work was performed on the Columbia Zuckerman Institute Axon GPU cluster and via generous access to Cloud TPUs from Google’s TPU Research Cloud (TRC). JWL was supported by the DOE CSGF (DE-SC0020347). EBI was supported by a Klingenstein-Simons fellowship, Sloan Foundation fellowship, and Grossman-Kavli Scholar Award. We thank Erica Shook for comments on a previous version of the manuscript. The authors declare no competing interests.
Version history
- Preprint posted : August 4, 2023
- Sent for peer review: August 22, 2023
- Reviewed Preprint version 1 : February 2, 2024
- Reviewed Preprint version 2 : June 5, 2024
- Version of Record published : July 5, 2024
Cite all versions
You can cite all versions using the DOI https://doi.org/10.7554/eLife.91685 . This DOI represents all versions, and will always resolve to the latest one.
© 2024, Lindsey and Issa
This article is distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use and redistribution provided that the original author and source are credited.
- 69 downloads
- 0 citations
Views, downloads and citations are aggregated across all versions of this paper published by eLife.
Download links
Downloads (link to download the article as pdf).
- Article PDF
- Figures PDF
Open citations (links to open the citations from this article in various online reference manager services)
Cite this article (links to download the citations from this article in formats compatible with various reference manager tools), categories and tags.
- visual cortex
- deep neural networks
- neurophysiology
- visual scenes
- object recognition
Research organisms
- Rhesus macaque
Further reading
A ‘double-edged’ role for type-5 metabotropic glutamate receptors in pain disclosed by light-sensitive drugs.
We used light-sensitive drugs to identify the brain region-specific role of mGlu5 metabotropic glutamate receptors in the control of pain. Optical activation of systemic JF-NP-26, a caged, normally inactive, negative allosteric modulator (NAM) of mGlu5 receptors, in cingulate, prelimbic, and infralimbic cortices and thalamus inhibited neuropathic pain hypersensitivity. Systemic treatment of alloswitch-1, an intrinsically active mGlu5 receptor NAM, caused analgesia, and the effect was reversed by light-induced drug inactivation in the prelimbic and infralimbic cortices, and thalamus. This demonstrates that mGlu5 receptor blockade in the medial prefrontal cortex and thalamus is both sufficient and necessary for the analgesic activity of mGlu5 receptor antagonists. Surprisingly, when the light was delivered in the basolateral amygdala, local activation of systemic JF-NP-26 reduced pain thresholds, whereas inactivation of alloswitch-1 enhanced analgesia. Electrophysiological analysis showed that alloswitch-1 increased excitatory synaptic responses in prelimbic pyramidal neurons evoked by stimulation of presumed BLA input, and decreased BLA-driven feedforward inhibition of amygdala output neurons. Both effects were reversed by optical silencing and reinstated by optical reactivation of alloswitch-1. These findings demonstrate for the first time that the action of mGlu5 receptors in the pain neuraxis is not homogenous, and suggest that blockade of mGlu5 receptors in the BLA may limit the overall analgesic activity of mGlu5 receptor antagonists. This could explain the suboptimal effect of mGlu5 NAMs on pain in human studies and validate photopharmacology as an important tool to determine ideal target sites for systemic drugs.
Task-specific invariant representation in auditory cortex
Categorical sensory representations are critical for many behaviors, including speech perception. In the auditory system, categorical information is thought to arise hierarchically, becoming increasingly prominent in higher-order cortical regions. The neural mechanisms that support this robust and flexible computation remain poorly understood. Here, we studied sound representations in the ferret primary and non-primary auditory cortex while animals engaged in a challenging sound discrimination task. Population-level decoding of simultaneously recorded single neurons revealed that task engagement caused categorical sound representations to emerge in non-primary auditory cortex. In primary auditory cortex, task engagement caused a general enhancement of sound decoding that was not specific to task-relevant categories. These findings are consistent with mixed selectivity models of neural disentanglement, in which early sensory regions build an overcomplete representation of the world and allow neurons in downstream brain regions to flexibly and selectively read out behaviorally relevant, categorical information.
- Stem Cells and Regenerative Medicine
Circulating platelets modulate oligodendrocyte progenitor cell differentiation during remyelination
Revealing unknown cues that regulate oligodendrocyte progenitor cell (OPC) function in remyelination is important to optimise the development of regenerative therapies for multiple sclerosis (MS). Platelets are present in chronic non-remyelinated lesions of MS and an increase in circulating platelets has been described in experimental autoimmune encephalomyelitis (EAE) mice, an animal model for MS. However, the contribution of platelets to remyelination remains unexplored. Here we show platelet aggregation in proximity to OPCs in areas of experimental demyelination. Partial depletion of circulating platelets impaired OPC differentiation and remyelination, without altering blood-brain barrier stability and neuroinflammation. Transient exposure to platelets enhanced OPC differentiation in vitro, whereas sustained exposure suppressed this effect. In a mouse model of thrombocytosis ( Calr +/- ), there was a sustained increase in platelet aggregation together with a reduction of newly-generated oligodendrocytes following toxin-induced demyelination. These findings reveal a complex bimodal contribution of platelet to remyelination and provide insights into remyelination failure in MS.
Be the first to read new articles from eLife

Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Open access
- Published: 25 August 2024
A ventromedial visual cortical ‘Where’ stream to the human hippocampus for spatial scenes revealed with magnetoencephalography
- Edmund T. Rolls ORCID: orcid.org/0000-0003-3025-1292 1 , 2 , 3 na1 ,
- Xiaoqian Yan ORCID: orcid.org/0000-0003-4711-7428 3 na1 ,
- Gustavo Deco ORCID: orcid.org/0000-0002-8995-7583 4 , 5 ,
- Yi Zhang 3 ,
- Veikko Jousmaki ORCID: orcid.org/0000-0003-1963-5834 6 &
- Jianfeng Feng 2 , 3
Communications Biology volume 7 , Article number: 1047 ( 2024 ) Cite this article
Metrics details
- Hippocampus
- Network models
- Spatial memory
The primate including the human hippocampus implicated in episodic memory and navigation represents a spatial view, very different from the place representations in rodents. To understand this system in humans, and the computations performed, the pathway for this spatial view information to reach the hippocampus was analysed in humans. Whole-brain effective connectivity was measured with magnetoencephalography between 30 visual cortical regions and 150 other cortical regions using the HCP-MMP1 atlas in 21 participants while performing a 0-back scene memory task. In a ventromedial visual stream, V1–V4 connect to the ProStriate region where the retrosplenial scene area is located. The ProStriate region has connectivity to ventromedial visual regions VMV1–3 and VVC. These ventromedial regions connect to the medial parahippocampal region PHA1–3, which, with the VMV regions, include the parahippocampal scene area. The medial parahippocampal regions have effective connectivity to the entorhinal cortex, perirhinal cortex, and hippocampus. In contrast, when viewing faces, the effective connectivity was more through a ventrolateral visual cortical stream via the fusiform face cortex to the inferior temporal visual cortex regions TE2p and TE2a. A ventromedial visual cortical ‘Where’ stream to the hippocampus for spatial scenes was supported by diffusion topography in 171 HCP participants at 7 T.

Similar content being viewed by others
The medial occipital longitudinal tract supports early stage encoding of visuospatial information
White matter dissection and structural connectivity of the human vertical occipital fasciculus to link vision-associated brain cortex
Perirhinal circuits for memory processing
Introduction.
The human hippocampus (Hipp) is involved in episodic memory, our memory for past events 1 , 2 , and in navigation 3 , 4 , 5 . Much of our understanding of the Hippocampus has been based on the place cells found in rodents such as rats and mice, which encode the place where the rodent is located 3 , 4 , 5 , 6 , 7 , 8 . However, there is now mounting evidence in the primate including the human Hipp for neuronal spatial view representations for the location being viewed in spatial scenes 6 , 9 , 10 , 11 , 12 , 13 , 14 , 15 , 16 , 17 , 18 , 19 , 20 , 21 , 22 , 23 , 24 . Consistent with this, in human neuroimaging, a parahippocampal place area (PPA) is activated by viewed scenes, not the place where the individual is located 25 , 26 , 27 , 28 , 29 , 30 , 31 , 32 , 33 . Indeed, because it responds to viewed scenes, the region might better be known as the parahippocampal scene area (PSA) 6 , 34 , and is in ventromedial cortical regions VMV1–3 and medial parahippocampal regions PHA1–3 25 , 33 (see Figs. 1 and S1 ).

The cortical regions are shown on images of the human brain with the sulci expanded to show the regions within the sulci. Table S1 shows abbreviations for the cortical regions. For comparison, Fig. S1 part 5 shows the labels on the human brain without the sulci expanded. Fig. S1 parts 1–4 shows labelled coronal slices of the human brain (this figure was produced by Edmund T. Rolls and Chu-Chung Huang 63 from data made available as part of the HCP-MMP 55 and is available open access in ref. 38 ).
The difference between rodents and humans is potentially very important for understanding human hippocampal systems for memory and navigation, which are very different if view-based rather than place-based computations are being used. For example, navigation in humans may be performed in part by using viewed landmarks rather than self-motion update of place 35 , 36 . Further, the computations involved in setting up representations of scenes are likely to involve forming feature combinations based on visual inputs that define parts of scenes, and linking these viewed parts together 6 , 36 , 37 . Because of these issues and major differences between rodents and primates including hippocampal representations 36 , 38 , 39 , it is important to understand better the cortical pathways that are involved in building scene representations in humans, which in turn has implications for what computations are performed, and how 38 , 40 .
In addition to the Hippocampus and the parahippocampal place (scene) area, another key brain area involved in human scene perception is the retrosplenial complex 41 (also referred to as the medial place area 42 ), which is located in regions ProStriate cortex (ProS) and dorsal visual transitional cortex (DVT) in the Human Connectome Project Multimodal Parcellation (HCP-MMP) of the cerebral cortex 25 , 33 , 34 (see Figs. 1 and S1 ). There is also an occipital place area 43 (also known as a transverse occipital sulcus region 44 ) on the lateral occipital surface, which is located in or close to V3CD (including parts of IP0, V3B, V4, and LO1 in the HCP-MMP parcellation 25 , 34 . In addition, the caudal inferior parietal lobule is implicated in scene memory 42 , 45 . Functional connectivity analysis while participants were in the resting state or were looking at scenes or movies of scenes shows that these cortical regions are strongly connected with each other 6 , 29 , 33 , 34 , 42 , 45 , 46 , 47 , 48 , 49 , 50 . Further studies also show differences between posterior and anterior scene regions in connectivity, with the occipital scene area, ventromedial visual regions (VMV1–2 and VVC) and the retrosplenial scene area (ProS and DVT) having strong functional connectivity with early visual cortices such as V1–V4, while the PSA (especially PHA1–3) has strong connectivity with the Hipp 6 , 29 , 34 , 42 , 46 , 47 , 48 , 49 , 51 , 52 .
In this research, we present evidence on the cortical connectivity of the human Hippocampus to address these issues, and go beyond previous functional magnetic resonance imaging (fMRI)-based research on hippocampal system connectivity in humans 53 by utilising the fast neuroimaging method magnetoencephalography (MEG) which together with a machine learning approach to measuring effective connectivity enables the directionality of the connectivity 54 when scenes are being viewed to be measured; by utilising the Human Connectome Project Multimodal Parcellation atlas (HCP-MMP) which defines 360 cortical regions based on anatomy, functional connectivity, and task-related activations and so provides a framework for specifying which cortical regions have connectivity 55 ; by presenting quantitative evidence for the connectivity between all 360 cortical regions in a new approach to describing connectivity rather than the functional connectivity measured with fMRI from a few seed regions 53 ; and by complementing the MEG effective connectivity measurements with MEG functional connectivity measurements, and with diffusion tractography which uses high resolution 7-T MRI to following fibre pathways anatomically in the human brain 56 .
Previous research on the visual pathways that reach the Hippocampus has involved measuring effective, that is directed, connectivity with resting-state fMRI neuroimaging 48 , 57 . However, fMRI is inherently slow, with a time to measure a change in the BOLD signal to help in the calculation of effective connectivity in the order of 2 s 48 , 57 . For that reason, effective connectivity in the visual pathways was then measured with MEG 54 with the data sampled at 20 ms in 88 participants in the HCP 58 . Although visual stimuli were being shown during the collection of the MEG data 58 for that analysis 54 , the only visual stimuli used with MEG were faces and tools 58 . Because at least the functional connectivity can differ depending on which visual stimuli are being shown 33 , the new investigation described here was performed in which new MEG data were collected from 21 participants while scenes were being shown, and with 1 ms temporal resolution, for MEG data with scenes is not available from the HCP. The visual stimuli we used of scenes were in fact those used for fMRI data collection by the HCP 59 , and we have performed an analysis of these fMRI data which show activations and functional connectivity in medial temporal lobe regions in the HCP-MMP atlas 33 . In the present MEG investigation, the effective connectivity was also measured to faces, to provide a comparison of the pathways activated to those activated by scenes.
