Subscribe to the PwC Newsletter
Join the community, trending research, autocoder: enhancing code large language model with \textsc{aiev-instruct}.
bin123apple/autocoder • 23 May 2024
We introduce AutoCoder, the first Large Language Model to surpass GPT-4 Turbo (April 2024) and GPT-4o in pass@1 on the Human Eval benchmark test ($\mathbf{90. 9\%}$ vs. $\mathbf{90. 2\%}$).

FinRobot: An Open-Source AI Agent Platform for Financial Applications using Large Language Models
ai4finance-foundation/finrobot • 23 May 2024
As financial institutions and professionals increasingly incorporate Large Language Models (LLMs) into their workflows, substantial barriers, including proprietary data and specialized knowledge, persist between the finance sector and the AI community.
EasyAnimate: A High-Performance Long Video Generation Method based on Transformer Architecture

The motion module can be adapted to various DiT baseline methods to generate video with different styles.

YOLOv10: Real-Time End-to-End Object Detection
In this work, we aim to further advance the performance-efficiency boundary of YOLOs from both the post-processing and model architecture.

InstaDrag: Lightning Fast and Accurate Drag-based Image Editing Emerging from Videos
magic-research/instadrag • 22 May 2024
Accuracy and speed are critical in image editing tasks.
$\textit{S}^3$Gaussian: Self-Supervised Street Gaussians for Autonomous Driving
Photorealistic 3D reconstruction of street scenes is a critical technique for developing real-world simulators for autonomous driving.

Switch Transformers: Scaling to Trillion Parameter Models with Simple and Efficient Sparsity
We design models based off T5-Base and T5-Large to obtain up to 7x increases in pre-training speed with the same computational resources.

Looking Backward: Streaming Video-to-Video Translation with Feature Banks
This paper introduces StreamV2V, a diffusion model that achieves real-time streaming video-to-video (V2V) translation with user prompts.
LLaVA-UHD: an LMM Perceiving Any Aspect Ratio and High-Resolution Images
To address the challenges, we present LLaVA-UHD, a large multimodal model that can efficiently perceive images in any aspect ratio and high resolution.
FlashRAG: A Modular Toolkit for Efficient Retrieval-Augmented Generation Research
With the advent of Large Language Models (LLMs), the potential of Retrieval Augmented Generation (RAG) techniques have garnered considerable research attention.
machine learning Recently Published Documents
Total documents.
- Latest Documents
- Most Cited Documents
- Contributed Authors
- Related Sources
- Related Keywords
An explainable machine learning model for identifying geographical origins of sea cucumber Apostichopus japonicus based on multi-element profile
A comparison of machine learning- and regression-based models for predicting ductility ratio of rc beam-column joints, alexa, is this a historical record.
Digital transformation in government has brought an increase in the scale, variety, and complexity of records and greater levels of disorganised data. Current practices for selecting records for transfer to The National Archives (TNA) were developed to deal with paper records and are struggling to deal with this shift. This article examines the background to the problem and outlines a project that TNA undertook to research the feasibility of using commercially available artificial intelligence tools to aid selection. The project AI for Selection evaluated a range of commercial solutions varying from off-the-shelf products to cloud-hosted machine learning platforms, as well as a benchmarking tool developed in-house. Suitability of tools depended on several factors, including requirements and skills of transferring bodies as well as the tools’ usability and configurability. This article also explores questions around trust and explainability of decisions made when using AI for sensitive tasks such as selection.
Automated Text Classification of Maintenance Data of Higher Education Buildings Using Text Mining and Machine Learning Techniques
Data-driven analysis and machine learning for energy prediction in distributed photovoltaic generation plants: a case study in queensland, australia, modeling nutrient removal by membrane bioreactor at a sewage treatment plant using machine learning models, big five personality prediction based in indonesian tweets using machine learning methods.
<span lang="EN-US">The popularity of social media has drawn the attention of researchers who have conducted cross-disciplinary studies examining the relationship between personality traits and behavior on social media. Most current work focuses on personality prediction analysis of English texts, but Indonesian has received scant attention. Therefore, this research aims to predict user’s personalities based on Indonesian text from social media using machine learning techniques. This paper evaluates several machine learning techniques, including <a name="_Hlk87278444"></a>naive Bayes (NB), K-nearest neighbors (KNN), and support vector machine (SVM), based on semantic features including emotion, sentiment, and publicly available Twitter profile. We predict the personality based on the big five personality model, the most appropriate model for predicting user personality in social media. We examine the relationships between the semantic features and the Big Five personality dimensions. The experimental results indicate that the Big Five personality exhibit distinct emotional, sentimental, and social characteristics and that SVM outperformed NB and KNN for Indonesian. In addition, we observe several terms in Indonesian that specifically refer to each personality type, each of which has distinct emotional, sentimental, and social features.</span>
Compressive strength of concrete with recycled aggregate; a machine learning-based evaluation
Temperature prediction of flat steel box girders of long-span bridges utilizing in situ environmental parameters and machine learning, computer-assisted cohort identification in practice.
The standard approach to expert-in-the-loop machine learning is active learning, where, repeatedly, an expert is asked to annotate one or more records and the machine finds a classifier that respects all annotations made until that point. We propose an alternative approach, IQRef , in which the expert iteratively designs a classifier and the machine helps him or her to determine how well it is performing and, importantly, when to stop, by reporting statistics on a fixed, hold-out sample of annotated records. We justify our approach based on prior work giving a theoretical model of how to re-use hold-out data. We compare the two approaches in the context of identifying a cohort of EHRs and examine their strengths and weaknesses through a case study arising from an optometric research problem. We conclude that both approaches are complementary, and we recommend that they both be employed in conjunction to address the problem of cohort identification in health research.
Export Citation Format
Share document.

Frequently Asked Questions
JMLR Papers
Select a volume number to see its table of contents with links to the papers.
Volume 23 (January 2022 - Present)
Volume 22 (January 2021 - December 2021)
Volume 21 (January 2020 - December 2020)
Volume 20 (January 2019 - December 2019)
Volume 19 (August 2018 - December 2018)
Volume 18 (February 2017 - August 2018)
Volume 17 (January 2016 - January 2017)
Volume 16 (January 2015 - December 2015)
Volume 15 (January 2014 - December 2014)
Volume 14 (January 2013 - December 2013)
Volume 13 (January 2012 - December 2012)
Volume 12 (January 2011 - December 2011)
Volume 11 (January 2010 - December 2010)
Volume 10 (January 2009 - December 2009)
Volume 9 (January 2008 - December 2008)
Volume 8 (January 2007 - December 2007)
Volume 7 (January 2006 - December 2006)
Volume 6 (January 2005 - December 2005)
Volume 5 (December 2003 - December 2004)
Volume 4 (Apr 2003 - December 2003)
Volume 3 (Jul 2002 - Mar 2003)
Volume 2 (Oct 2001 - Mar 2002)
Volume 1 (Oct 2000 - Sep 2001)
Special Topics
Bayesian Optimization
Learning from Electronic Health Data (December 2016)
Gesture Recognition (May 2012 - present)
Large Scale Learning (Jul 2009 - present)
Mining and Learning with Graphs and Relations (February 2009 - present)
Grammar Induction, Representation of Language and Language Learning (Nov 2010 - Apr 2011)
Causality (Sep 2007 - May 2010)
Model Selection (Apr 2007 - Jul 2010)
Conference on Learning Theory 2005 (February 2007 - Jul 2007)
Machine Learning for Computer Security (December 2006)
Machine Learning and Large Scale Optimization (Jul 2006 - Oct 2006)
Approaches and Applications of Inductive Programming (February 2006 - Mar 2006)
Learning Theory (Jun 2004 - Aug 2004)
Special Issues
In Memory of Alexey Chervonenkis (Sep 2015)
Independent Components Analysis (December 2003)
Learning Theory (Oct 2003)
Inductive Logic Programming (Aug 2003)
Fusion of Domain Knowledge with Data for Decision Support (Jul 2003)
Variable and Feature Selection (Mar 2003)
Machine Learning Methods for Text and Images (February 2003)
Eighteenth International Conference on Machine Learning (ICML2001) (December 2002)
Computational Learning Theory (Nov 2002)
Shallow Parsing (Mar 2002)
Kernel Methods (December 2001)
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- My Account Login
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Open access
- Published: 01 February 2021
An open source machine learning framework for efficient and transparent systematic reviews
- Rens van de Schoot ORCID: orcid.org/0000-0001-7736-2091 1 ,
- Jonathan de Bruin ORCID: orcid.org/0000-0002-4297-0502 2 ,
- Raoul Schram 2 ,
- Parisa Zahedi ORCID: orcid.org/0000-0002-1610-3149 2 ,
- Jan de Boer ORCID: orcid.org/0000-0002-0531-3888 3 ,
- Felix Weijdema ORCID: orcid.org/0000-0001-5150-1102 3 ,
- Bianca Kramer ORCID: orcid.org/0000-0002-5965-6560 3 ,
- Martijn Huijts ORCID: orcid.org/0000-0002-8353-0853 4 ,
- Maarten Hoogerwerf ORCID: orcid.org/0000-0003-1498-2052 2 ,
- Gerbrich Ferdinands ORCID: orcid.org/0000-0002-4998-3293 1 ,
- Albert Harkema ORCID: orcid.org/0000-0002-7091-1147 1 ,
- Joukje Willemsen ORCID: orcid.org/0000-0002-7260-0828 1 ,
- Yongchao Ma ORCID: orcid.org/0000-0003-4100-5468 1 ,
- Qixiang Fang ORCID: orcid.org/0000-0003-2689-6653 1 ,
- Sybren Hindriks 1 ,
- Lars Tummers ORCID: orcid.org/0000-0001-9940-9874 5 &
- Daniel L. Oberski ORCID: orcid.org/0000-0001-7467-2297 1 , 6
Nature Machine Intelligence volume 3 , pages 125–133 ( 2021 ) Cite this article
73k Accesses
228 Citations
165 Altmetric
Metrics details
- Computational biology and bioinformatics
- Computer science
- Medical research
A preprint version of the article is available at arXiv.
To help researchers conduct a systematic review or meta-analysis as efficiently and transparently as possible, we designed a tool to accelerate the step of screening titles and abstracts. For many tasks—including but not limited to systematic reviews and meta-analyses—the scientific literature needs to be checked systematically. Scholars and practitioners currently screen thousands of studies by hand to determine which studies to include in their review or meta-analysis. This is error prone and inefficient because of extremely imbalanced data: only a fraction of the screened studies is relevant. The future of systematic reviewing will be an interaction with machine learning algorithms to deal with the enormous increase of available text. We therefore developed an open source machine learning-aided pipeline applying active learning: ASReview. We demonstrate by means of simulation studies that active learning can yield far more efficient reviewing than manual reviewing while providing high quality. Furthermore, we describe the options of the free and open source research software and present the results from user experience tests. We invite the community to contribute to open source projects such as our own that provide measurable and reproducible improvements over current practice.
Similar content being viewed by others