In summary, the aim of the present investigation was to trace the cortical regions through which spatial scene information reaches the Hippocampus in humans, by using the fast neuroimaging modality MEG with fast sampling at 1 ms during the presentation of spatial scene visual stimuli. New MEG data were collected, as the HCP did not use scene stimuli with MEG.
In the present investigation, the effective connectivities were measured between 30 visual cortical regions in the HCP-MMP 55 . The HCP-MMP atlas is a detailed parcellation of the human cortical regions, with its 360 regions defined using structural measures (cortical thickness and cortical myelin), functional connectivity, and task-related fMRI 55 . This parcellation is very useful for the human cerebral cortex as it utilises multimodal information 55 with the definitions and boundaries set out in Glasser_2016_SuppNeuroanatomy.pdf 55 , and as it is being used for much new research on cortical function and connectivity, which can all be placed in the same framework 25 , 33 , 38 , 48 , 49 , 57 , 60 , 61 , 62 , 63 , 64 , 65 , 66 , 67 , 68 , 69 . The boundaries, tractography, functional connectivity and task-related activations of visual cortical regions with the HCP-MMP atlas are available 55 , 70 , 71 , but the effective and functional connectivity measures here are new, as they are based on presenting visual stimuli of spatial scenes and faces in a new set of participants with MEG data with sampling at 1 ms.
In the present investigation, effective connectivity was measured utilising correlations between the signals between different brain regions measured with delays, as in previous investigations 48 , 54 , 57 . A whole-brain Hopf model of the simultaneous and delayed correlations between cortical regions produces what we term a generative effective connectivity matrix, as it can generate the functional connectivities and the delayed functional connectivities 57 , 72 , 73 as described in the Methods. It is highly relevant to this MEG investigation that the characteristic timescale for the computations performed by a cortical region is approximately 15 ms 38 , 74 , given that this is the timescale for the recurrent collateral connections between nearby pyramidal cells to operate for local attractor dynamics 38 , 75 , 76 , 77 , so analysis with MEG which provides data on the scale of 1–10 ms is very useful.
The results focus on the key new findings of this investigation, which are about the directionality of the effective connectivity when scenes are being viewed from V1 via the several stages of the ventromedial visual stream including the ventromedial visual regions VMV1–3 and the medial PSAs in PHA1–3 to the Hippocampus, for this has not previously been investigated with MEG. This is an important issue, for MEG is sufficiently fast, with the 1 ms acquisition used here, to follow the progression through visual cortical regions of the signal produced when scenes are shown. The use of MEG is important, for the directionality of the effective connectivity when measured with resting-state fMRI for faces, places, tools and body parts shows as the reverse of what is expected 48 , and this is probably related to the slow time course of fMRI which means that much of what is measured with fMRI resting state effective connectivity is the top–down effects from the top of the visual hierarchy where short-term memory keeps representations active 54 , 78 . The previous magnetoencephalography available from the HCP is available with the visual stimuli only for faces and tools 54 , and that is why we performed the investigation described here, to measure the directionality of the effective connectivity for scenes as that has not been measured before with MEG. We ran a new group of participants especially for the present investigation in which scenes were the stimuli and MEG was used, and that is thus the focus of the results described in the present investigation, and detailed statistical analyses for directionality were performed for scenes with MEG. For comparison, MEG responses to faces were included in the present study, but there is less emphasis on this in the results here, for the effective connectivity to faces has been measured previously with MEG, with HCP data 54 . We note that the localisation of signal into particular cortical regions is likely to be more accurate with fMRI than with MEG, and so rely on an fMRI study with faces, scenes, tools and body parts in 956 HCP participants for more accurate measures of the exact cortical regions in the HCP-MMP parcellation that are selectively activated by faces, scenes, tools, and body parts 33 .
MEG Effective connectivity of visual cortical regions when viewing spatial scenes vs faces
Mean effective connectivity when viewing spatial scenes.
The effective connectivity of 30 visual cortical regions in the HCP-MMP atlas when viewing spatial scenes with all cortical regions is shown in Fig. 2 , and for comparison when viewing faces in Fig. S2 . The visual cortical regions are grouped for convenience as shown in Fig. 2 and as described in the Methods. The effective connectivities shown in Fig. 2 are the mean for both directions, i.e. the mean of the column-to-row effective connectivity and of the row-to-column effective connectivity. The differences in the connectivity in the two directions are then shown in Fig. 3 , where yellow/red/brown colour indicates greater effective connectivity from column to row. Figure 2 shows that when visual scenes are being viewed in the 0-back memory task, in the early visual cortical regions, there is effective connectivity of V1 with V2, V3, VMV1, VMV2, and PHA2. V2 has effective connectivity with V3, POS1, VMV1, and VMV2. V3 has effective connectivity with V4, POS1, VMV1, VMV2, PHA3, FFC, and PIT. V4 has effective connectivity with ProS, VMV1, VMV3, VVC, PeEc (perirhinal cortex), TF (lateral parahippocampal), and V8.

The effective connectivities are the mean across both directions for every pair of cortical regions. Effective connectivities of <0.03 are shown as white to help reveal the main effective connectivities between the cortical regions. The effective connectivity map is scaled with 0.14 as the maximum. The effective connectivity in the top panel is for the first set of 90 cortical regions; and in the lower panel for the second set of 90 cortical regions. The abbreviations for cortical regions are shown in Table S1 . Horizontal red lines separate the groups of visual cortex regions. Group 1: (top) early visual cortical areas V1–V4 in the HCP-MMP atlas; Group 2: cortical regions in the retrosplenial complex; Group 3: ventromedial visual cortical regions; Group 4: parahippocampal cortex regions; Group 5: hippocampal and related regions; Group 6: intermediate ventrolateral cortical visual regions FFC (fusiform face cortex), PIT (posterior inferior temporal cortex), and V8. Group 7: inferior temporal visual cortex regions TE2p and TE1p. Group 8: anterior temporal lobe multimodal regions including the temporal pole TGd and TGv. The coloured labelled bars indicate the cortical divisions in the HCP-MMP atlas 55 . The order of the cortical regions on the horizontal axes is that in Huang, ref. 120 .

For a given link, the effective connectivity difference is shown as positive when the connectivity is stronger in the direction from column to row. For a link, the effective connectivity difference is shown as negative when the connectivity is weaker in the direction from column to row. The threshold value for any effective connectivity difference to be included is 0.0005 for the connectivities shown in Fig. 2 . This threshold was chosen to help show which differences were greater than or lesser than zero. Table S1 shows the abbreviations for the cortical regions, and the cortical regions are shown in Figs. 1 and S1 . The effective connectivity difference in the top panel is for the first set of 90 cortical regions; and in the lower panel for the second set of 90 cortical regions. The conventions are as in Fig. 2 .
Figure 2 also shows that when visual scenes are being viewed, for the Retrosplenial Regions, ProS has effective connectivity with V4, VMV1–3, and VVC, linking ProS to ventromedial visual cortical stream processing. Interestingly, ProS also has some effective connectivity with MT+ complex regions with visual motion sensitivity, especially LO1 and V4t. ProS also has effective connectivity with the medial parahippocampal cortex PHA1 and PHA2, the entorhinal cortex (EC), and the Hipp. In contrast, the DVT region has effective connectivity with Dorsal Stream Regions IPSI, V3A, and V6; and with superior parietal 7Pm, 7Pl, and MIP, suggesting that DVT is involved in visual motion analysis. POS1, which is close to ProS, and which can be activated by scenes 33 , has effective connectivity with V2 and V3, with DVT and v23ab, and with V3A and V6. v23ab, which is in the same general retrosplenial region, does have interesting effective connectivity not only with POS1, but also with V1, V2, V3, and ventromedial visual regions VMV1 and VMV2 (Fig. 2 ).
The ventromedial visual regions VMV1–3 and VVC have effective connectivity with V1, V2, and more with V3 and V4, and with each other (Fig. 2 ). The ventromedial visual regions also have effective connectivity with medial parahippocampal regions PHA1–3, and further with the PeEc, EC, and Hipp, when viewing spatial scenes (Fig. 2 ). The ventromedial visual regions thus are in a route from early visual regions to parahippocampal and hippocampal regions when viewing spatial scenes. There is also effective connectivity with FFC, V8, PIT, and TE2p (Fig. 2 ).
Although fMRI may allow more accurate localization of activations and connectivities than MEG, these findings with MEG are in fact well supported by the findings with activations and functional connectivities selective for scenes when measured with fMRI 33 .
Mean effective connectivity when viewing faces
The effective connectivity of 30 visual cortical regions in the HCP-MMP atlas when viewing faces is shown in Fig. S2 . Overall, these mean effective connectivities when viewing faces (Fig. S2 ) are rather similar to those when viewing spatial scenes (Fig. 2 ), and it is when the differences in the directions of effective connectivity between every pair of cortical regions are considered that the effective connectivities for spatial scenes and faces are found to be different (see next section, and Figs. 3 and 4 ). However, comparison of Figs. 2 and S2 does provide some indication that when viewing faces there is higher effective connectivity in inferior temporal cortex TE1p and TE1a, and the anterior temporal and temporal pole regions than when viewing scenes.

For a given link, the effective connectivity difference is shown as positive when the connectivity is stronger in the direction from column to row. For a link, the effective connectivity difference is shown as negative when the connectivity is weaker in the direction from column to row. The threshold value for any effective connectivity difference to be included is 0.0005 for the connectivities shown in Fig. 2 . Table S1 shows the abbreviations for the cortical regions, and the cortical regions are shown in Figs. 1 and S1 . The effective connectivity difference in the top panel is for the first set of 90 cortical regions; and in the lower panel for the second set of 90 cortical regions. The conventions are as in Fig. 2 .
These MEG results with faces are provided just for comparison with the results with scenes. For accurate localization of activations and functional connectivities that are selective for faces vs scenes, this is better provided by the fMRI analyses in 956 HCP participants 33 .
The directionality of the effective connectivities when viewing spatial scenes (Fig. 3 ) compared to faces (Fig. 4 )
The directionalities of the effective connectivities of 30 visual cortical regions in the HCP-MMP atlas when viewing spatial scenes are shown in Fig. 3 , with yellow/red/brown indicating higher effective connectivity from a column to a row than vice versa. The directionality differences from early visual regions such as V2, V3, and V4 to retrosplenial visual regions such as ProS and POS1, and to ventromedial visual regions VMV1–3 and VVC, are higher during viewing of scenes (Fig. 3 ) than faces (Fig. 4 ). Further, the directionality differences from the parahippocampal cortex regions PHA1–3 to hippocampal regions including the Hipp, EC, and PeEc are higher during viewing of scenes (Fig. 3 ) than faces (Fig. 4 ). In addition, the directionality differences from posterior cingulate regions implicated in memory such as 31pd, 32pv, d23ab, and v23ab to retrosplenial regions such as ProS, POS1, and POS2 are higher during viewing of scenes (Fig. 3 ) than faces (Fig. 4 ).
The most important statistical comparisons are for the directionality of effective connectivity between cortical regions in the ventromedial visual cortical stream when scenes are being viewed, as that is a key aim of this paper using MEG, and to support what is shown in Fig. 3 . The statistical comparisons were based on two-tailed paired t -tests performed across the 21 participants of whether there was higher effective connectivity in one direction than another between sets of cortical regions in preplanned comparisons. It was found that there was stronger effective connectivity from V3-V4 to the ProS, the key region in the retrosplenial scene area, than in the backward direction ( t = 3.51, p = 0.0022, d f = 20). There was stronger effective connectivity from V3–V4 to the ventromedial visual regions VMV1, VMV2, VMV3, and VVC than in the backward direction (for example, V3–V4 to VMV1 t = 2.62, p = 0.016, d f = 20). There was stronger effective connectivity from V3 to the parahippocampal visual regions PHA1, PHA2 and PHA3 than in the backward direction ( t = 4.09, p = 0.0006, d f = 20). There was also stronger effective connectivity when viewing scenes from the PSA regions PHA1-PHA3 to the Hipp than in the backward direction ( t = 2.97, p = 0.008, d f =20). Thus this MEG investigation provided new evidence that when viewing scenes, the effective connectivity is from early visual cortical regions such as V2–V4 to a retrosplenial region the ProS; to ventromedial visual cortical regions VMV1, VMV2, VMV3, and VVC; and to medial parahippocampal cortex regions PHA1, PHA2, and PHA3. Interestingly, it was also possible to show onward effective connectivity from the medial parahippocampal regions PHA1–3 to the Hipp when scenes were being viewed in the 0-back memory task. The directionality was made evident by using a delay of tau = 20 ms when measuring the effective connectivity, and this is in the order of time that it takes for visual information to cross one or two stages in a visual processing hierarchy 38 , 54 , 75 , 79 . With tau = 10 ms, the directionality was similar, but the effect size for the directionality was smaller, as expected.
Similarly, when viewing faces (Fig. 4 ), there was stronger effective connectivity in the ventrolateral ‘What’ (face and object) pathway in the direction from early visual cortical regions (e.g. V3) to PIT (posterior inferior temporal) and V8; and from PIT, V8 and fusiform face cortex (FFC) to TE2p, which is at the highest level in the inferior temporal visual cortex where the processing is mainly unimodal, as well as to the more anterior multimodal semantic regions such as TE2a 54 . Some key statistical comparisons are as follows: The effective connectivity from FFC, PIT and V8 to TE2p is stronger in that direction than vice versa ( t = 2.19, p = 0.04, d f = 20). The effective connectivity from FFC, PIT and V8 to TE2a is stronger in that direction than vice versa ( t = 6.85 p = 10 −5 , d f = 20).
The new evidence from magnetoencephalography is thus consistent with the directionality of the flow of information based on differences in effective connectivity in a ventromedial visual cortical stream from early visual cortical regions (V2–V4), to retrosplenial regions such as ProS and POS1, to ventromedial visual cortical regions VMV1–3 and VVC, and via parahippocampal regions PHA1–3, which in turn have connectivity to the Hippocampus and related regions such as the entorhinal and PeEc (Fig. 3 ).
Further, when viewing faces, the flow of information is from early visual cortical regions (and interestingly from visual motion regions in the MT+ complex even though the faces were stationary) to the regions close to the level of FFC such as PIT and V8, and from the FFC, PIT, and V8 to inferior temporal cortex TE2p and anterior temporal cortex TE2a (Fig. 4 ).
The functional connectivities when viewing spatial scenes (Fig. S3 )
For completeness, the MEG functional connectivities of the visual cortical regions are shown when viewing spatial scenes in Fig. S3 . The threshold has been set so that the proportion of connectivities shown is the same as for the effective connectivities in Fig. 2 , 0.087, to facilitate comparison between the two. The functional connectivity matrix (Fig. S3 ) is rather similar to the mean effective connectivity matrix (Fig. 2 ), which also does not capture the directionality of the connectivity. What is quite interesting about the similarity is that this may imply that a functional connectivity matrix does reflect considerably the effective connectivity matrix, and that might have implications for the interpretation of what a functional connectivity matrix can show. However, it must be remembered that in the Hopf effective connectivity algorithm, the effective connectivity matrix is being optimized that will best generate the functional connectivity matrix and the functional connectivity matrix delayed by tau = 20 ms, so the similarity is not surprising. A real difference though that makes the effective connectivity matrix useful is that it generates the set of connection strengths that can best generate the functional connectivity matrices without and with a delay, setting the other connectivities to zero, and thereby the mean effective connectivity matrix allows a threshold to be set in the functional connectivity matrix about what may be relevant, and this is useful for the functional connectivity matrix has continuous values in the range −1 to +1, and it is otherwise difficult to know what range of values may be useful.
The tractography of the visual cortical regions
The diffusion tractography matrix for these visual cortical regions is shown in Fig. 5 , and may be useful in providing evidence about which of the effective connectivities in Figs. 2 – 4 may be mediated by direct vs trans-synaptic connections. The tractography provides evidence for direct connections from V1, V2, and V3 to ProS and most of the other retrosplenial regions in Fig. 5 . There is also evidence for some direct connection between V1 and V4 and the ventromedial visual cortical regions, but much less with parahippocampal regions PHA1–3. ProS is shown as having connections with VMV1. The ventromedial visual cortical regions VMV1–3 and VVC have connections with the parahippocampal regions PHA1–3. The medial parahippocampal regions PHA1–3 have connections with the Hipp, and to a lesser extent with the PeEc and EC. These connections are consistent with what has been described by the effective connectivities shown in Figs. 2 and 3 in providing evidence for a staged hierarchically organised ventromedial cortical visual stream involving connectivity from early visual cortical regions V1–V4, to retrosplenial regions including ProS, to ventromedial visual cortical regions VMV1–3 and VVC, to parahippocampal regions PHA1–3, to the Hipp. Moreover, this ventromedial visual cortical pathway is implicated in spatial scene processing in that its directed effective connectivity is greater when viewing scenes than faces (Figs. 2 and 3 ); by the activations of retrosplenial regions ProS and POS1 when scenes are viewed 25 , 33 in what is termed the retrosplenial scene area; by the activations of ventromedial visual cortical regions VMV1–3 and VVC, and of parahippocampal regions PHA1–3, when spatial scenes are viewed in what is termed the parahippocampal place or scene area 25 , 33 ; and by the presence of spatial view cells in the macaque parahippocampal gyrus (as well as Hipp) 6 , 9 , 10 , 11 , 12 .

The layout is the same as in Figs. 2 – 4 . The number of streamlines shown was thresholded at 50; values less than this are shown as white to reveal the main connections. The colour bar was thresholded at 1000 streamlines. Table S1 shows the abbreviations. The conventions are as in Fig. 2 .
The tractography also shows in a ventrolateral visual cortical stream (Fig. 5 ) connections between V4 and V3 (and to a smaller extent V2 and V1) and FFC, V8 and PIT; with FFC having connections with inferior temporal visual cortex TE1p and TE2p; which in turn have connections with anterior temporal lobe regions including TE1a, TE1m, and TE2a. These anterior temporal lobe regions also have connections with regions in the STS (STSGa to STSvp). The ventrolateral pathway via FFC to the inferior temporal cortex is implicated in face processing, partly by the evidence of greater directional effective connectivity to faces (Fig. 3 ) but by a wealth of other evidence 34 . The visual stream to the STS is also implicated inter alia in responses to facial expressions and to moving heads that make or break social interactions 34 , 80 , 81 , 82 , 83 .
Laterality differences in the functional connectivity of the visual cortical regions
Laterality differences in the functional connectivity when viewing scenes might provide further evidence on the specialization of different visual cortical processing streams, and are shown in Fig. 6 . In these participants (who were Chinese), the functional connectivity of the ventromedial cortical regions VMV1–3 and VVC with ProS, with parahippocampal region PHA1–3, and with the Hippocampus and EC was stronger on the left. This is not inconsistent with earlier results on laterality when places/scenes are viewed, which show less lateralisation for scenes than for faces and that the PSA can have higher functional connectivity in the left hemisphere in 956 HCP participants with fMRI 33 , and this supports the current MEG analyses.

Differences in functional connectivity of less than 0.075 are shown as blank in order to reveal the main differences.
In contrast, the functional connectivity of the FFC, V8, and PIT, parts of the ventrolateral cortical visual stream involved in face and object processing 33 , 34 , 48 , 54 , had higher functional connectivity on the right, as does the STS system involved in face expression and movement 34 , 81 , 82 , 83 (Fig. 6 ). The consistency with earlier results with fMRI on face laterality 33 , 84 , 85 supports the current MEG analyses.
This evidence for a dissociation of pathways based on laterality supports the hypothesis that there is a ventromedial cortical visual stream for scene information to reach via ProS, the ventromedial visual cortical regions VMV1–3 and VVC, the medial parahippocampal cortex PHA1–3, and Hipp that is distinct from a ventrolateral pathway via FFC, V8, and PIT for information about faces and objects to reach the inferior temporal visual cortex in TE1p and TE1p, and more anterior temporal lobe regions including TE2a.
This research used MEG during the presentation of images of spatial scenes and showed (see Fig. 7 ) with the HCP-MMP atlas that MEG effective connectivity reveals a ventromedial cortical visual stream from V1–V4 to the ProS where the retrosplenial scene area is located; then to the ventromedial visual cortical regions VMV1–3 and VMV; then to the parahippocampal cortex PHA1–3; and then to the Hippocampus. The parahippocampal place or scene area is located in the VMV and PHA regions 25 , 33 . This ventromedial cortical visual stream was supported by analysis of diffusion tractography in 171 HCP participants. For comparison, when faces were being viewed, the effective connectivity was directed from V1–V4 more to the FFC, and then to the inferior temporal cortex regions TE2p and TE2a, in a ventrolateral visual cortical stream.

At a first level, after V1, V2–V4 have connectivity to the ProS and POS1 which are where in humans the retrosplenial scene area is located. At a second level, ProS has connectivity to the ventromedial visual cortical regions (VMV1–3 and VVC). These ventromedial visual cortical regions also receive effective connectivity from MT and MST etc in the dorsal visual cortical stream. At a third level, the ventromedial visual cortical regions have effective connectivity to the medial parahippocampal cortex regions PHA1–3. The medial parahippocampal cortex regions PHA1–3 also have effective connectivity from the ventrolateral visual cortical stream region FFC. The PSA is located at the intersection of the ventromedial visual regions (VMV1–3 and VVC) and medial parahippocampal regions PHA1–3. At a fourth level, the medial parahippocampal regions PHA1–3 have effective connectivity to the hippocampal memory system (arrow in green). The line widths of the arrowhead's size indicate the magnitude and direction of the effective connectivity (this figure was produced by Edmund T. Rolls using the cortical region template shown in Fig. 1 ).
These results provide an important addition to previous research 48 , 54 for the following reasons. First, the direction of the effective connectivity is easier to establish with MEG with its fast time resolution of 1–20 ms than with fMRI, perhaps because of the long time delays in the order of 2 s inherent with the development of the BOLD fMRI signal which may provide time for the signal to propagate upwards, and then back down again to earlier regions in a hierarchy 54 , 78 . Thus although fMRI can give useful results for effective connectivity, care is needed in interpretation of the direction of the effective connectivity measured with fMRI 54 . Second, previous fMRI analyses of effective connectivity with the HCP-MMP atlas were with the resting state 48 , 57 , whereas the present analysis measured effective connectivity while spatial scenes were being viewed and remembered. Third, a previous MEG investigation that used MEG data provided by the HCP was for face and tool visual stimuli 54 , whereas here we acquired new MEG data in which the visual stimuli were spatial scenes. This is important, for which pairs of regions show directed effective connectivity is different for spatial scenes vs faces (see Figs. 3 and 4 ).
Some interesting points arise from the findings with MEG described here.
First, the ProS appears to be a key region in the ventromedial visual cortical stream for spatial scenes, because it has effective connectivity from V3 and V4, to the ventromedial cortical visual areas VMV1–3 and VVC, and with parahippocampal regions PHA1–2 (Fig. 2 ). In contrast, the dorsal transitional visual region DVT has less of this connectivity, and instead has effective connectivity with some dorsal visual division regions IPS1, V3A, V6, and MT+ division region PH (Fig. 2 ). This is an indication that DVT is more concerned with visual motion analysis than with spatial scene analysis.
Second, POS1 and v23ab are both in the general retrosplenial area, when viewing scenes have strong effective connectivity with each other and v23ab has effective connectivity with ventromedial visual cortical regions (Fig. 2 ), and POS1 is activated in the HCP fMRI task-related data when viewing spatial scenes 33 . This provides an indication that there may be quite an extended part of the retrosplenial area including ProS, and POS1, and perhaps v23ab given its connectivity, that is related to spatial scene processing.
Third, the differences in effective connectivity in the two directions between any pair of regions may be smaller with MEG than with fMRI. The magnitude that is measured with either depends on the tau value used for the time delay between the two FC correlations used to calculate the effective connectivity: if tau is short, then there is less time for the time-delayed connectivity to become different. Given that a tau of 20 ms was used for the MEG analyses used here and in a previous investigation 54 , and of 2 s for the fMRI analyses of effective connectivity 48 , the small difference for the two directions with MEG is expected.
Fourth, we did not attempt to measure the magnitude of the activations of different cortical regions with MEG, because the magnitude of the MEG signals can depend on the orientation of a cortical region with respect to the sensors. Instead, we rely on activations measured with fMRI to allow the activations of different cortical regions to be compared more accurately to stimuli such as faces, scenes, tools, and body parts e.g. ref. 33 .
Fifth, the interestingly different functional connectivities shown in Fig. 6 with higher values for faces in the right hemisphere and scenes in the left hemisphere were found with Chinese participants, and these particular laterality differences might not be found in all populations, relating to differences in the organisation of language systems in the cortex in different populations. The laterality differences shown in Fig. 6 though do provide useful extra evidence that a ventromedial visual cortical pathway activated by scenes is different from a ventrolateral cortical visual pathway activated by faces etc.