Testing theory of mind in large language models and humans

Highly accurate protein structure prediction with AlphaFold

A guide to artificial intelligence for cancer researchers
With the emergence of online publishing, the number of scientific manuscripts on many topics is skyrocketing 1 . All of these textual data present opportunities to scholars and practitioners while simultaneously confronting them with new challenges. Scholars often develop systematic reviews and meta-analyses to develop comprehensive overviews of the relevant topics 2 . The process entails several explicit and, ideally, reproducible steps, including identifying all likely relevant publications in a standardized way, extracting data from eligible studies and synthesizing the results. Systematic reviews differ from traditional literature reviews in that they are more replicable and transparent 3 , 4 . Such systematic overviews of literature on a specific topic are pivotal not only for scholars, but also for clinicians, policy-makers, journalists and, ultimately, the general public 5 , 6 , 7 .
Given that screening the entire research literature on a given topic is too labour intensive, scholars often develop quite narrow searches. Developing a search strategy for a systematic review is an iterative process aimed at balancing recall and precision 8 , 9 ; that is, including as many potentially relevant studies as possible while simultaneously limiting the total number of studies retrieved. The vast number of publications in the field of study often leads to a relatively precise search, with the risk of missing relevant studies. The process of systematic reviewing is error prone and extremely time intensive 10 . In fact, if the literature of a field is growing faster than the amount of time available for systematic reviews, adequate manual review of this field then becomes impossible 11 .
The rapidly evolving field of machine learning has aided researchers by allowing the development of software tools that assist in developing systematic reviews 11 , 12 , 13 , 14 . Machine learning offers approaches to overcome the manual and time-consuming screening of large numbers of studies by prioritizing relevant studies via active learning 15 . Active learning is a type of machine learning in which a model can choose the data points (for example, records obtained from a systematic search) it would like to learn from and thereby drastically reduce the total number of records that require manual screening 16 , 17 , 18 . In most so-called human-in-the-loop 19 machine-learning applications, the interaction between the machine-learning algorithm and the human is used to train a model with a minimum number of labelling tasks. Unique to systematic reviewing is that not only do all relevant records (that is, titles and abstracts) need to seen by a researcher, but an extremely diverse range of concepts also need to be learned, thereby requiring flexibility in the modelling approach as well as careful error evaluation 11 . In the case of systematic reviewing, the algorithm(s) are interactively optimized for finding the most relevant records, instead of finding the most accurate model. The term researcher-in-the-loop was introduced 20 as a special case of human-in-the-loop with three unique components: (1) the primary output of the process is a selection of the records, not a trained machine learning model; (2) all records in the relevant selection are seen by a human at the end of the process 21 ; (3) the use-case requires a reproducible workflow and complete transparency is required 22 .
Existing tools that implement such an active learning cycle for systematic reviewing are described in Table 1 ; see the Supplementary Information for an overview of all of the software that we considered (note that this list was based on a review of software tools 12 ). However, existing tools have two main drawbacks. First, many are closed source applications with black box algorithms, which is problematic as transparency and data ownership are essential in the era of open science 22 . Second, to our knowledge, existing tools lack the necessary flexibility to deal with the large range of possible concepts to be learned by a screening machine. For example, in systematic reviews, the optimal type of classifier will depend on variable parameters, such as the proportion of relevant publications in the initial search and the complexity of the inclusion criteria used by the researcher 23 . For this reason, any successful system must allow for a wide range of classifier types. Benchmark testing is crucial to understand the real-world performance of any machine learning-aided system, but such benchmark options are currently mostly lacking.
In this paper we present an open source machine learning-aided pipeline with active learning for systematic reviews called ASReview. The goal of ASReview is to help scholars and practitioners to get an overview of the most relevant records for their work as efficiently as possible while being transparent in the process. The open, free and ready-to-use software ASReview addresses all concerns mentioned above: it is open source, uses active learning, allows multiple machine learning models. It also has a benchmark mode, which is especially useful for comparing and designing algorithms. Furthermore, it is intended to be easily extensible, allowing third parties to add modules that enhance the pipeline. Although we focus this paper on systematic reviews, ASReview can handle any text source.
In what follows, we first present the pipeline for manual versus machine learning-aided systematic reviews. We then show how ASReview has been set up and how ASReview can be used in different workflows by presenting several real-world use cases. We subsequently demonstrate the results of simulations that benchmark performance and present the results of a series of user-experience tests. Finally, we discuss future directions.
Pipeline for manual and machine learning-aided systematic reviews
The pipeline of a systematic review without active learning traditionally starts with researchers doing a comprehensive search in multiple databases 24 , using free text words as well as controlled vocabulary to retrieve potentially relevant references. The researcher then typically verifies that the key papers they expect to find are indeed included in the search results. The researcher downloads a file with records containing the text to be screened. In the case of systematic reviewing it contains the titles and abstracts (and potentially other metadata such as the authors’s names, journal name, DOI) of potentially relevant references into a reference manager. Ideally, two or more researchers then screen the records’s titles and abstracts on the basis of the eligibility criteria established beforehand 4 . After all records have been screened, the full texts of the potentially relevant records are read to determine which of them will be ultimately included in the review. Most records are excluded in the title and abstract phase. Typically, only a small fraction of the records belong to the relevant class, making title and abstract screening an important bottleneck in systematic reviewing process 25 . For instance, a recent study analysed 10,115 records and excluded 9,847 after title and abstract screening, a drop of more than 95% 26 . ASReview therefore focuses on this labour-intensive step.
The research pipeline of ASReview is depicted in Fig. 1 . The researcher starts with a search exactly as described above and subsequently uploads a file containing the records (that is, metadata containing the text of the titles and abstracts) into the software. Prior knowledge is then selected, which is used for training of the first model and presenting the first record to the researcher. As screening is a binary classification problem, the reviewer must select at least one key record to include and exclude on the basis of background knowledge. More prior knowledge may result in improved efficiency of the active learning process.
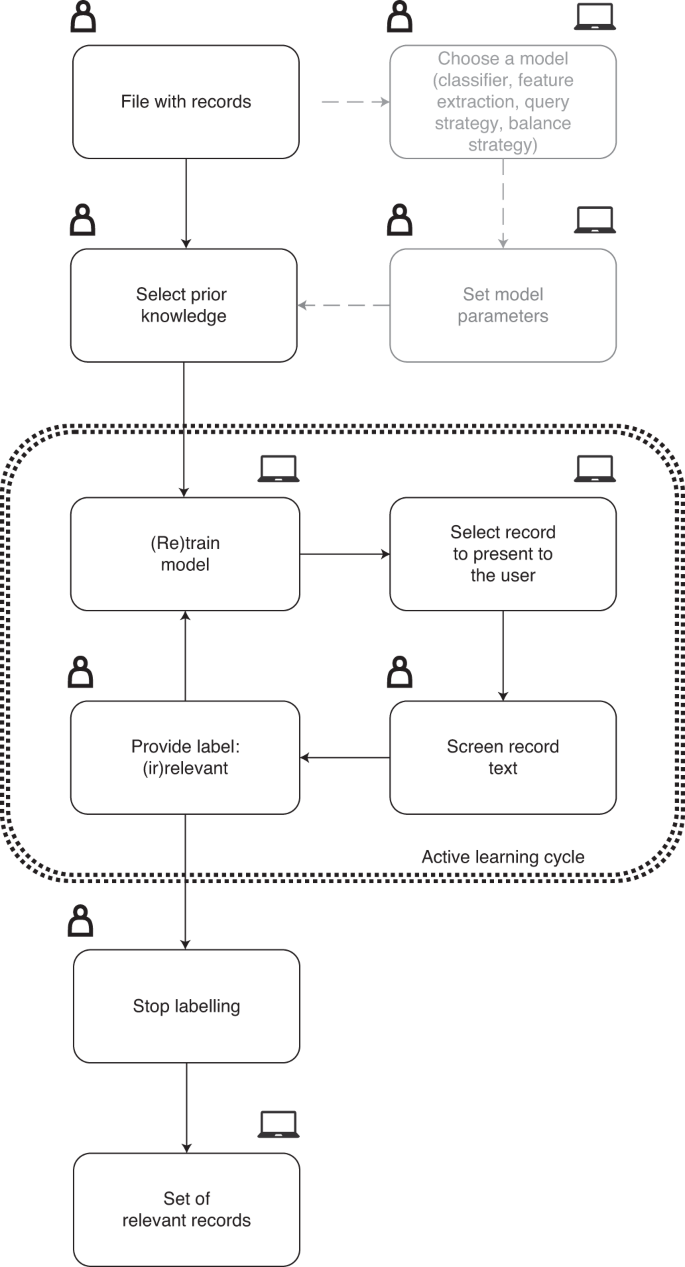
The symbols indicate whether the action is taken by a human, a computer, or whether both options are available.
A machine learning classifier is trained to predict study relevance (labels) from a representation of the record-containing text (feature space) on the basis of prior knowledge. We have purposefully chosen not to include an author name or citation network representation in the feature space to prevent authority bias in the inclusions. In the active learning cycle, the software presents one new record to be screened and labelled by the user. The user’s binary label (1 for relevant versus 0 for irrelevant) is subsequently used to train a new model, after which a new record is presented to the user. This cycle continues up to a certain user-specified stopping criterion has been reached. The user now has a file with (1) records labelled as either relevant or irrelevant and (2) unlabelled records ordered from most to least probable to be relevant as predicted by the current model. This set-up helps to move through a large database much quicker than in the manual process, while the decision process simultaneously remains transparent.
Software implementation for ASReview
The source code 27 of ASReview is available open source under an Apache 2.0 license, including documentation 28 . Compiled and packaged versions of the software are available on the Python Package Index 29 or Docker Hub 30 . The free and ready-to-use software ASReview implements oracle, simulation and exploration modes. The oracle mode is used to perform a systematic review with interaction by the user, the simulation mode is used for simulation of the ASReview performance on existing datasets, and the exploration mode can be used for teaching purposes and includes several preloaded labelled datasets.