It is useful to compare the approach taken here and in related recent studies on connectivity of the human medial temporal lobe using the HCP-MMP atlas 33 , 34 , 48 , 49 , 54 , 57 , 64 , 65 , 66 , 68 , 86 with another recent investigation which used resting state individualized fMRI to measure functional connectivity from 3 seed regions in four participants, and described the results in terms of, for example, “the human parahippocampal area TH is preferably associated with the retrosplenial cortex rather than with the posterior cingulate cortex” 53 (What was described as TH probably corresponds to PHA1–3 here.) In contrast, here we measured functional and effective connectivity between 30 visual cortical and medial temporal lobe regions with each other and with 150 other cortical regions in 21 participants who were viewing spatial scenes or faces for there is clear evidence with fMRI in 956 HCP participants that this affects the functional and effective connectivity and reveals different pathways 33 ; expressed the results in terms of connectivity matrices using the HCP-MMP atlas which provides a foundational basis for defining the cortical regions involved and by being surface-based helps to resolve problems with the measurement of activation peaks in volume space when defined cortical regions may be in different locations with respect to sulci in different individuals; and complemented these effective and functional connectivity analyses with diffusion tractography performed in the same HCP-MMP space with high resolution 7-T MRI. We suggest that the use of connection matrices to provide quantitative values for the connectivity between large numbers of well-defined cortical regions as used here and elsewhere 33 , 34 , 38 , 48 , 49 , 54 , 56 , 57 , 64 , 65 , 66 , 67 , 68 , 69 , 87 may provide an important development in the way that anatomical/connectivity evidence can be presented in future, with part of its attractiveness that cortical activations etc can be shown in the same well-defined cortical space, allowing comparison between different studies and obtained with different methods.
A key conceptual point made by the findings described here is that they provide support for the hypothesis that in humans (and other primates) a key spatial pathway for ‘Where’ information to reach the hippocampal memory and navigation system is involved in a ventral stream type of processing in which images on the retina are analysed in terms of spatial visual features being looked at in the world 36 . The coding is for information about spatial scenes, as shown by fMRI for the retrosplenial and PSAs in humans that are activated by what the human looks at, not the place where the individual is located 25 , 26 , 27 , 28 , 29 , 30 , 31 , 32 , 33 . Moreover, the spatial view information in the parahippocampal gyrus and Hippocampus is about the location in the scene, as shown by neuronal recordings in non-human primates from spatial view cells 6 , 9 , 10 , 11 , 12 , 13 , 14 , 15 , 16 , 17 , 18 , 24 , 39 , 88 , 89 , 90 . The representation of the location in a scene being looked at encoded by these spatial view neurons provides a ‘Where’ representation for hippocampal episodic memory in primates including humans, which typically involves associations between what face or object is being looked at, and where it is in a spatial scene 6 , 13 , 88 , 89 , 90 , 91 . In great contrast, in rodents, the place where the individual is located is represented in the Hipp, and update may be by path integration over places based on body movements 5 , 6 , 7 , 8 , 92 . This underlines the great importance of analysing the ‘Where’ cortical pathways in primates including humans as in the present paper, for what is found in rodents may be considerably different 36 , 38 .
This ventromedial cortical ‘Where’ visual pathway for scenes is hierarchically organised as shown by the following. First, the connectivity is hierarchical with for example early visual cortical regions V1–V4 having prominent effective connectivity with VMV regions; then VMV regions having prominent effective connectivity with PHA regions; and then PHA regions having prominent effective connectivity with the Hippocampus (Fig. 3 ). Second, the effective connectivity has directionality that is generally in this forward direction through these stages from early visual cortical regions to the Hippocampus (Fig. 3 ). Third, as noted above, the early visual cortical regions have relatively small receptive fields that are in retinotopic coordinates and that respond to stimuli such as bars and curves 93 , 94 , whereas by the medial parahippocampal gyrus (PHA1–3 in humans) and the Hippocampus spatial view neurons have allocentric representations to locations being viewed in scenes 6 , 9 , 10 , 11 , 12 , 39 , 95 , and in the Hippocampus can combine this with object and reward information to encode episodic memories 36 , 40 , 89 , 90 , 96 , 97 .
The evidence that the ventromedial cortical visual stream for scenes leading to the medial parahippocampal cortex (PHA1–3) is distinct from the ventrolateral cortical visual stream for faces and objects leading to the lateral parahippocampal cortex (TF) and the anterior temporal lobe semantic systems includes the following. First, the effective connectivities for VMV and PHA regions and PHA connectivity to the Hippocampus tend to be higher for scenes (Fig. 2 ) than for faces (Fig. S2 ), whereas the effective connectivities for faces tend to be higher than for scenes in FFC, V8, and PIT cortical regions (Figs. 2 and S2 ). This particular point is greatly strengthened by the analysis of scene-selective and face-selective activations and functional connectivities when measured with fMRI in a much larger group of 956 participants 33 . Second, the identification of the scene pathway as a separate visual pathway is confirmed by cluster and multidimensional scaling analyses based on resting-state functional connectivity 98 . Third, we have developed a dynamical graph approach to the analysis of cortical streams that enables whole networks rather than pairwise effective connectivities to be analysed, and this provides evidence from our effective connectivity investigations with 171 HCP participants imaged at 7 T 48 , 49 , 57 and from 956 HCP participants performing a working memory task with scenes, faces, tools or body parts as the stimuli for separate pathways and indeed whole networks to the Hippocampus for scenes (via VMV and PHA regions), and for faces via the FFC and TF regions 78 . Fourth, in the laterality section of the Results of this paper, the functional connectivity of the ventromedial cortical regions VMV1–3 and VVC with ProS, with parahippocampal region PHA1–3, and with the Hippocampus and EC was stronger on the left (Fig. 6 ). In contrast, the functional connectivity of the FFC, V8, and PIT, parts of the ventrolateral cortical visual stream involved in face and object processing, had higher functional connectivity on the right (Fig. 6 ). This evidence for a dissociation of pathways based on laterality supports the hypothesis that there is a ventromedial cortical visual stream for scene information to reach via ProS, the ventromedial visual cortical regions VMV1–3 and VVC, the parahippocampal cortex, and Hippocampus, that is distinct from a ventrolateral pathway via FFC, V8, and PIT for information about faces and objects to reach the inferior temporal visual cortex in TE1p and TE1p, and more anterior temporal lobe regions. Fifth, this ‘Where’ ventromedial cortical visual stream is more activated when performing episodic memory tasks with scenes than with word-pairs 91 .
This ventromedial pathway for spatial scenes has important implications for what is computed for ‘Where’ representations in the parahippocampal cortex and Hippocampus of primates including humans. The implication is that the spatial view representations are built by combinations of spatial features that are nearby in a scene, that is which fall within the primate fovea 6 , 99 , 100 . It is proposed that then these feature combination neurons are locked together in the correct spatial arrangement in a continuous attractor network built because nearby locations in a scene will have co-firing neurons that support associative synaptic modification 36 , 37 , 101 . This raises the interesting issue of how the spatial representations change up through the ventromedial visual cortical stream for spatial scene representations. In V1–V4, the visual receptive fields are relatively small, a few degrees across, and encode simple feature combinations such as lines and edges and in V4 combinations of these may form curves 93 , 94 , 102 , 103 , 104 , 105 , and are retinotopic. In contrast, in the macaque parahippocampal cortex and Hippocampus, spatial view cells have larger receptive fields that may subtend many degrees of visual angle, respond to a location in a world-based scene, are relatively invariant with respect to eye position and head direction, and even of the place at which the individual is located provided of course that the spatial view field can be viewed, and can thus be described as allocentric, world-based, and not egocentric 6 , 9 , 10 , 11 , 12 , 13 , 106 . The implication is that along the primate (including human) ventromedial visual stream, in regions such as the retrosplenial regions and ventromedial visual cortical regions, the representations will gradually become larger, less retinotopic and more head or even eye-direction-based in world coordinates, and later more independent of the place where the individual is located provided of course that the view field in the scene can be seen. Indeed, with wide-field retinotopic mapping, the ProS, where the retrosplenial scene area is located 25 , 33 , is found to have a complete representation of the visual field, different from adjacent area peripheral V1 and dorsal V2 107 , 108 , exhibits large receptive fields that can extend 30–50° in diameter 109 , respond to very fast visual motion 110 , and is associated with peripheral scene monitoring 42 , 107 , 108 , 111 . The mechanisms by which these transformations may be learned include gain modulation of neuronal responses by for example eye position to convert from retinotopic to head-based coordinates, facilitated by slow learning 95 , 101 , and then even beyond to world eye-direction-based coordinates 112 . The proposed mechanisms are analogous to learning of invariant object and face representations in the ventrolateral visual cortical stream via the FFC to the inferior temporal visual cortex 36 , 38 , 101 , 113 .
The implications of this ventromedial visual cortical ‘Where’ stream for spatial scenes for understanding hippocampal function in memory and navigation in humans and other primates are developed further elsewhere 6 , 13 , 33 , 34 , 36 , 38 , 96 .
The visual stimuli and task
The scene and face visual stimuli were those provided by the HCP that were used in their fMRI data collection 59 , and the 0-back task used by the HCP in their MEG data collection for faces and tools 58 was implemented (details are available at https://www.humanconnectome.org/hcp-protocols-ya-task-fmri and https://db.humanconnectome.org/app/action/ChooseDownloadResources?project=HCP_Resources&resource=Scripts&filePath=HCP_TFMRI_scripts.zip .) A 0-back task was used to ensure that the participants looked at and processed the visual stimuli by remembering the cue stimulus and indicating whether it appeared in the next ten trials of a run. Within each task block or run, first, a cue image was presented for 2.5 s to indicate the 0-back cue stimulus, and then ten trials were run for a given stimulus type, with each stimulus shown for 2.0 s followed by an interstimulus interval of 0.5 s in which the screen was blank. The ten stimuli in each block thus lasted for 25 s. During the experiment, participants received 8 blocks of scenes and eight blocks of faces in random order, with no repetition of any image. Examples of the scene and face stimuli provided by the HCP and used here are available 33 .
Twenty-four participants from Fudan University aged 19–30 years (13 females) participated in the experiment. Three of them had to be excluded because of MEG artefacts in the data, leaving 21 datasets for the final analysis. All participants were reported to be right-handed and have normal or corrected-to-normal vision. They reported to have no history of neurological disorders. The study received ethical approval from the Ethics Committee of the Institute of Science and Technology for Brain-Inspired Intelligence at Fudan University (reference number AF/SC-115/20230822). All participants provided informed written consent. All ethical regulations relevant to human research participants were followed.
MEG data acquisition and preprocessing
MEG data were acquired on a TRIUX neo (MEGIN Oy, Finland) system at Zhangjiang Imaging Center (ZIC), containing 306 MEG sensors (102 magnetometers and 204 gradiometers). The sampling rate during data acquisition was 1000 Hz and an online band pass filter 0.03–330 Hz was applied. Prior to data recording, five Head Position Indicator coils that were attached to the forehead of the subjects, as well as three anatomical fiducial points (two preauricular points and one nasion) were digitized using the FASTRAK Digitizer system for later co-registration with MRI data.
High-resolution structural T1-weighted MRI images were acquired in a 3-T Siemens Prisma scanner at ZIC with a 3D T1 MPRAGE sequence, field of view = 192 × 240 × 256 mm, 1 mm isotropic resolution, repetition time = 2.5 s, echo time = 2.15 ms, and flip angle = 8°.
The MEG preprocessing was performed with the MNE-Python version 1.5.1 software package ( https://zenodo.org/records/8322569 ) 114 . First, the environmental noise, recorded every day in the morning before testing, was suppressed from the raw MEG data with the spatio-temporal signal-space separation (SSP) method that is implemented in MNE-Python. A notch filter at 50 Hz and 100 Hz was then applied, followed by a band-pass filter between 1 Hz and 140 Hz. We further used the “find_bad_channels_maxwell” function from the MNE-Python software, plus visual inspection to identify bad channels, and further used the “maxwell_filter” function to implement bad channel reconstruction, movement compensation, and temporal spatiotemporal signal space separation. On average, five channels were interpolated per participant. To suppress eye movement and cardio artefacts, we used the SSP method to remove one eye and one cardio component as provided by the MNE-Python software.
The mean evoked responses (baseline-corrected) in the time domain were computed for each of the categories places (scenes) and faces, separately, with a window from −200 ms to 2500 ms after stimulus onset. The mean signals were then baseline-corrected with a window from −200 ms to 0 ms relative to stimulus onset.