The oracle mode presents records to the researcher and the researcher classifies these. Multiple file formats are supported: (1) RIS files are used by digital libraries such as IEEE Xplore, Scopus and ScienceDirect; the citation managers Mendeley, RefWorks, Zotero and EndNote support the RIS format too. (2) Tabular datasets with the .csv, .xlsx and .xls file extensions. CSV files should be comma separated and UTF-8 encoded; the software for CSV files accepts a set of predetermined labels in line with the ones used in RIS files. Each record in the dataset should hold the metadata on, for example, a scientific publication. Mandatory metadata is text and can, for example, be titles or abstracts from scientific papers. If available, both are used to train the model, but at least one is needed. An advanced option is available that splits the title and abstracts in the feature-extraction step and weights the two feature matrices independently (for TF–IDF only). Other metadata such as author, date, DOI and keywords are optional but not used for training the models. When using ASReview in the simulation or exploration mode, an additional binary variable is required to indicate historical labelling decisions. This column, which is automatically detected, can also be used in the oracle mode as background knowledge for previous selection of relevant papers before entering the active learning cycle. If unavailable, the user has to select at least one relevant record that can be identified by searching the pool of records. At least one irrelevant record should also be identified; the software allows to search for specific records or presents random records that are most likely to be irrelevant due to the extremely imbalanced data.
The software has a simple yet extensible default model: a naive Bayes classifier, TF–IDF feature extraction, a dynamic resampling balance strategy 31 and certainty-based sampling 17 , 32 for the query strategy. These defaults were chosen on the basis of their consistently high performance in benchmark experiments across several datasets 31 . Moreover, the low computation time of these default settings makes them attractive in applications, given that the software should be able to run locally. Users can change the settings, shown in Table 2 , and technical details are described in our documentation 28 . Users can also add their own classifiers, feature extraction techniques, query strategies and balance strategies.
ASReview has a number of implemented features (see Table 2 ). First, there are several classifiers available: (1) naive Bayes; (2) support vector machines; (3) logistic regression; (4) neural networks; (5) random forests; (6) LSTM-base, which consists of an embedding layer, an LSTM layer with one output, a dense layer and a single sigmoid output node; and (7) LSTM-pool, which consists of an embedding layer, an LSTM layer with many outputs, a max pooling layer and a single sigmoid output node. The feature extraction techniques available are Doc2Vec 33 , embedding LSTM, embedding with IDF or TF–IDF 34 (the default is unigram, with the option to run n -grams while other parameters are set to the defaults of Scikit-learn 35 ) and sBERT 36 . The available query strategies for the active learning part are (1) random selection, ignoring model-assigned probabilities; (2) uncertainty-based sampling, which chooses the most uncertain record according to the model (that is, closest to 0.5 probability); (3) certainty-based sampling (max in ASReview), which chooses the record most likely to be included according to the model; and (4) mixed sampling, which uses a combination of random and certainty-based sampling.
There are several balance strategies that rebalance and reorder the training data. This is necessary, because the data is typically extremely imbalanced and therefore we have implemented the following balance strategies: (1) full sampling, which uses all of the labelled records; (2) undersampling the irrelevant records so that the included and excluded records are in some particular ratio (closer to one); and (3) dynamic resampling, a novel method similar to undersampling in that it decreases the imbalance of the training data 31 . However, in dynamic resampling, the number of irrelevant records is decreased, whereas the number of relevant records is increased by duplication such that the total number of records in the training data remains the same. The ratio between relevant and irrelevant records is not fixed over interactions, but dynamically updated depending on the number of labelled records, the total number of records and the ratio between relevant and irrelevant records. Details on all of the described algorithms can be found in the code and documentation referred to above.
By default, ASReview converts the records’s texts into a document-term matrix, terms are converted to lowercase and no stop words are removed by default (but this can be changed). As the document-term matrix is identical in each iteration of the active learning cycle, it is generated in advance of model training and stored in the (active learning) state file. Each row of the document-term matrix can easily be requested from the state-file. Records are internally identified by their row number in the input dataset. In oracle mode, the record that is selected to be classified is retrieved from the state file and the record text and other metadata (such as title and abstract) are retrieved from the original dataset (from the file or the computer’s memory). ASReview can run on your local computer, or on a (self-hosted) local or remote server. Data (all records and their labels) remain on the users’s computer. Data ownership and confidentiality are crucial and no data are processed or used in any way by third parties. This is unique by comparison with some of the existing systems, as shown in the last column of Table 1 .
Real-world use cases and high-level function descriptions
Below we highlight a number of real-world use cases and high-level function descriptions for using the pipeline of ASReview.
ASReview can be integrated in classic systematic reviews or meta-analyses. Such reviews or meta-analyses entail several explicit and reproducible steps, as outlined in the PRISMA guidelines 4 . Scholars identify all likely relevant publications in a standardized way, screen retrieved publications to select eligible studies on the basis of defined eligibility criteria, extract data from eligible studies and synthesize the results. ASReview fits into this process, particularly in the abstract screening phase. ASReview does not replace the initial step of collecting all potentially relevant studies. As such, results from ASReview depend on the quality of the initial search process, including selection of databases 24 and construction of comprehensive searches using keywords and controlled vocabulary. However, ASReview can be used to broaden the scope of the search (by keyword expansion or omitting limitation in the search query), resulting in a higher number of initial papers to limit the risk of missing relevant papers during the search part (that is, more focus on recall instead of precision).
Furthermore, many reviewers nowadays move towards meta-reviews when analysing very large literature streams, that is, systematic reviews of systematic reviews 37 . This can be problematic as the various reviews included could use different eligibility criteria and are therefore not always directly comparable. Due to the efficiency of ASReview, scholars using the tool could conduct the study by analysing the papers directly instead of using the systematic reviews. Furthermore, ASReview supports the rapid update of a systematic review. The included papers from the initial review are used to train the machine learning model before screening of the updated set of papers starts. This allows the researcher to quickly screen the updated set of papers on the basis of decisions made in the initial run.
As an example case, let us look at the current literature on COVID-19 and the coronavirus. An enormous number of papers are being published on COVID-19. It is very time consuming to manually find relevant papers (for example, to develop treatment guidelines). This is especially problematic as urgent overviews are required. Medical guidelines rely on comprehensive systematic reviews, but the medical literature is growing at breakneck pace and the quality of the research is not universally adequate for summarization into policy 38 . Such reviews must entail adequate protocols with explicit and reproducible steps, including identifying all potentially relevant papers, extracting data from eligible studies, assessing potential for bias and synthesizing the results into medical guidelines. Researchers need to screen (tens of) thousands of COVID-19-related studies by hand to find relevant papers to include in their overview. Using ASReview, this can be done far more efficiently by selecting key papers that match their (COVID-19) research question in the first step; this should start the active learning cycle and lead to the most relevant COVID-19 papers for their research question being presented next. A plug-in was therefore developed for ASReview 39 , which contained three databases that are updated automatically whenever a new version is released by the owners of the data: (1) the Cord19 database, developed by the Allen Institute for AI, with over all publications on COVID-19 and other coronavirus research (for example SARS, MERS and so on) from PubMed Central, the WHO COVID-19 database of publications, the preprint servers bioRxiv and medRxiv and papers contributed by specific publishers 40 . The CORD-19 dataset is updated daily by the Allen Institute for AI and updated also daily in the plugin. (2) In addition to the full dataset, we automatically construct a daily subset of the database with studies published after December 1st, 2019 to search for relevant papers published during the COVID-19 crisis. (3) A separate dataset of COVID-19 related preprints, containing metadata of preprints from over 15 preprints servers across disciplines, published since January 1st, 2020 41 . The preprint dataset is updated weekly by the maintainers and then automatically updated in ASReview as well. As this dataset is not readily available to researchers through regular search engines (for example, PubMed), its inclusion in ASReview provided added value to researchers interested in COVID-19 research, especially if they want a quick way to screen preprints specifically.
Simulation study
To evaluate the performance of ASReview on a labelled dataset, users can employ the simulation mode. As an example, we ran simulations based on four labelled datasets with version 0.7.2 of ASReview. All scripts to reproduce the results in this paper can be found on Zenodo ( https://doi.org/10.5281/zenodo.4024122 ) 42 , whereas the results are available at OSF ( https://doi.org/10.17605/OSF.IO/2JKD6 ) 43 .
First, we analysed the performance for a study systematically describing studies that performed viral metagenomic next-generation sequencing in common livestock such as cattle, small ruminants, poultry and pigs 44 . Studies were retrieved from Embase ( n = 1,806), Medline ( n = 1,384), Cochrane Central ( n = 1), Web of Science ( n = 977) and Google Scholar ( n = 200, the top relevant references). After deduplication this led to 2,481 studies obtained in the initial search, of which 120 were inclusions (4.84%).
A second simulation study was performed on the results for a systematic review of studies on fault prediction in software engineering 45 . Studies were obtained from ACM Digital Library, IEEExplore and the ISI Web of Science. Furthermore, a snowballing strategy and a manual search were conducted, accumulating to 8,911 publications of which 104 were included in the systematic review (1.2%).
A third simulation study was performed on a review of longitudinal studies that applied unsupervised machine learning techniques to longitudinal data of self-reported symptoms of the post-traumatic stress assessed after trauma exposure 46 , 47 ; 5,782 studies were obtained by searching Pubmed, Embase, PsychInfo and Scopus and through a snowballing strategy in which both the references and the citation of the included papers were screened. Thirty-eight studies were included in the review (0.66%).
A fourth simulation study was performed on the results for a systematic review on the efficacy of angiotensin-converting enzyme inhibitors, from a study collecting various systematic review datasets from the medical sciences 15 . The collection is a subset of 2,544 publications from the TREC 2004 Genomics Track document corpus 48 . This is a static subset from all MEDLINE records from 1994 through 2003, which allows for replicability of results. Forty-one publications were included in the review (1.6%).
Performance metrics
We evaluated the four datasets using three performance metrics. We first assess the work saved over sampling (WSS), which is the percentage reduction in the number of records needed to screen achieved by using active learning instead of screening records at random; WSS is measured at a given level of recall of relevant records, for example 95%, indicating the work reduction in screening effort at the cost of failing to detect 5% of the relevant records. For some researchers it is essential that all relevant literature on the topic is retrieved; this entails that the recall should be 100% (that is, WSS@100%). We also propose the amount of relevant references found after having screened the first 10% of the records (RRF10%). This is a useful metric for getting a quick overview of the relevant literature.
For every dataset, 15 runs were performed with one random inclusion and one random exclusion (see Fig. 2 ). The classical review performance with randomly found inclusions is shown by the dashed line. The average work saved over sampling at 95% recall for ASReview is 83% and ranges from 67% to 92%. Hence, 95% of the eligible studies will be found after screening between only 8% to 33% of the studies. Furthermore, the number of relevant abstracts found after reading 10% of the abstracts ranges from 70% to 100%. In short, our software would have saved many hours of work.
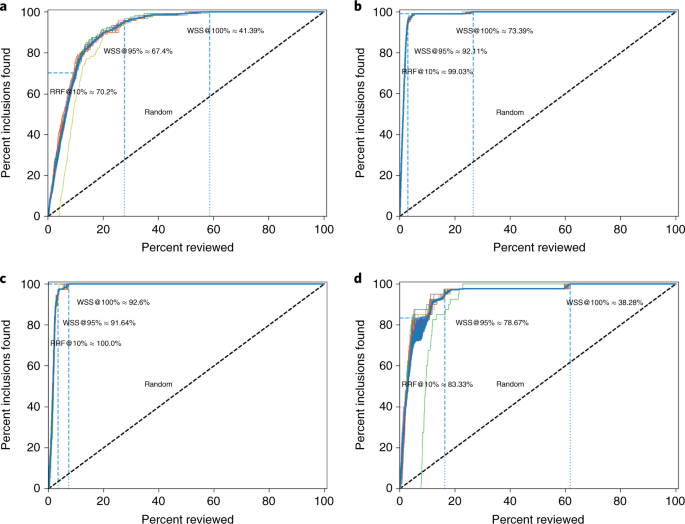
a – d , Results of the simulation study for the results for a study systematically review studies that performed viral metagenomic next-generation sequencing in common livestock ( a ), results for a systematic review of studies on fault prediction in software engineering ( b ), results for longitudinal studies that applied unsupervised machine learning techniques on longitudinal data of self-reported symptoms of posttraumatic stress assessed after trauma exposure ( c ), and results for a systematic review on the efficacy of angiotensin-converting enzyme inhibitors ( d ). Fiteen runs (shown with separate lines) were performed for every dataset, with only one random inclusion and one random exclusion. The classical review performances with randomly found inclusions are shown by the dashed lines.
Usability testing (user experience testing)
We conducted a series of user experience tests to learn from end users how they experience the software and implement it in their workflow. The study was approved by the Ethics Committee of the Faculty of Social and Behavioral Sciences of Utrecht University (ID 20-104).
Unstructured interviews
The first user experience (UX) test—carried out in December 2019—was conducted with an academic research team in a substantive research field (public administration and organizational science) that has conducted various systematic reviews and meta-analyses. It was composed of three university professors (ranging from assistant to full) and three PhD candidates. In one 3.5 h session, the participants used the software and provided feedback via unstructured interviews and group discussions. The goal was to provide feedback on installing the software and testing the performance on their own data. After these sessions we prioritized the feedback in a meeting with the ASReview team, which resulted in the release of v.0.4 and v.0.6. An overview of all releases can be found on GitHub 27 .
A second UX test was conducted with four experienced researchers developing medical guidelines based on classical systematic reviews, and two experienced reviewers working at a pharmaceutical non-profit organization who work on updating reviews with new data. In four sessions, held in February to March 2020, these users tested the software following our testing protocol. After each session we implemented the feedback provided by the experts and asked them to review the software again. The main feedback was about how to upload datasets and select prior papers. Their feedback resulted in the release of v.0.7 and v.0.9.
Systematic UX test
In May 2020 we conducted a systematic UX test. Two groups of users were distinguished: an unexperienced group and an experienced user who already used ASReview. Due to the COVID-19 lockdown the usability tests were conducted via video calling where one person gave instructions to the participant and one person observed, called human-moderated remote testing 49 . During the tests, one person (SH) asked the questions and helped the participant with the tasks, the other person observed and made notes, a user experience professional at the IT department of Utrecht University (MH).
To analyse the notes, thematic analysis was used, which is a method to analyse data by dividing the information in subjects that all have a different meaning 50 using the Nvivo 12 software 51 . When something went wrong the text was coded as showstopper, when something did not go smoothly the text was coded as doubtful, and when something went well the subject was coded as superb. The features the participants requested for future versions of the ASReview tool were discussed with the lead engineer of the ASReview team and were submitted to GitHub as issues or feature requests.
The answers to the quantitative questions can be found at the Open Science Framework 52 . The participants ( N = 11) rated the tool with a grade of 7.9 (s.d. = 0.9) on a scale from one to ten (Table 2 ). The unexperienced users on average rated the tool with an 8.0 (s.d. = 1.1, N = 6). The experienced user on average rated the tool with a 7.8 (s.d. = 0.9, N = 5). The participants described the usability test with words such as helpful, accessible, fun, clear and obvious.
The UX tests resulted in the new release v0.10, v0.10.1 and the major release v0.11, which is a major revision of the graphical user interface. The documentation has been upgraded to make installing and launching ASReview more straightforward. We made setting up the project, selecting a dataset and finding past knowledge is more intuitive and flexible. We also added a project dashboard with information on your progress and advanced settings.
Continuous input via the open source community
Finally, the ASReview development team receives continuous feedback from the open science community about, among other things, the user experience. In every new release we implement features listed by our users. Recurring UX tests are performed to keep up with the needs of users and improve the value of the tool.