The mean evoked responses (baseline corrected) in the time domain were used for source-space analysis at the surface level. Source estimation was performed using L2-minimum-norm estimation (depth weighting coefficients: 0.8, loose orientation constraint: 0.2) on an ico-5 source space (10242 sources per hemisphere). We used individual MRI images for head modelling. The MRI data were pre-processed in Freesurfer V.7.1.4 (Fischel, 2012), and the head model (1-layer boundary element model, BEM, with the default conductivity of 0.3) was created in MNE-Python. Source estimation was used with the standard ‘whitening’ approach implemented in the MNE-Python software to combine data from different sensor types.
The MEG data were converted into the HCP-MMP surface space 55 using the MNE-Python functions ‘fetch_hcp_mmp_parcellation’ and ‘extract_label_time_course’ to produce for each participant a 1 ms MEG time-series in HCP-MMP space. As described previously 54 , although the spatial resolution of the MEG data may not be sufficient to provide an independent signal for each of the 360 cortical regions in the HCP-MMP atlas, the use of the atlas is potentially valuable because the cortical regions in the atlas are themselves well-defined 55 , and use of this atlas provides a framework for comparing findings from different investigations of the cortex 25 , 38 , 48 , 49 , 57 , 63 , 64 , 65 , 67 , 68 , 69 , 87 , 115 , 116 , 117 , 118 , 119 . Also, the spatial resolution of MEG may be poorer for visual cortical regions far from the skull, due to the inverse problem, though this is unlikely to account for the findings described here, for many regions high in the hierarchy such as the TE regions are as close to the skull as early visual cortical regions (see Figs. S1 – 5 ).
The effective connectivity between the 360 cortical regions was computed with the same Hopf generative effective connectivity algorithm used for the fMRI data 48 . This is important to note, for the use of the same algorithm that the directionality with the MEG and fMRI data can be compared. Because the MEG time series could be very long, the effective connectivity was calculated with an analytic version of the Hopf algorithm, rather than the simulation approach used for fMRI data 48 . The analytic approach is described in the Supplementary Material, and when tested with fMRI data, produced very similar results to the simulation approach ( r > 0.95 for a comparison of the effective connectivities calculated with the simulation and analytic methods).
Brain atlas and region selection
To construct the effective connectivity for the cortical regions of interest in this investigation with other cortical regions in the human brain, the parcellation of human cortical regions was used that is provided by the HCP-MMP which has 360 cortical regions 55 . The cortical regions in this parcellation 55 are shown in Figs. 1 and S1 , and a list of the cortical regions and the divisions into which they are placed are provided in Table S1 in the reordered form used in the extended volumetric HCPex atlas 120 .
The 30 visual cortical regions selected for connectivity analysis here were as follows, with reference to Fig. 1 useful in showing where these regions are in the human brain. The regions are grouped based on earlier evidence 48 , 55 purely to simplify the description of the connectivity, and the groups are separated by red lines in Figs. 2 – 6 .
Group 1: early visual cortical areas V1, V2, V3, and V4 of the HCP-MMP atlas.
Group 2: the retrosplenial group includes the ProStriate region and the DVT region which are retrosplenial regions in the HCP-MMP atlas where viewing scenes produces activations with fMRI which sometimes include POS1 25 , 33 .
Group 3: the ventromedial group VMV1–3 and VVC, and Group 4: the parahippocampal PHA1–3, are regions in the HCP-MMP atlas where viewing scenes produces activations with fMRI 25 , 33 in what corresponds to the PPA 29 , 30 , which might better be termed the PSA because it is where the individual looks in scenes, not the place where the individual is located, that produces thise activations 6 .
Group 5: includes the Hipp and related regions including the EC and PeEc.
Group 6: includes the FFC, PIT and V8.
Group 7: TE1p and TE2p are the last mainly unimodal visual cortical regions, and correspond to the macaque inferior temporal visual cortex 48 , 54 .
Group 8: the anterior temporal lobe group includes TE1a, TE1m and TE2a, and the temporal pole regions TGd and TGv, which are multimodal semantic regions 48 , 54 , 63 .
Measurement of effective connectivity
The effective connectivity between all pairs of the 360 cortical regions was computed with the same Hopf generative effective connectivity algorithm used for the fMRI data 48 , and the text that follows are similar to that used in some earlier descriptions, which helps to show that the same effective connectivity algorithm was used as in the earlier investigations 48 , 54 , 63 , 65 , 66 , 68 , 69 , 87 . Effective connectivity measures the effect of one brain region on another, and utilizes differences detected at different times in the signals in each connected pair of brain regions to infer the effects of one brain region on another. One such approach is dynamic causal modelling, but it applies most easily to activation studies, and is typically limited to measuring the effective connectivity between just a few brain areas 121 , 122 , 123 , though there have been moves to extend it to resting state studies and more brain areas 124 , 125 . The method used here in refs. 57 , 64 was developed from a Hopf algorithm to enable the measurement of effective connectivity between many brain areas, as described by Deco et al. 73 . A principle is that the functional connectivity is measured at time t and time t + tau , where tau was set to 20 ms.
To infer effective connectivity, we use a whole-brain model that allows us to analyse the MEG signal across all brain regions and time. We use the so-called Hopf computational model, which integrates the dynamics of Stuart–Landau oscillators, expressing the activity of each brain region, by the underlying anatomical connectivity 72 . As mentioned above, we include in the model 360 cortical brain areas 120 . The local dynamics of each brain area (node) is given by Stuart–Landau oscillators which express the normal form of a supercritical Hopf bifurcation, describing the transition from noisy to oscillatory dynamics 126 . During the last years, numerous studies have been able to show how the Hopf whole-brain model successfully simulates empirical electrophysiology 127 , 128 , MEG 129 , and fMRI 72 , 130 , 131 , 132 .
The Hopf whole-brain model can be expressed mathematically as follows:
Equations 1 and 2 describe the coupling of Stuart–Landau oscillators through an effective connectivity matrix C. The \({x}_{i}\left(t\right)\) term represents the simulated BOLD signal data of brain area i . The values of \({y}_{i}\left(t\right)\) are relevant to the dynamics of the system but are not part of the information read out from the system. These equations, \({{{{\rm{\eta }}}}}_{i}\left(t\right)\) provides additive Gaussian noise with standard deviation β. The Stuart–Landau oscillators for each brain area i express a Hopf normal form that has a supercritical bifurcation at \({a}_{i}\) = 0, so that if \({a}_{i}\) > 0 the system has a stable limit cycle with frequency \({f}_{i}\) = \({{{{\rm{\omega }}}}}_{i}\) /2π (where \({{{{\rm{\omega }}}}}_{i}\) is the angular velocity); and when \({a}_{i}\) < 0 the system has a stable fixed point representing a low activity noisy state. The intrinsic frequencies are fitted from the data, as given by the averaged peak frequency of the narrowband BOLD signals of each brain region. The intrinsic frequency \({f}_{i}\) of each Stuart–Landau oscillator corresponding to a brain area i was in the 0.5–2 Hz band ( i = 1, …, 360) for the HCP MEG data used here, which was sampled at 20 ms and not further filtered. The mean power spectrum across participants from the time series of the MEG signal for each of the 360 cortical regions used in the analyses described here is shown in Fig. S3 . The coupling term representing the input received in node i from every other node j , is weighted by the corresponding effective connectivity \({C}_{{ij}}\) . The coupling is the canonical diffusive coupling, which approximates the simplest (linear) part of a general coupling function. G denotes the global coupling weight, scaling equally the total input received in each brain area. While the oscillators are weakly coupled, the periodic orbit of the uncoupled oscillators is preserved.
The effective connectivity ( \(C\) ) matrix is derived by optimizing the conductivity of each connection in the matrix in order to fit the empirical functional connectivity ( \({{{{\boldsymbol{FC}}}}}^{{{\rm{empirical}}}}\) ) pairs and the lagged normalised covariance, the \({{{\boldsymbol{F}}}}{{{{\boldsymbol{S}}}}}^{{{\rm{empirical}}}}\) pairs. By this, we are able to infer a non-symmetric Effective Connectivity matrix (see ref. 133 ). We refer to this as a generative effective connectivity model approach because the \(C\) matrix is used to generate the functional connectivity and lagged normalised covariance matrices, and the \(C\) matrix is optimised so that the simulated matrices match the empirically measured matrices. Note that \({{{\boldsymbol{F}}}}{{{{\boldsymbol{S}}}}}^{{{\rm{empirical}}}}\) , i.e. the normalised lagged covariance of the functional connectivity between pairs, lagged at \(\tau\) , breaks the symmetry and thus is fundamental for our purpose. Specifically, we compute the distance between the model functional connectivity \({{{{\boldsymbol{FC}}}}}^{{{\rm{model}}}}\) calculated analytically from the current estimate of the effective connectivity and the empirical data \({{{{\boldsymbol{FC}}}}}^{{{\rm{empirical}}}}\) , as well as the calculated model \({{{{\boldsymbol{FS}}}}}^{{{\rm{model}}}}\) and empirical data \({{{\boldsymbol{F}}}}{{{{\boldsymbol{S}}}}}^{{{\rm{empirical}}}}\) and adjust each effective connection (entry in the effective connectivity matrix) separately with a gradient-descent approach. The model is run repeatedly with the updated effective connectivity until the fit converges towards a stable value.
We start with the anatomical connectivity obtained with probabilistic tractography from dMRI (or from an initial zero C ij matrix) and use a pseudo gradient procedure for updating the effective connectivity matrix (see Eq. 11 in the Supplementary Material). The effective connectivity matrices shown here were those computed without the structural connection matrix, as use of the structural connectivity matrix limited the connectivity to fewer links than were otherwise found with these MEG data, probably because the DTI analysis missed some connections. However, the correlation between the matrices produced with these different methods was reasonable (0.80).
Effective connectome
Whole-brain effective connectivity ( EC ) analysis was performed between the 30 visual cortical regions described above (see Fig. 1 and S1 ) and the 360 regions defined in the surface-based HCP-MMP atlas 55 in their reordered form provided in Table S1 , described in the Supplementary Material 120 . This EC was computed across all 21 participants, and the reliability was checked by a data split, which showed a correlation of 0.85 between the two halves. For each participant, the mean for the 2700-point long time series was calculated for each trial type (places/scenes and faces). From this, the functional connectivity FC for the 360 cortical regions and the covariance COV of the connectivity for the 360 cortical regions were calculated from the time series and the time series delayed by tau (where tau =20 ms) was calculated for each participant, and then the FC and COV matrices were averaged across participants. These provided the inputs FC emp and COV tauemp to the effective connectivity algorithm ( COV tauemp refers to the \({{{\boldsymbol{F}}}}{{{{\boldsymbol{S}}}}}^{{{\rm{empirical}}}}\) defined above.) Because effective connectivity measured in the way described utilises functional connectivity and what can be viewed as a lagged functional connectivity with a lag of tau, effective connectivity is not limited to measuring direct neuronal connections, but may reflect connectivity over perhaps 1–3 synapses (based on the evidence for example that V1 does not have effective connectivity with this approach with all visual cortical regions). We emphasize though that the generative effective connectivity algorithm is non-linear with respect to the functional connectivity, in that the generative effective connectivity algorithm sets to zero those connectivities that are not useful in generating the FC and COV matrices.
Statistics and reproducibility
The key statistical analyses for the new investigation described here are on the directionality of the effective connectivities between key sets of cortical regions when scenes are being viewed, with the data illustrated in Fig. 3 . Paired within-subjects comparisons were performed to test the differences in the effective connectivities in the two directions calculated for each participant. The degrees of freedom reflect the number of participants (21). The tests were two-tailed, and exact values were provided for the t and p values to provide evidence about the reliability of each test performed. Only a few preplanned tests were performed. Similar tests were performed for key cortical regions in the ventrolateral ‘What’ stream for faces and objects 34 based on the data shown in Fig. 4 when faces were the stimuli. Further, the effective connectivity was computed across all 21 participants, and the reliability was checked by a data split, which showed a correlation of 0.85 between the two halves.
We note that the use of MEG is important, for the directionality of the effective connectivity when measured with resting-state fMRI for faces, places, tools and body parts shows as the reverse of what is expected and of what is found with MEG 48 , and this is probably related to the slow timecourse of fMRI which means that much of what is measured with fMRI resting state effective connectivity is the top–down effects from the top of the visual hierarchy where short-term memory keeps representations active 54 . All of our previous publications on effective connectivity measured with this algorithm with fMRI in different brain systems do show the effective connectivity in the direction found with MEG, and that would be expected for the initial flow of signal up through a sensory cortical hierarchy starting for example with V1 and progressing to the temporal lobe 48 , 49 , 57 , 63 , 64 , 65 , 67 , 68 , 69 , 87 .