We designed a system to accelerate the step of screening titles and abstracts to help researchers conduct a systematic review or meta-analysis as efficiently and transparently as possible. Our system uses active learning to train a machine learning model that predicts relevance from texts using a limited number of labelled examples. The classifier, feature extraction technique, balance strategy and active learning query strategy are flexible. We provide an open source software implementation, ASReview with state-of-the-art systems across a wide range of real-world systematic reviewing applications. Based on our experiments, ASReview provides defaults on its parameters, which exhibited good performance on average across the applications we examined. However, we stress that in practical applications, these defaults should be carefully examined; for this purpose, the software provides a simulation mode to users. We encourage users and developers to perform further evaluation of the proposed approach in their application, and to take advantage of the open source nature of the project by contributing further developments.
Drawbacks of machine learning-based screening systems, including our own, remain. First, although the active learning step greatly reduces the number of manuscripts that must be screened, it also prevents a straightforward evaluation of the system’s error rates without further onerous labelling. Providing users with an accurate estimate of the system’s error rate in the application at hand is therefore a pressing open problem. Second, although, as argued above, the use of such systems is not limited in principle to reviewing, no empirical benchmarks of actual performance in these other situations yet exist to our knowledge. Third, machine learning-based screening systems automate the screening step only; although the screening step is time-consuming and a good target for automation, it is just one part of a much larger process, including the initial search, data extraction, coding for risk of bias, summarizing results and so on. Although some other works, similar to our own, have looked at (semi-)automating some of these steps in isolation 53 , 54 , to our knowledge the field is still far removed from an integrated system that would truly automate the review process while guaranteeing the quality of the produced evidence synthesis. Integrating the various tools that are currently under development to aid the systematic reviewing pipeline is therefore a worthwhile topic for future development.
Possible future research could also focus on the performance of identifying full text articles with different document length and domain-specific terminologies or even other types of text, such as newspaper articles and court cases. When the selection of past knowledge is not possible based on expert knowledge, alternative methods could be explored. For example, unsupervised learning or pseudolabelling algorithms could be used to improve training 55 , 56 . In addition, as the NLP community pushes forward the state of the art in feature extraction methods, these are easily added to our system as well. In all cases, performance benefits should be carefully evaluated using benchmarks for the task at hand. To this end, common benchmark challenges should be constructed that allow for an even comparison of the various tools now available. To facilitate such a benchmark, we have constructed a repository of publicly available systematic reviewing datasets 57 .
The future of systematic reviewing will be an interaction with machine learning algorithms to deal with the enormous increase of available text. We invite the community to contribute to open source projects such as our own, as well as to common benchmark challenges, so that we can provide measurable and reproducible improvement over current practice.
Data availability
The results described in this paper are available at the Open Science Framework ( https://doi.org/10.17605/OSF.IO/2JKD6 ) 43 . The answers to the quantitative questions of the UX test can be found at the Open Science Framework (OSF.IO/7PQNM) 52 .
Code availability
All code to reproduce the results described in this paper can be found on Zenodo ( https://doi.org/10.5281/zenodo.4024122 ) 42 . All code for the software ASReview is available under an Apache 2.0 license ( https://doi.org/10.5281/zenodo.3345592 ) 27 , is maintained on GitHub 63 and includes documentation ( https://doi.org/10.5281/zenodo.4287120 ) 28 .
Bornmann, L. & Mutz, R. Growth rates of modern science: a bibliometric analysis based on the number of publications and cited references. J. Assoc. Inf. Sci. Technol. 66 , 2215–2222 (2015).
Article Google Scholar
Gough, D., Oliver, S. & Thomas, J. An Introduction to Systematic Reviews (Sage, 2017).
Cooper, H. Research Synthesis and Meta-analysis: A Step-by-Step Approach (SAGE Publications, 2015).
Liberati, A. et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. J. Clin. Epidemiol. 62 , e1–e34 (2009).
Boaz, A. et al. Systematic Reviews: What have They Got to Offer Evidence Based Policy and Practice? (ESRC UK Centre for Evidence Based Policy and Practice London, 2002).
Oliver, S., Dickson, K. & Bangpan, M. Systematic Reviews: Making Them Policy Relevant. A Briefing for Policy Makers and Systematic Reviewers (UCL Institute of Education, 2015).
Petticrew, M. Systematic reviews from astronomy to zoology: myths and misconceptions. Brit. Med. J. 322 , 98–101 (2001).
Lefebvre, C., Manheimer, E. & Glanville, J. in Cochrane Handbook for Systematic Reviews of Interventions (eds. Higgins, J. P. & Green, S.) 95–150 (John Wiley & Sons, 2008); https://doi.org/10.1002/9780470712184.ch6 .
Sampson, M., Tetzlaff, J. & Urquhart, C. Precision of healthcare systematic review searches in a cross-sectional sample. Res. Synth. Methods 2 , 119–125 (2011).
Wang, Z., Nayfeh, T., Tetzlaff, J., O’Blenis, P. & Murad, M. H. Error rates of human reviewers during abstract screening in systematic reviews. PLoS ONE 15 , e0227742 (2020).
Marshall, I. J. & Wallace, B. C. Toward systematic review automation: a practical guide to using machine learning tools in research synthesis. Syst. Rev. 8 , 163 (2019).
Harrison, H., Griffin, S. J., Kuhn, I. & Usher-Smith, J. A. Software tools to support title and abstract screening for systematic reviews in healthcare: an evaluation. BMC Med. Res. Methodol. 20 , 7 (2020).
O’Mara-Eves, A., Thomas, J., McNaught, J., Miwa, M. & Ananiadou, S. Using text mining for study identification in systematic reviews: a systematic review of current approaches. Syst. Rev. 4 , 5 (2015).
Wallace, B. C., Trikalinos, T. A., Lau, J., Brodley, C. & Schmid, C. H. Semi-automated screening of biomedical citations for systematic reviews. BMC Bioinf. 11 , 55 (2010).
Cohen, A. M., Hersh, W. R., Peterson, K. & Yen, P.-Y. Reducing workload in systematic review preparation using automated citation classification. J. Am. Med. Inform. Assoc. 13 , 206–219 (2006).
Kremer, J., Steenstrup Pedersen, K. & Igel, C. Active learning with support vector machines. WIREs Data Min. Knowl. Discov. 4 , 313–326 (2014).
Miwa, M., Thomas, J., O’Mara-Eves, A. & Ananiadou, S. Reducing systematic review workload through certainty-based screening. J. Biomed. Inform. 51 , 242–253 (2014).
Settles, B. Active Learning Literature Survey (Minds@UW, 2009); https://minds.wisconsin.edu/handle/1793/60660
Holzinger, A. Interactive machine learning for health informatics: when do we need the human-in-the-loop? Brain Inform. 3 , 119–131 (2016).
Van de Schoot, R. & De Bruin, J. Researcher-in-the-loop for Systematic Reviewing of Text Databases (Zenodo, 2020); https://doi.org/10.5281/zenodo.4013207
Kim, D., Seo, D., Cho, S. & Kang, P. Multi-co-training for document classification using various document representations: TF–IDF, LDA, and Doc2Vec. Inf. Sci. 477 , 15–29 (2019).
Nosek, B. A. et al. Promoting an open research culture. Science 348 , 1422–1425 (2015).
Kilicoglu, H., Demner-Fushman, D., Rindflesch, T. C., Wilczynski, N. L. & Haynes, R. B. Towards automatic recognition of scientifically rigorous clinical research evidence. J. Am. Med. Inform. Assoc. 16 , 25–31 (2009).
Gusenbauer, M. & Haddaway, N. R. Which academic search systems are suitable for systematic reviews or meta‐analyses? Evaluating retrieval qualities of Google Scholar, PubMed, and 26 other resources. Res. Synth. Methods 11 , 181–217 (2020).
Borah, R., Brown, A. W., Capers, P. L. & Kaiser, K. A. Analysis of the time and workers needed to conduct systematic reviews of medical interventions using data from the PROSPERO registry. BMJ Open 7 , e012545 (2017).
de Vries, H., Bekkers, V. & Tummers, L. Innovation in the Public Sector: a systematic review and future research agenda. Public Adm. 94 , 146–166 (2016).
Van de Schoot, R. et al. ASReview: Active Learning for Systematic Reviews (Zenodo, 2020); https://doi.org/10.5281/zenodo.3345592
De Bruin, J. et al. ASReview Software Documentation 0.14 (Zenodo, 2020); https://doi.org/10.5281/zenodo.4287120
ASReview PyPI Package (ASReview Core Development Team, 2020); https://pypi.org/project/asreview/
Docker container for ASReview (ASReview Core Development Team, 2020); https://hub.docker.com/r/asreview/asreview
Ferdinands, G. et al. Active Learning for Screening Prioritization in Systematic Reviews—A Simulation Study (OSF Preprints, 2020); https://doi.org/10.31219/osf.io/w6qbg
Fu, J. H. & Lee, S. L. Certainty-enhanced active learning for improving imbalanced data classification. In 2011 IEEE 11th International Conference on Data Mining Workshops 405–412 (IEEE, 2011).
Le, Q. V. & Mikolov, T. Distributed representations of sentences and documents. Preprint at https://arxiv.org/abs/1405.4053 (2014).
Ramos, J. Using TF–IDF to determine word relevance in document queries. In Proc. 1st Instructional Conference on Machine Learning Vol. 242, 133–142 (ICML, 2003).
Pedregosa, F. et al. Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 12 , 2825–2830 (2011).
MathSciNet MATH Google Scholar
Reimers, N. & Gurevych, I. Sentence-BERT: sentence embeddings using siamese BERT-networks Preprint at https://arxiv.org/abs/1908.10084 (2019).
Smith, V., Devane, D., Begley, C. M. & Clarke, M. Methodology in conducting a systematic review of systematic reviews of healthcare interventions. BMC Med. Res. Methodol. 11 , 15 (2011).
Wynants, L. et al. Prediction models for diagnosis and prognosis of COVID-19: systematic review and critical appraisal. Brit. Med. J . 369 , 1328 (2020).
Van de Schoot, R. et al. Extension for COVID-19 Related Datasets in ASReview (Zenodo, 2020). https://doi.org/10.5281/zenodo.3891420 .
Lu Wang, L. et al. CORD-19: The COVID-19 open research dataset. Preprint at https://arxiv.org/abs/2004.10706 (2020).
Fraser, N. & Kramer, B. Covid19_preprints (FigShare, 2020); https://doi.org/10.6084/m9.figshare.12033672.v18
Ferdinands, G., Schram, R., Van de Schoot, R. & De Bruin, J. Scripts for ‘ASReview: Open Source Software for Efficient and Transparent Active Learning for Systematic Reviews’ (Zenodo, 2020); https://doi.org/10.5281/zenodo.4024122
Ferdinands, G., Schram, R., van de Schoot, R. & de Bruin, J. Results for ‘ASReview: Open Source Software for Efficient and Transparent Active Learning for Systematic Reviews’ (OSF, 2020); https://doi.org/10.17605/OSF.IO/2JKD6
Kwok, K. T. T., Nieuwenhuijse, D. F., Phan, M. V. T. & Koopmans, M. P. G. Virus metagenomics in farm animals: a systematic review. Viruses 12 , 107 (2020).
Hall, T., Beecham, S., Bowes, D., Gray, D. & Counsell, S. A systematic literature review on fault prediction performance in software engineering. IEEE Trans. Softw. Eng. 38 , 1276–1304 (2012).
van de Schoot, R., Sijbrandij, M., Winter, S. D., Depaoli, S. & Vermunt, J. K. The GRoLTS-Checklist: guidelines for reporting on latent trajectory studies. Struct. Equ. Model. Multidiscip. J. 24 , 451–467 (2017).
Article MathSciNet Google Scholar
van de Schoot, R. et al. Bayesian PTSD-trajectory analysis with informed priors based on a systematic literature search and expert elicitation. Multivar. Behav. Res. 53 , 267–291 (2018).
Cohen, A. M., Bhupatiraju, R. T. & Hersh, W. R. Feature generation, feature selection, classifiers, and conceptual drift for biomedical document triage. In Proc. 13th Text Retrieval Conference (TREC, 2004).
Vasalou, A., Ng, B. D., Wiemer-Hastings, P. & Oshlyansky, L. Human-moderated remote user testing: orotocols and applications. In 8th ERCIM Workshop, User Interfaces for All Vol. 19 (ERCIM, 2004).
Joffe, H. in Qualitative Research Methods in Mental Health and Psychotherapy: A Guide for Students and Practitioners (eds Harper, D. & Thompson, A. R.) Ch. 15 (Wiley, 2012).
NVivo v. 12 (QSR International Pty, 2019).
Hindriks, S., Huijts, M. & van de Schoot, R. Data for UX-test ASReview - June 2020. OSF https://doi.org/10.17605/OSF.IO/7PQNM (2020).
Marshall, I. J., Kuiper, J. & Wallace, B. C. RobotReviewer: evaluation of a system for automatically assessing bias in clinical trials. J. Am. Med. Inform. Assoc. 23 , 193–201 (2016).
Nallapati, R., Zhou, B., dos Santos, C. N., Gulcehre, Ç. & Xiang, B. Abstractive text summarization using sequence-to-sequence RNNs and beyond. In Proc. 20th SIGNLL Conference on Computational Natural Language Learning 280–290 (Association for Computational Linguistics, 2016).
Xie, Q., Dai, Z., Hovy, E., Luong, M.-T. & Le, Q. V. Unsupervised data augmentation for consistency training. Preprint at https://arxiv.org/abs/1904.12848 (2019).
Ratner, A. et al. Snorkel: rapid training data creation with weak supervision. VLDB J. 29 , 709–730 (2020).
Systematic Review Datasets (ASReview Core Development Team, 2020); https://github.com/asreview/systematic-review-datasets
Wallace, B. C., Small, K., Brodley, C. E., Lau, J. & Trikalinos, T. A. Deploying an interactive machine learning system in an evidence-based practice center: Abstrackr. In Proc. 2nd ACM SIGHIT International Health Informatics Symposium 819–824 (Association for Computing Machinery, 2012).
Cheng, S. H. et al. Using machine learning to advance synthesis and use of conservation and environmental evidence. Conserv. Biol. 32 , 762–764 (2018).
Yu, Z., Kraft, N. & Menzies, T. Finding better active learners for faster literature reviews. Empir. Softw. Eng . 23 , 3161–3186 (2018).
Ouzzani, M., Hammady, H., Fedorowicz, Z. & Elmagarmid, A. Rayyan—a web and mobile app for systematic reviews. Syst. Rev. 5 , 210 (2016).
Przybyła, P. et al. Prioritising references for systematic reviews with RobotAnalyst: a user study. Res. Synth. Methods 9 , 470–488 (2018).
ASReview: Active learning for Systematic Reviews (ASReview Core Development Team, 2020); https://github.com/asreview/asreview
Download references
Acknowledgements
We would like to thank the Utrecht University Library, focus area Applied Data Science, and departments of Information and Technology Services, Test and Quality Services, and Methodology and Statistics, for their support. We also want to thank all researchers who shared data, participated in our user experience tests or who gave us feedback on ASReview in other ways. Furthermore, we would like to thank the editors and reviewers for providing constructive feedback. This project was funded by the Innovation Fund for IT in Research Projects, Utrecht University, the Netherlands.
Author information
Authors and affiliations.
Department of Methodology and Statistics, Faculty of Social and Behavioral Sciences, Utrecht University, Utrecht, the Netherlands
Rens van de Schoot, Gerbrich Ferdinands, Albert Harkema, Joukje Willemsen, Yongchao Ma, Qixiang Fang, Sybren Hindriks & Daniel L. Oberski
Department of Research and Data Management Services, Information Technology Services, Utrecht University, Utrecht, the Netherlands
Jonathan de Bruin, Raoul Schram, Parisa Zahedi & Maarten Hoogerwerf
Utrecht University Library, Utrecht University, Utrecht, the Netherlands
Jan de Boer, Felix Weijdema & Bianca Kramer
Department of Test and Quality Services, Information Technology Services, Utrecht University, Utrecht, the Netherlands
Martijn Huijts
School of Governance, Faculty of Law, Economics and Governance, Utrecht University, Utrecht, the Netherlands
Lars Tummers
Department of Biostatistics, Data management and Data Science, Julius Center, University Medical Center Utrecht, Utrecht, the Netherlands
Daniel L. Oberski
You can also search for this author in PubMed Google Scholar
Contributions
R.v.d.S. and D.O. originally designed the project, with later input from L.T. J.d.Br. is the lead engineer, software architect and supervises the code base on GitHub. R.S. coded the algorithms and simulation studies. P.Z. coded the very first version of the software. J.d.Bo., F.W. and B.K. developed the systematic review pipeline. M.Huijts is leading the UX tests and was supported by S.H. M.Hoogerwerf developed the architecture of the produced (meta)data. G.F. conducted the simulation study together with R.S. A.H. performed the literature search comparing the different tools together with G.F. J.W. designed all the artwork and helped with formatting the manuscript. Y.M. and Q.F. are responsible for the preprocessing of the metadata under the supervision of J.d.Br. R.v.d.S, D.O. and L.T. wrote the paper with input from all authors. Each co-author has written parts of the manuscript.
Corresponding author
Correspondence to Rens van de Schoot .
Ethics declarations
Competing interests.
The authors declare no competing interests.
Additional information
Peer review information Nature Machine Intelligence thanks Jian Wu and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information.
Overview of software tools supporting systematic reviews.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/ .
Reprints and permissions
About this article
Cite this article.
van de Schoot, R., de Bruin, J., Schram, R. et al. An open source machine learning framework for efficient and transparent systematic reviews. Nat Mach Intell 3 , 125–133 (2021). https://doi.org/10.1038/s42256-020-00287-7
Download citation
Received : 04 June 2020
Accepted : 17 December 2020
Published : 01 February 2021
Issue Date : February 2021
DOI : https://doi.org/10.1038/s42256-020-00287-7
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
This article is cited by
A systematic review, meta-analysis, and meta-regression of the prevalence of self-reported disordered eating and associated factors among athletes worldwide.
- Hadeel A. Ghazzawi
- Lana S. Nimer
- Haitham Jahrami
Journal of Eating Disorders (2024)
Systematic review using a spiral approach with machine learning
- Amirhossein Saeidmehr
- Piers David Gareth Steel
- Faramarz F. Samavati
Systematic Reviews (2024)
The spatial patterning of emergency demand for police services: a scoping review
- Samuel Langton
- Stijn Ruiter
- Linda Schoonmade
Crime Science (2024)
The SAFE procedure: a practical stopping heuristic for active learning-based screening in systematic reviews and meta-analyses
- Josien Boetje
- Rens van de Schoot
Effect of capacity building interventions on classroom teacher and early childhood educator perceived capabilities, knowledge, and attitudes relating to physical activity and fundamental movement skills: a systematic review and meta-analysis
- Matthew Bourke
- Ameena Haddara
- Patricia Tucker
BMC Public Health (2024)
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing: AI and Robotics newsletter — what matters in AI and robotics research, free to your inbox weekly.
Machine Learning
Ieee account.
- Change Username/Password
- Update Address
Purchase Details
- Payment Options
- Order History
- View Purchased Documents
Profile Information
- Communications Preferences
- Profession and Education
- Technical Interests
- US & Canada: +1 800 678 4333
- Worldwide: +1 732 981 0060
- Contact & Support
- About IEEE Xplore
- Accessibility
- Terms of Use
- Nondiscrimination Policy
- Privacy & Opting Out of Cookies
A not-for-profit organization, IEEE is the world's largest technical professional organization dedicated to advancing technology for the benefit of humanity. © Copyright 2024 IEEE - All rights reserved. Use of this web site signifies your agreement to the terms and conditions.
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- Springer Nature - PMC COVID-19 Collection