Functional connectivity
For comparison with the effective connectivity, the functional connectivity was also measured from the MEG signals with the identical set of participants and data. The functional connectivity was measured by the Pearson correlation between the MEG signal time series for each pair of brain regions, and is the FC emp referred to above. A threshold was used for the presentation of the findings in Fig. S3 , to set the sparseness of what is shown to a level commensurate with the effective connectivity, to facilitate comparison between the functional and the effective connectivity. The functional connectivity can provide evidence that may relate to interactions between brain regions, while providing no evidence about causal direction-specific effects. High functional connectivity may in this scenario thus reflect strong physiological interactions between areas, and provide a different type of evidence to effective connectivity. The effective connectivity is non-linearly related to the functional connectivity, with effective connectivities being identified (i.e. greater than zero) only for the links with relatively high functional connectivity.
Connections shown with diffusion tractography
Diffusion tractography can provide evidence about fibre pathways linking different brain regions with a method that is completely different to the ways in which effective and functional connectivity are measured. Diffusion tractography shows only direct connections, so comparison with effective connectivity can help to suggest which effective connectivities may be mediated directly or indirectly. Diffusion tractography does not provide evidence about the direction of connections. Diffusion tractography was performed in a set of 171 HCP participants imaged at 7 T with methods described in detail elsewhere 56 . Some of the results are provided elsewhere 48 , 56 , but are shown in Fig. 5 for exactly the visual cortical regions investigated here with MEG, to facilitate comparison.
The major parameters were: 1.05 mm isotropic voxels; a two-shell acquisition scheme with b-values = 1000 s/mm 2 , 2000 s/mm 2 , repetition time/echo time = 7000/71 ms, 65 unique diffusion gradient directions and 6 b0 images obtained for each phase encoding direction pair (AP and PA pairs). Pre-processing steps included distortion correction, eddy-current correction, motion correction, and gradient non-linearity correction. In brief, whole-brain tractography was reconstructed for each subject in native space. To improve the tractography termination accuracy in GM, MRtrix3’s 5ttgen command was used to generate multi-tissue segment images (5tt) using T1 images, the segmented tissues were then co-registered with the b0 image in diffusion space. For multi-shell data, tissue response functions in GM, WM, and CSF were estimated by the MRtrix3’ dwi2response function with the Dhollander algorithm 134 . A multi-shell multi-tissue constrained spherical deconvolution model with lmax = 8 and prior co-registered 5tt image was used on the preprocessed multi-shell DWI data to obtain the fibre orientation distribution (FOD) function 135 , 136 . Based on the voxel-wise FOD, anatomically-constrained tractography using the probabilistic tracking algorithm: iFOD2 (2nd order integration based on FOD) with dynamic seeding was applied to generate the initial tractogram (1 million streamlines with maximum tract length = 250 mm and minimal tract length = 5 mm). To quantify the number of streamlines connecting pairs of regions, the updated version of the spherical-deconvolution informed filtering of the tractograms method was applied, which provides more biologically meaningful estimates of structural connection density 137 .
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The MEG data which are very large are available from the corresponding author on reasonable request.
Code availability
Code for the Hopf generative effective connectivity algorithm is available at https://github.com/decolab/gec . Matlab 2023; Freesurfer V.7.1.4; and MNE-Python Version 1.5.1 were used.
Moscovitch, M., Cabeza, R., Winocur, G. & Nadel, L. Episodic memory and beyond: the hippocampus and neocortex in transformation. Annu. Rev. Psychol. 67 , 105–134 (2016).
Article PubMed PubMed Central Google Scholar
Squire, L. R. & Wixted, J. T. The cognitive neuroscience of human memory since H.M. Annu. Rev. Neurosci. 34 , 259–288 (2011).
Article CAS PubMed PubMed Central Google Scholar
Burgess, N., Jackson, A., Hartley, T. & O’Keefe, J. Predictions derived from modelling the hippocampal role in navigation. Biol. Cybern. 83 , 301–312 (2000).
Article CAS PubMed Google Scholar
O’Keefe, J., Burgess, N., Donnett, J. G., Jeffery, K. J. & Maguire, E. A. Place cells, navigational accuracy, and the human hippocampus. Philos. Trans. R. Soc. B 353 , 1333–1340 (1998).
Article Google Scholar
Burgess, N. & O’Keefe, J. Neuronal computations underlying the firing of place cells and their role in navigation. Hippocampus 6 , 749–762 (1996).
Rolls, E. T. Hippocampal spatial view cells for memory and navigation, and their underlying connectivity in humans. Hippocampus 33 , 533–572 (2023).
Article PubMed Google Scholar
O’Keefe, J. A review of the hippocampal place cells. Prog. Neurobiol. 13 , 419–439 (1979).
Moser, E. I., Moser, M. B. & McNaughton, B. L. Spatial representation in the hippocampal formation: a history. Nat. Neurosci. 20 , 1448–1464 (2017).
Georges-François, P., Rolls, E. T. & Robertson, R. G. Spatial view cells in the primate hippocampus: allocentric view not head direction or eye position or place. Cereb. Cortex 9 , 197–212 (1999).
Rolls, E. T., Treves, A., Robertson, R. G., Georges-François, P. & Panzeri, S. Information about spatial view in an ensemble of primate hippocampal cells. J. Neurophysiol. 79 , 1797–1813 (1998).
Robertson, R. G., Rolls, E. T. & Georges-François, P. Spatial view cells in the primate hippocampus: Effects of removal of view details. J. Neurophysiol. 79 , 1145–1156 (1998).
Rolls, E. T., Robertson, R. G. & Georges-François, P. Spatial view cells in the primate hippocampus. Eur. J. Neurosci. 9 , 1789–1794 (1997).
Rolls, E. T. Hippocampal spatial view cells, place cells, and concept cells: view representations. Hippocampus 33 , 667–687 (2023).
Rolls, E. T. et al. Hippocampal neurons in the monkey with activity related to the place in which a stimulus is shown. J. Neurosci. 9 , 1835–1845 (1989).
Rolls, E. T. & O’Mara, S. M. View-responsive neurons in the primate hippocampal complex. Hippocampus 5 , 409–424 (1995).
Wirth, S., Baraduc, P., Plante, A., Pinede, S. & Duhamel, J. R. Gaze-informed, task-situated representation of space in primate hippocampus during virtual navigation. PLoS Biol. 15 , e2001045 (2017).
Zhu, S. L., Lakshminarasimhan, K. J. & Angelaki, D. E. Computational cross-species views of the hippocampal formation. Hippocampus 33 , 586–599 (2023).
Mao, D. et al. Spatial modulation of hippocampal activity in freely moving macaques. Neuron 109 , 3521–3534.e3526 (2021).
Tsitsiklis, M. et al. Single-neuron representations of spatial targets in humans. Curr. Biol. 30 , 245–253.e244 (2020).
Donoghue, T. et al. Single neurons in the human medial temporal lobe flexibly shift representations across spatial and memory tasks. Hippocampus 33 , 600–615 (2023).
Qasim, S. E. et al. Memory retrieval modulates spatial tuning of single neurons in the human entorhinal cortex. Nat. Neurosci. 22 , 2078–2086 (2019).
Qasim, S. E., Fried, I. & Jacobs, J. Phase precession in the human hippocampus and entorhinal cortex. Cell 184 , 3242–3255.e3210 (2021).
Ison, M. J., Quian Quiroga, R. & Fried, I. Rapid encoding of new memories by individual neurons in the human brain. Neuron 87 , 220–230 (2015).
Piza, D. B. et al. Primacy of vision shapes behavioral strategies and neural substrates of spatial navigation in marmoset hippocampus. Nat. Commun. 15 , 4053 (2024).
Sulpizio, V., Galati, G., Fattori, P., Galletti, C. & Pitzalis, S. A common neural substrate for processing scenes and egomotion-compatible visual motion. Brain Struct. Funct. 225 , 2091–2110 (2020).
Natu, V. S. et al. Sulcal depth in the medial ventral temporal cortex predicts the location of a place-selective region in macaques, children, and adults. Cereb. Cortex 31 , 48–61 (2021).
Kamps, F. S., Julian, J. B., Kubilius, J., Kanwisher, N. & Dilks, D. D. The occipital place area represents the local elements of scenes. Neuroimage 132 , 417–424 (2016).
Epstein, R. & Kanwisher, N. A cortical representation of the local visual environment. Nature 392 , 598–601 (1998).
Epstein, R. A. & Baker, C. I. Scene perception in the human brain. Annu Rev. Vis. Sci. 5 , 373–397 (2019).
Epstein, R. A. & Julian, J. B. Scene areas in humans and macaques. Neuron 79 , 615–617 (2013).
Epstein, R. A. Parahippocampal and retrosplenial contributions to human spatial navigation. Trends Cogn. Sci. 12 , 388–396 (2008).
Epstein, R. The cortical basis of visual scene processing. Vis. Cogn. 12 , 954–978 (2005).
Rolls, E. T., Feng, J. & Zhang, R. Selective activations and functional connectivities to the sight of faces, scenes, body parts and tools in visual and non-visual cortical regions leading to the human hippocampus. Brain Struct. Funct. 229 , 1471–1493 (2024).
Rolls, E. T. Two what, two where, visual cortical streams in humans. Neurosci. Biobehav. Rev. 160 , 105650 (2024).
Rolls, E. T. Neurons including hippocampal spatial view cells, and navigation in primates including humans. Hippocampus 31 , 593–611 (2021).
Rolls, E. T. Hippocampal discoveries: spatial view cells, and computations for memory and navigation, in primates including humans. Hippocampus (2024).
Stringer, S. M., Rolls, E. T. & Trappenberg, T. P. Self-organizing continuous attractor network models of hippocampal spatial view cells. Neurobiol. Learn. Mem. 83 , 79–92 (2005).
Rolls, E. T. Brain Computations and Connectivity (Oxford University Press, 2023).
Rolls, E. T. & Wirth, S. Spatial representations in the primate hippocampus, and their functions in memory and navigation. Prog. Neurobiol. 171 , 90–113 (2018).
Rolls, E. T. The memory systems of the human brain and generative artificial intelligence. Heliyon 10 , e31965 (2024).
Maguire, E. A. The retrosplenial contribution to human navigation: a review of lesion and neuroimaging findings. Scand. J. Psychol. 42 , 225–238 (2001).
Silson, E. H., Steel, A. D. & Baker, C. I. Scene-selectivity and retinotopy in medial parietal cortex. Front. Hum. Neurosci. 10 , 412 (2016).
Dilks, D. D., Julian, J. B., Paunov, A. M. & Kanwisher, N. The occipital place area is causally and selectively involved in scene perception. J. Neurosci. 33 , 1331–1336a (2013).
Hasson, U., Harel, M., Levy, I. & Malach, R. Large-scale mirror-symmetry organization of human occipito-temporal object areas. Neuron 37 , 1027–1041 (2003).
Baldassano, C., Esteva, A., Fei-Fei, L. & Beck, D. M. Two distinct scene-processing networks connecting vision and memory. eNeuro 3 , e0178–0116.2016 (2016).
Nasr, S., Devaney, K. J. & Tootell, R. B. Spatial encoding and underlying circuitry in scene-selective cortex. Neuroimage 83 , 892–900 (2013).
Watson, D. M. & Andrews, T. J. Mapping the functional and structural connectivity of the scene network. Hum. Brain Mapp. 45 , e26628 (2024).
Rolls, E. T., Deco, G., Huang, C.-C. & Feng, J. Multiple cortical visual streams in humans. Cereb. Cortex 33 , 3319–3349 (2023).
Rolls, E. T., Wirth, S., Deco, G., Huang, C.-C. & Feng, J. The human posterior cingulate, retrosplenial and medial parietal cortex effective connectome, and implications for memory and navigation. Hum. Brain Mapp. 44 , 629–655 (2023).
Libby, L. A., Ekstrom, A. D., Ragland, J. D. & Ranganath, C. Differential connectivity of perirhinal and parahippocampal cortices within human hippocampal subregions revealed by high-resolution functional imaging. J. Neurosci. 32 , 6550–6560 (2012).
Steel, A., Billings, M. M., Silson, E. H. & Robertson, C. E. A network linking scene perception and spatial memory systems in posterior cerebral cortex. Nat. Commun. 12 , 2632 (2021).
Kahn, I., Andrews-Hanna, J. R., Vincent, J. L., Snyder, A. Z. & Buckner, R. L. Distinct cortical anatomy linked to subregions of the medial temporal lobe revealed by intrinsic functional connectivity. J. Neurophysiol. 100 , 129–139 (2008).