Machine Learning-Based Research for COVID-19 Detection, Diagnosis, and Prediction: A Survey
Yassine meraihi.
1 LIST Laboratory, University of M’Hamed Bougara Boumerdes, Avenue of Independence, 35000 Boumerdes, Algeria

Asma Benmessaoud Gabis
2 Ecole nationale Supérieure d’Informatique, Laboratoire des Méthodes de Conception des Systèmes, BP 68 M, 16309 Oued-Smar, Alger Algeria
Seyedali Mirjalili
3 Centre for Artificial Intelligence Research and Optimisation, Torrens University Australia, Fortitude Valley, Brisbane, QLD 4006 Australia
4 Yonsei Frontier Lab, Yonsei University, Seoul, Korea
Amar Ramdane-Cherif
5 LISV Laboratory, University of Versailles St-Quentin-en-Yvelines, 10-12 Avenue of Europe, 78140 Velizy, France
Fawaz E. Alsaadi
6 Information Technology Department, Faculty of Computing and Information Technology, King Abdulaziz University, Jeddah, Saudi Arabia
The year 2020 experienced an unprecedented pandemic called COVID-19, which impacted the whole world. The absence of treatment has motivated research in all fields to deal with it. In Computer Science, contributions mainly include the development of methods for the diagnosis, detection, and prediction of COVID-19 cases. Data science and Machine Learning (ML) are the most widely used techniques in this area. This paper presents an overview of more than 160 ML-based approaches developed to combat COVID-19. They come from various sources like Elsevier, Springer, ArXiv, MedRxiv, and IEEE Xplore. They are analyzed and classified into two categories: Supervised Learning-based approaches and Deep Learning-based ones. In each category, the employed ML algorithm is specified and a number of used parameters is given. The parameters set for each of the algorithms are gathered in different tables. They include the type of the addressed problem (detection, diagnosis, or detection), the type of the analyzed data (Text data, X-ray images, CT images, Time series, Clinical data,...) and the evaluated metrics (accuracy, precision, sensitivity, specificity, F1-Score, and AUC). The study discusses the collected information and provides a number of statistics drawing a picture about the state of the art. Results show that Deep Learning is used in 79% of cases where 65% of them are based on the Convolutional Neural Network (CNN) and 17% use Specialized CNN. On his side, supervised learning is found in only 16% of the reviewed approaches and only Random Forest, Support Vector Machine (SVM) and Regression algorithms are employed.
Introduction
COVID-19 has led to one of the most disruptive disasters in the current century and is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The health system and economy of a large number of countries have been impacted. As per World Health Organization (WHO) data, there have been 225,024,781 confirmed cases of COVID-19, including 4,636,153 deaths as of 14 September 2021. Immediately, after its outbreak, several studies are conducted to understand the characteristics of this coronavirus.
It is argued that human-to-human transmission of SARS-CoV-2 is typically done via direct contacts and respiratory droplets [ 1 ]. On the other side, the incubation of the infection is estimated to a period of 2–14 days. This helps in controlling it and preventing the spread of COVID-19 is the primary intervention being used. Moreover, studies on clinical forms reveal the presence of asymptomatic carriers in the population and the most affected age groups [ 2 ]. After almost a year in this situation, and the high number of researches conducted in different disciplines to bring a relief, a huge amount of data is generated. Computer science researchers find themselves involved to provide their help. One of the first registered contributions is the visualization of data. The latter was mapped and/or plotted in graphs which allows to: (i) better track the propagation of the virus over the globe in general and country by country in particular (Fig. 1 );