Reznik, D., Trampel, R., Weiskopf, N., Witter, M. P. & Doeller, C. F. Dissociating distinct cortical networks associated with subregions of the human medial temporal lobe using precision neuroimaging. Neuron 111 , 2756–2772.e2757 (2023).
Rolls, E. T., Deco, G., Zhang, Y. & Feng, J. Hierarchical organization of the human ventral visual streams revealed with magnetoencephalography. Cereb. Cortex 33 , 10686–10701 (2023).
Glasser, M. F. et al. A multi-modal parcellation of human cerebral cortex. Nature 536 , 171–178 (2016).
Huang, C.-C., Rolls, E. T., Hsu, C.-C. H., Feng, J. & Lin, C.-P. Extensive cortical connectivity of the human hippocampal memory system: beyond the “what” and “where” dual-stream model. Cereb. Cortex 31 , 4652–4669 (2021).
Rolls, E. T., Deco, G., Huang, C. C. & Feng, J. The effective connectivity of the human hippocampal memory system. Cereb. Cortex 32 , 3706–3725 (2022).
Larson-Prior, L. J. et al. Adding dynamics to the human connectome project with MEG. Neuroimage 80 , 190–201 (2013).
Barch, D. M. et al. Function in the human connectome: task-fMRI and individual differences in behavior. Neuroimage 80 , 169–189 (2013).
Yokoyama, C. et al. Comparative connectomics of the primate social brain. Neuroimage 245 , 118693 (2021).
Van Essen, D. C. & Glasser, M. F. Parcellating cerebral cortex: how invasive animal studies inform noninvasive mapmaking in humans. Neuron 99 , 640–663 (2018).
Colclough, G. L. et al. The heritability of multi-modal connectivity in human brain activity. Elife 6 , e20178 (2017).
Rolls, E. T., Deco, G., Huang, C.-C. & Feng, J. The human language effective connectome. Neuroimage 258 , 119352 (2022).
Rolls, E. T., Deco, G., Huang, C. C. & Feng, J. The human orbitofrontal cortex, vmPFC, and anterior cingulate cortex effective connectome: emotion, memory, and action. Cereb. Cortex 33 , 330–356 (2022).
Rolls, E. T., Deco, G., Huang, C. C. & Feng, J. The human posterior parietal cortex: effective connectome, and its relation to function. Cereb. Cortex 33 , 3142–3170 (2023).
Ma, Q., Rolls, E. T., Huang, C.-C., Cheng, W. & Feng, J. Extensive cortical functional connectivity of the human hippocampal memory system. Cortex 147 , 83–101 (2022).
Rolls, E. T., Deco, G., Huang, C.-C. & Feng, J. Human amygdala compared to orbitofrontal cortex connectivity, and emotion. Prog. Neurobiol. 220 , 102385 (2023).
Rolls, E. T., Deco, G., Huang, C. C. & Feng, J. Prefrontal and somatosensory-motor cortex effective connectivity in humans. Cereb. Cortex 33 , 4939–4963 (2023).
Rolls, E. T., Rauschecker, J. P., Deco, G., Huang, C. C. & Feng, J. Auditory cortical connectivity in humans. Cereb. Cortex 33 , 6207–6227 (2023).
Baker, C. M. et al. A connectomic atlas of the human cerebrum-chapter 7: the lateral parietal lobe. Oper. Neurosurg. 15 , S295–S349 (2018).
Baker, C. M. et al. A connectomic atlas of the human cerebrum-chapter 6: the temporal lobe. Oper. Neurosurg. 15 , S245–S294 (2018).
Deco, G., Kringelbach, M. L., Jirsa, V. K. & Ritter, P. The dynamics of resting fluctuations in the brain: metastability and its dynamical cortical core. Sci. Rep. 7 , 3095 (2017).
Deco, G. et al. Awakening: predicting external stimulation to force transitions between different brain states. Proc. Natl. Acad. Sci. 116 , 18088–18097 (2019).
Wallis, G. & Rolls, E. T. Invariant face and object recognition in the visual system. Prog. Neurobiol. 51 , 167–194 (1997).
Panzeri, S., Rolls, E. T., Battaglia, F. & Lavis, R. Speed of feedforward and recurrent processing in multilayer networks of integrate-and-fire neurons. Network 12 , 423–440 (2001).
Rolls, E. T. Cerebral Cortex: Principles of Operation (Oxford University Press, 2016).
Battaglia, F. P. & Treves, A. Stable and rapid recurrent processing in realistic auto-associative memories. Neural Comput. 10 , 431–450 (1998).
Rolls, E. T. & Turova, T. S. Visual cortical networks for ‘What’ and ‘Where’ to the human hippocampus revealed with dynamical graphs (2024).
Rolls, E. T. Neurophysiological mechanisms underlying face processing within and beyond the temporal cortical visual areas. Philos. Trans. R. Soc. Lond. B 335 , 11–21 (1992).
Article CAS Google Scholar
Baylis, G. C., Rolls, E. T. & Leonard, C. M. Functional subdivisions of the temporal lobe neocortex. J. Neurosci. 7 , 330–342 (1987).
Hasselmo, M. E., Rolls, E. T., Baylis, G. C. & Nalwa, V. Object-centred encoding by face-selective neurons in the cortex in the superior temporal sulcus of the monkey. Exp. Brain Res. 75 , 417–429 (1989).
Hasselmo, M. E., Rolls, E. T. & Baylis, G. C. The role of expression and identity in the face-selective responses of neurons in the temporal visual cortex of the monkey. Behav. Brain Res. 32 , 203–218 (1989).
Pitcher, D. & Ungerleider, L. G. Evidence for a third visual pathway specialized for social perception. Trends Cogn. Sci. 25 , 100–110 (2021).
Scherf, K. S., Behrmann, M., Humphreys, K. & Luna, B. Visual category-selectivity for faces, places and objects emerges along different developmental trajectories. Dev. Sci. 10 , F15–F30 (2007).
Kanwisher, N., McDermott, J. & Chun, M. M. The fusiform face area: a module in human extrastriate cortex specialized for face perception. J. Neurosci. 17 , 4302–4311 (1997).
Rolls, E. T. The hippocampus, ventromedial prefrontal cortex, and episodic and semantic memory. Prog. Neurobiol. 217 , 102334 (2022).
Rolls, E. T., Deco, G., Huang, C. C. & Feng, J. The connectivity of the human frontal pole cortex, and a theory of its involvement in exploit versus explore. Cereb. Cortex 34 , 1–19 (2024).
Rolls, E. T. & Xiang, J.-Z. Spatial view cells in the primate hippocampus, and memory recall. Rev. Neurosci. 17 , 175–200 (2006).
Rolls, E. T., Xiang, J.-Z. & Franco, L. Object, space and object-space representations in the primate hippocampus. J. Neurophysiol. 94 , 833–844 (2005).
Rolls, E. T. & Xiang, J.-Z. Reward-spatial view representations and learning in the hippocampus. J. Neurosci. 25 , 6167–6174 (2005).
Rolls, E. T., Zhang, R., Deco, G., Vatansever, D. & Feng, J. Selective brain activations and connectivities related to the storage and recall of human object-location, reward-location, and word-pair episodic memories. Hum. Brain Mapp . (2024).
McNaughton, B. L. et al. Deciphering the hippocampal polyglot: the hippocampus as a path integration system. J. Exp. Biol. 199 , 173–185 (1996).
Jiang, R., Andolina, I. M., Li, M. & Tang, S. Clustered functional domains for curves and corners in cortical area V4. Elife 10 , e63798 (2021).
Kim, T., Bair, W. & Pasupathy, A. Neural coding for shape and texture in macaque area V4. J. Neurosci. 39 , 4760–4774 (2019).
Rolls, E. T. Spatial coordinate transforms linking the allocentric hippocampal and egocentric parietal primate brain systems for memory, action in space, and navigation. Hippocampus 30 , 332–353 (2020).
Rolls, E. T. & Treves, A. A theory of hippocampal function: new developments. Prog. Neurobiol. 238 , 102636 (2024).
Rolls, E. T., Zhang, C. & Feng, J. Hippocampal storage and recall of neocortical ‘What’–‘Where’ representations. Hippocampus https://doi.org/10.1002/hipo.23636 (2024).
Haak, K. V. & Beckmann, C. F. Objective analysis of the topological organization of the human cortical visual connectome suggests three visual pathways. Cortex 98 , 73–83 (2018).
De Araujo, I. E. T., Rolls, E. T. & Stringer, S. M. A view model which accounts for the spatial fields of hippocampal primate spatial view cells and rat place cells. Hippocampus 11 , 699–706 (2001).
Rolls, E. T. A theory and model of scene representations with hippocampal spatial view cells (2024).
Rolls, E. T. Learning invariant object and spatial view representations in the brain using slow unsupervised learning. Front. Comput. Neurosci. 15 , 686239 (2021).
Hubel, D. H. & Wiesel, T. N. Ferrier lecture. Functional architecture of macaque monkey visual cortex. Proc. R. Soc. Lond. B. Biol. Sci. 198 , 1–59 (1977).
Wei, H., Dong, Z. & Wang, L. V4 shape features for contour representation and object detection. Neural Netw. 97 , 46–61 (2018).
Nandy, A. S., Sharpee, T. O., Reynolds, J. H. & Mitchell, J. F. The fine structure of shape tuning in area V4. Neuron 78 , 1102–1115 (2013).
Roe, A. W. et al. Toward a unified theory of visual area V4. Neuron 74 , 12–29 (2012).
Feigenbaum, J. D. & Rolls, E. T. Allocentric and egocentric spatial information processing in the hippocampal formation of the behaving primate. Psychobiology 19 , 21–40 (1991).
Nasr, S. et al. Scene-selective cortical regions in human and nonhuman primates. J. Neurosci. 31 , 13771–13785 (2011).
Elshout, J. A., van den Berg, A. V. & Haak, K. V. Human V2A: a map of the peripheral visual hemifield with functional connections to scene-selective cortex. J. Vis. 18 , 22 (2018).
Yu, H. H., Chaplin, T. A., Davies, A. J., Verma, R. & Rosa, M. G. A specialized area in limbic cortex for fast analysis of peripheral vision. Curr. Biol. 22 , 1351–1357 (2012).
Mikellidou, K. et al. Area prostriata in the human brain. Curr. Biol. 27 , 3056–3060.e3053 (2017).
Solomon, S. G. & Rosa, M. G. A simpler primate brain: the visual system of the marmoset monkey. Front. Neural. Circuits 8 , 96 (2014).
Snyder, L. H., Grieve, K. L., Brotchie, P. & Andersen, R. A. Separate body- and world-referenced representations of visual space in parietal cortex. Nature 394 , 887–891 (1998).
Rolls, E. T. Invariant visual object and face recognition: neural and computational bases, and a model, VisNet. Front. Comput. Neurosci. 6 , 1–70 (2012).
Gramfort, A. et al. MEG and EEG data analysis with MNE-Python. Front. Neurosci. 7 , 267 (2013).
Rolls, E. T., Feng, R., Cheng, W. & Feng, J. Orbitofrontal cortex connectivity is associated with food reward and body weight in humans. Soc. Cogn. Affect. Neurosci. 18 , nsab083 (2023).
Wan, Z., Rolls, E. T., Cheng, W. & Feng, J. Brain functional connectivities that mediate the association between childhood traumatic events and adult mental health and cognition. EBioMedicine 79 , 104002 (2022).
Zhang, R., Rolls, E. T., Cheng, W. & Feng, J. Different cortical connectivities in human females and males relate to differences in strength and body composition, reward and emotional systems, and memory. Brain Struct. Funct. 229 , 47–61 (2024).
Rolls, E. T., Feng, R. & Feng, J. Lifestyle risks associated with brain functional connectivity and structure. Hum. Brain Mapp. 44 , 2479–2492 (2023).
Rolls, E. T. Emotion, motivation, decision-making, the orbitofrontal cortex, anterior cingulate cortex, and the amygdala. Brain Struct. Funct. 228 , 1201–1257 (2023).
Huang, C. C., Rolls, E. T., Feng, J. & Lin, C. P. An extended human connectome project multimodal parcellation atlas of the human cortex and subcortical areas. Brain Struct. Funct. 227 , 763–778 (2022).
Valdes-Sosa, P. A., Roebroeck, A., Daunizeau, J. & Friston, K. Effective connectivity: influence, causality and biophysical modeling. Neuroimage 58 , 339–361 (2011).
Bajaj, S., Adhikari, B. M., Friston, K. J. & Dhamala, M. Bridging the gap: dynamic causal modeling and granger causality analysis of resting state functional magnetic resonance imaging. Brain Connect. 6 , 652–661 (2016).
Friston, K. Causal modelling and brain connectivity in functional magnetic resonance imaging. PLoS Biol. 7 , e33 (2009).
Razi, A. et al. Large-scale DCMs for resting-state fMRI. Netw. Neurosci. 1 , 222–241 (2017).
Frassle, S. et al. Regression DCM for fMRI. Neuroimage 155 , 406–421 (2017).
Kuznetsov, Y. A. (ed) Elements of applied bifurcation theory (Springer Science and Business Media, 2013).
Freyer, F. et al. Biophysical mechanisms of multistability in resting-state cortical rhythms. J. Neurosci. 31 , 6353–6361 (2011).
Freyer, F., Roberts, J. A., Ritter, P. & Breakspear, M. A canonical model of multistability and scale-invariance in biological systems. PLoS Comput. Biol. 8 , e1002634 (2012).
Deco, G. et al. Single or multiple frequency generators in on-going brain activity: a mechanistic whole-brain model of empirical MEG data. Neuroimage 152 , 538–550 (2017).
Kringelbach, M. L., McIntosh, A. R., Ritter, P., Jirsa, V. K. & Deco, G. The rediscovery of slowness: exploring the timing of cognition. Trends Cogn. Sci. 19 , 616–628 (2015).
Kringelbach, M. L. & Deco, G. Brain states and transitions: insights from computational neuroscience. Cell Rep. 32 , 108128 (2020).
Kringelbach, M. L., Perl, Y. S., Tagliazucchi, E. & Deco, G. Toward naturalistic neuroscience: Mechanisms underlying the flattening of brain hierarchy in movie-watching compared to rest and task. Sci. Adv. 9 , eade6049 (2023).
Gilson, M., Moreno-Bote, R., Ponce-Alvarez, A., Ritter, P. & Deco, G. Estimation of directed effective connectivity from fMRI functional connectivity hints at asymmetries in the cortical connectome. PLoS Comput. Biol. 12 , e1004762 (2016).
Dhollander, T., Raffelt, D. & Connelly, A. Unsupervised 3-tissue response function estimation from single-shell or multi-shell diffusion MR data without a co-registered T1 image. In ISMRM Workshop on Breaking the Barriers of Diffusion MRI 5 (ISMRM, Lisbon, 2016).
Jeurissen, B., Tournier, J. D., Dhollander, T., Connelly, A. & Sijbers, J. Multi-tissue constrained spherical deconvolution for improved analysis of multi-shell diffusion MRI data. Neuroimage 103 , 411–426 (2014).
Smith, S. M. Fast robust automated brain extraction. Hum. Brain Mapp. 17 , 143–155 (2002).
Smith, R. E., Tournier, J. D., Calamante, F. & Connelly, A. SIFT2: Enabling dense quantitative assessment of brain white matter connectivity using streamlines tractography. Neuroimage 119 , 338–351 (2015).
Download references
Acknowledgements
The neuroimaging data used for the diffusion tractography were provided by the HCP, WU-Minn Consortium (Principal Investigators: David Van Essen and Kamil Ugurbil; 1U54MH091657) funded by the 16 NIH Institutes and Centres that support the NIH Blueprint for Neuroscience Research; and by the McDonnell Center for Systems Neuroscience at Washington University. The research described here was supported by the following grants. Professor J. Feng: National Key R&D Programme of China (No. 2019YFA0709502); 111 Project (No. B18015); Shanghai Municipal Science and Technology Major Project (No. 2018SHZDZX01), ZJLab, and Shanghai Center for Brain Science and Brain-Inspired Technology; and National Key R&D Programme of China (No. 2018YFC1312904). G.D. is supported by a Spanish national research project (ref. PID2019-105772GB-I00 MCIU AEI) funded by the Spanish Ministry of Science, Innovation and Universities (MCIU), State Research Agency (AEI); HBP SGA3 Human Brain Project Specific Grant Agreement 3 (grant agreement no. 945539), funded by the EU H2020 FET Flagship programme; SGR Research Support Group support (ref. 2017 SGR 1545), funded by the Catalan Agency for Management of University and Research Grants (AGAUR); Neurotwin Digital twins for model-driven non-invasive electrical brain stimulation (grant agreement ID: 101017716) funded by the EU H2020 FET Proactive programme; euSNN European School of Network Neuroscience (grant agreement ID: 860563) funded by the EU H2020 MSCA-ITN Innovative Training Networks; CECH The Emerging Human Brain Cluster (Id. 001-P-001682) within the framework of the European Research Development Fund Operational Programme of Catalonia 2014-2020; Brain-Connects: Brain Connectivity during Stroke Recovery and Rehabilitation (id. 201725.33) funded by the Fundacio La Marato TV3; Corticity, FLAG˙˙ERA JTC 2017, (ref. PCI2018-092891) funded by the Spanish MCIU, State Research Agency (AEI). The funding sources had no role in the study design; in the collection, analysis and interpretation of data; in the writing of the report; and in the decision to submit the article for publication.
Author information
These authors contributed equally: Edmund T. Rolls, Xiaoqian Yan.
Authors and Affiliations
Oxford Centre for Computational Neuroscience, Oxford, UK
Edmund T. Rolls
Department of Computer Science, University of Warwick, Coventry, UK
Edmund T. Rolls & Jianfeng Feng
Institute of Science and Technology for Brain Inspired Intelligence, Fudan University, Shanghai, China
Edmund T. Rolls, Xiaoqian Yan, Yi Zhang & Jianfeng Feng
Department of Information and Communication Technologies, Center for Brain and Cognition, Computational Neuroscience Group, Universitat Pompeu Fabra, Barcelona, Spain
Gustavo Deco
Institució Catalana de la Recerca i Estudis Avançats (ICREA), Universitat Pompeu Fabra, Passeig Lluís Companys 23, Barcelona, Spain
Aalto NeuroImaging, Department of Neuroscience and Biomedical Engineering, Aalto University, Espoo, Finland
Veikko Jousmaki
You can also search for this author in PubMed Google Scholar
Contributions
Edmund Rolls designed the research, took part in the data acquisition, performed the analyses, made the Figures, and wrote the paper. Xiaoqian Yan took part in the design of the research and data acquisition and writing, and performed the preprocessing. Gustavo Deco provided the generative effective connectivity algorithm. Yi Zhang adapted the MNE preprocessing pipeline for this research. Veikko Jousmaki provided advice on the MEG data acquisition. Jianfeng Feng performed the funding acquisition. All authors approved the paper.
Corresponding author
Correspondence to Edmund T. Rolls .
Ethics declarations
Competing interests.
The authors declare no competing interests.
Ethical permissions
The study received ethical approval from the Ethics Committee of the Institute of Science and Technology for Brain-Inspired Intelligence at Fudan University (reference number AF/SC-115/20230822).
Peer review
Peer review information.
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Christina Karlsson Rosenthal.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplemental material, reporting summary, rights and permissions.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ .
Reprints and permissions
About this article
Cite this article.
Rolls, E.T., Yan, X., Deco, G. et al. A ventromedial visual cortical ‘Where’ stream to the human hippocampus for spatial scenes revealed with magnetoencephalography. Commun Biol 7 , 1047 (2024). https://doi.org/10.1038/s42003-024-06719-z
Download citation
Received : 20 March 2024
Accepted : 12 August 2024
Published : 25 August 2024
DOI : https://doi.org/10.1038/s42003-024-06719-z
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
By submitting a comment you agree to abide by our Terms and Community Guidelines . If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing newsletter — what matters in science, free to your inbox daily.

IMAGES
COMMENTS
• Model: The visual representation of a biological system. • mEPN: Modified Edinburgh Pathway Notation scheme is a graphical notation system based on the concepts of the process diagram.
Visualization for network biology. a Timeline of the emergence of relevant technologies and concepts.b A simple drawing of an undirected unweighted graph.c A 2D representation of a yeast protein-protein interaction network visualized in Cytoscape (left) and potential protein complexes identified by the MCL algorithm from that network (right).d A 3D view of a protein-protein interaction network ...
However, a significant aspect of biological engineering is the visual representation of the components (from parts to devices and finally systems), ability to simulate different components in silico and tools to support such graphical representation and simulation. In §3.2, we overview the different efforts in the systems biology community for ...
The use of visual representations (i.e., photographs, diagrams, models) has been part of science, and their use makes it possible for scientists to interact with and represent complex phenomena, not observable in other ways. Despite a wealth of research in science education on visual representations, the emphasis of such research has mainly been on the conceptual understanding when using ...
1 INTRODUCTION. Networks are used ubiquitously throughout biology to represent the relationships between genes and gene products. Perhaps due to their simple discrete nature and to their amenability to visual representation, biological networks allow for the most salient properties of complex systems to be highlighted in a succinct and powerful manner.
MONGKIE provides the sophisticated data models for visualization of biological networks with advanced graph drawing techniques, and therefore can represent different types of biological entities and interactions between them with out-of-the-box visual styles, shown in Fig. 4.1.Both nodes and edges differ in their style according to their biological meaning.
High-throughput studies of biological systems are rapidly accumulating a wealth of 'omics'-scale data. ... it can be important to use compact visual representations and pathway layouts that reduce ...
The graphical representation of biological networks is a topic that has been largely neglected, and its importance has only recently been recognized because of the growing need to understand large ...
1. Introduction. Design and biology can interact and mutually benefit each other in many ways: from designing and using graphical representations and visualisations of biological information [1, 2] to biomimicry, a type of bio-inspired design [] to biodesign which aims to incorporate living organisms as components into design [].As Michael Gross writes: "Nature inspires art, but conversely ...
Yet, biological systems are chaotic and complex with interactions that span different size and time scales. In a series of presentations "Media for Thinking the Unthinkable", Bret Victor, a computer scientist and interface designer, argues that dynamic representations and new media are needed in order to explore and understand such complex data efficiently.
We follow another layout strategy that spatially arranges the visual representations of the published articles according to similarity. ... Due to the sheer size, complexity, and heterogeneity of biological data, truly consolidated systems—in which several visualization techniques for a number of data types are integrated—are still rare ...
visual representation of the proof's execution. 3 Analyzing biological networks As we have noted, the graphical models users produce are formally represented using Qualitative Networks (QN) [15]. The tool automatically translates the graphical models to a QNs. The QNs include variables representing the con-
Artificial deep neural networks (DNNs) are the most predictive models of neural responses to images in the primate high-level visual cortex 1, 2. Many studies have reported that DNNs trained to perform image classification produce internal feature representations broadly similar to those in areas V4 and IT of the primate cortex, and that this ...
Recently, a review of the classification systems employed for the characterization of visual representations of biological entities led to the proposal of a three-dimensional classificational ...
Metrics. Networks offer a powerful visual representation of complex systems. Cartographs introduce a diverse set of network layouts for highlighting and visually inspecting chosen characteristics ...
View a PDF of the paper titled Universal dimensions of visual representation, by Zirui Chen and Michael F. Bonner ... These results suggest that the underlying similarities between artificial and biological vision are primarily governed by a core set of universal image representations that are convergently learned by diverse systems. Subjects ...
Author Summary One of the primary obstacles to understanding the computational underpinnings of biological vision is its sheer scale—the visual system is a massively parallel computer, comprised of billions of elements. While this scale has historically been beyond the reach of even the fastest super-computing systems, recent advances in commodity graphics processors (such as those found in ...
Biological visual systems cannot measure the properties that define the physical world. Nonetheless, visually guided behaviors of humans and other animals are routinely successful. The purpose of this article is to consider how this feat is accomplished. Most concepts of vision propose, explicitly or implicitly, that visual behavior depends on ...
Visual representations or diagrammatic illustrations occupy a significant part of science textbooks, especially biology. ... organs and organ systems. Iconic Diagrams in Tamil Nadu Biology Textbooks need to be improved in terms of size and clarity. ... understanding and categorization of visual representations used in biological textbooks for ...
Biological vision systems are unparalleled in their ability to learn visual representations without supervision. In machine learning, contrastive learning (CL) has led to major advances in forming object representations in an unsupervised fashion. These systems learn representations invariant to augmentation operations over images, like cropping or flipping. In contrast, biological vision ...
Implications for the biological visual system. ... Our work adds further support for the viability of this hypothesis of visual system representation, by demonstrating an emergent correspondence ...
High-level visual cortex and leading neural network models of the visual system retain information about multiple visual scene variables in independent, non-interfering dimensions of their population codes. ... In this work, we provide evidence that factorization is a normative principle of biological visual representations. In the monkey ...
Networks offer a powerful visual representation of complex systems. This study introduces network visualizations that are easy to interpret and can help explore large datasets, such as the map of ...
Indeed, with wide-field retinotopic mapping, the ProS, where the retrosplenial scene area is located 25,33, is found to have a complete representation of the visual field, different from adjacent ...