Propagation of COVID-19 over the world
ii) better track the propagation of the pandemic over the time; iii) better estimate the number of confirmed cases and the number of deaths (Fig. (Fig.2a, 2 a, b). Later, more advanced techniques based essentially on Artificial Intelligence (AI) are employed. Bringing AI to go against COVID-19 has served in the prevention and monitoring of infectious patients. In fact, by using geographical coordinates of people, some governments were able to limit their movements and locate people with whom they were in contact. The second aspect in which AI benefits is the ability to classify individuals whether they are affected or not. Finally, AI offers the ability to make a prediction on possible future contaminations. To this purpose, Machine Learning (ML), which is often confused with AI, is precisely used. Beyond the different ML algorithms, Neural Network (NN) is one of the most used to solve real-world problems which gives the emergence of Deep Learning (DL).

Data-visualization for tracking COVID-19 progress
Deep learning is particularly suited to contexts where the data is complex and where there are large datasets available as it is the case with COVID-19.
In this context, the present paper gives an overview of the Machine Learning researches performed to handle COVID-19 data. It specifies for each of them the targeted objectives and the type of data used to achieve them.
To accomplish this study, we use Google scholar by employing the following search strings to build a database of COVID-19 related articles:
- COVID-19 detection using Machine learning;
- COVID-19 detection using Deep learning;
- COVID-19 detection using Artificial intelligence;
- COVID-19 diagnosis using Machine learning;
- COVID-19 diagnosis using Deep learning;
- COVID-19 diagnosis using Artificial intelligence;
- COVID-19 prediction using Machine learning;
- Deep learning for COVID-19 prediction;
- Artificial intelligence for COVID-19 prediction.
We retain all articles in this field which:
- Are published in scientific journals;
- Propose new algorithms to deal with COVID-19;
- Have more than 4 pages;
- Are written in English;
- Represent complete versions when several are available;
- Do not report the statistical tests used to assess the significance of the presented results.
- Do not report details on the source of their data sets.
The result is impressive. In fact, since February 2020, several papers are published in this area every month. As we can see in Fig. 3 , India and China seem to be those having the highest number of COVID-19 publications. However, many other countries showed a strong activity in the number of contributions. This is expected as the situation affects the entire world. The different papers appeared from various well-known publishers such as IEEE, Elsevier, Springer, ArXIv and many others as shown in Fig. 4 .

Number of COVID-19 published articles by countries

Percentage of identified COVID-19 papers in different scientific publishers
In this paper, the surveyed approaches are presented according to the Machine Learning classification given in Fig. 8 . Techniques highlighted in yellow color are those employed in the different propositions to go against COVID-19. We show that most of them are based on Convolutional Neural Networks (CNN) which allows making Deep Learning. Almost half of these techniques use X-ray images. Nevertheless, several other data sources are used at different proportions as shown in Fig. 5 . They include Computed Tomography (CT) images, Text data, Time series, Sounds, Coughing/Breathing videos, and even Blood Samples world cloud of the works we have summarized, reviewed, and analyzed in this paper can be seen in Fig. 6 .

Proportion of the different data sources used in COVID-19 publications

A world cloud of the works we have summarized, reviewed, and analyzed in this paper

Classification of Machine Learning Algorithms
There are similar surveys on AI and COVID-19 (e.g. in the works of Rasheed et al. [ 3 ], Shah et al. [ 4 ], Mehta et al. [ 5 ], Shinde et al. [ 6 ] and Chiroma et al. [ 7 ]). What makes this survey different is the focus on specialized Machine Learning techniques proposed globally to detect, diagnose, and predict COVID-19.
The remainder of this paper is organized as follows. In the second section, the definition of Deep Learning and its connection with AI and Machine Learning is given with descriptions of the most used algorithms. The third section presents a classification of the different approaches proposed to deal with COVID-19. They are illustrated by multiple tables highlighting the most important parameters of each of them. The fourth section discusses the results revealed from the conducted study in regard to the techniques used and their evaluation. It notes the limitations encountered and possible solutions to overcome them. The last section concludes the present article.
Artificial Intelligence, Machine Learning and Deep Learning
Artificial Intelligence (AI) as it is traditionally known is considered weak. Making it stronger results in making it capable of reproducing human behavior with consciousness, sensitivity and spirit. The appearance of Machine Learning (ML) was the means that made it possible to take a step towards achieving this objective. By definition, Machine Learning is a subfield of AI concerned with giving computers the ability to learn without being explicitly programmed. It is based on the principle of reproducing a behavior thanks to algorithms, themselves fed by a large amount of data. Faced with many situations, the algorithm learns which decision to make and creates a model. The machine can therefore automate the tasks according to the situations. The general process to carry out a Machine Learning requires a training dataset, a test dataset and an algorithm to generate a predictive model (Fig. 7 ). Four types of ML can be distinguished as we can see in Fig. 8 .

Machine learning prediction process
Supervised Learning
It is a form of machine learning that falls under artificial intelligence. The idea is to “guide” the algorithm on the way of learning based on pre-labeled examples of expected results. Artificial intelligence then learns from each example by adjusting its parameters to reduce the gap between the results obtained and the expected ones. The margin of error is thus reduced over the training sessions, with the aim of being able to generalize learning in the objective to predict the result of new cases [ 8 , 9 ]. The output is called classification if labels are like discrete classes or regression if they are like continuous quantities. Within each category, there exists several algorithms [ 10 , 11 ]. We define below those which was applied in the detection/prediction of COVID-19.
Linear Regression
Linea regression can be considered as one of the most conventional machine learning techniques [ 12 ], in which the best fit line/hyperplane for the available training data is determined using the minimum mean squared error function. This algorithm considers the predictive function as linear. Its general form is as follows: Y = a ∗ X + b + ϵ with a and b two constants. Y is the variable to be predicted, X the variable used to predict, a is the slope of the regression and b is the intercept, that is, the value of Y when X is zero.
Logistic Regression
Despite its name, Logistic Regression [ 13 ] can be employed to perform regression as classification. It is based on the sigmoid predictive function defined as: h ( z ) = 1 1 + e - z where z is a linear function. The function returns a probability score P between 0 and 1. In order to map this to two discrete classes ( 0 or 1), a threshold value θ is fixed. The predicted class is equal to 1 if P ≥ θ , to 0 otherwise.
Support Vector Machine (SVM)
Similar to the previously defined algorithms, the idea behind SVM [ 14 , 15 ] is to distinctly classifies data points by finding an hyperplane in an N-dimensional space. Since there are several possibilities to choose the hyperplane, in SVM a margin distance is calculated between data points of the two classes to separate. The objective is to maximize the value of this margin to get a clear decision boundary helping in the classification of future data points.
Decision Tree
A Decision Tree [ 16 ] is an algorithm that seeks to partition the individuals into groups of individuals as similar as possible from the point of view of the variable to be predicted. The result of the algorithm produces a tree that reveals hierarchical relationships between the variables. An iterative process is used where at each iteration a sub-population of individuals is obtained by choosing the explanatory variable which allows the best separation of individuals. The algorithm stops when no more split is possible.
Random Forest Algorithms
Random Forest Algorithms are methods that provide predictive models for classification and regression [ 17 , 18 ]. They are composed of a large number of Decision Tree blocks used as individual predictors. The fundamental idea behind the method is that instead of trying to get an optimized method all at once, several predictors are generated and their different predictions are pooled. The final predicted class is the one having the most votes.
Artificial Neural Network (ANN)
Artificial Neural Networks is a popular Supervised classification algorithm trying to mimic the way human brain works. It is often used whenever there is abundant labeled training data with many features [ 19 ]. The network calculates from the input a score (or a probability) to belong to each class. The class assigned to the input object corresponds to the one with the highest score. A Neural Network is a system made up of neurons. It is divided into several layers connected to each other where the output of one layer corresponds to the input of the next one [ 20 , 21 ]. The calculation of the final score is based on the calculation of a linear function from the layers weights and an activation function. The weights values are randomly assigned to each input at the beginning and then are learned (updated) by backpropagation of the gradient to minimize the loss function associated with the final layer. The optimization is done with a gradient descent technique [ 22 ].
Unsupervised Learning
Unsupervised learning is a type of self-organized learning that learns and creates models from unlabeled training datasets (unlike Supervised Learning). There are two practices in Unsupervised Learning. The first one is the clustering, which is the fact of gathering similar data in homogeneous groups. It is performed by applying one of the many existing clustering algorithms [ 23 ]: K-means, Hierarchical clustering, Hidden Markov, etc. The second practice is the dimensionality reduction [ 24 ] which consists of the reduction of features in highly dimensional data. The purpose is to extract new features and to find the best linear transformation representing maximum data points by guaranteeing a minimum loss of information.
Deep Learning
As illustrated in Fig. 9 , Deep Learning [ 25 , 26 ] is a branch of AI that focuses on creating large Neural Network models that are capable of making decision based on Machine Learning models, it is a Neural Networks with many hidden neural layers. Indeed, it has been observed that the addition of layers of neurons has a great impact on the quality of the results obtained.

Classification of Machine Learning Approaches
There are many different deep learning algorithms other than ANN. In the following we define the most used ones and which are applied in the context of COVID-19.
Convolutional Neural Network (CNN)
Convolutional Neural Networks or ConvNets [ 27 , 28 ] is a type of ANN used to make a Deep Learning that is able to categorize information from the simplest to the most complex one. They consist of a multilayer stack of neurons as well as mathematical functions with several adjustable parameters, which preprocess small amounts of information. Convolutional networks are characterized by their first convolutional layers (usually one to three). They seek to identify the presence of a basic and abstract pattern in an object. Successive layers can use this information to distinguish objects from each other (classification / recognition).
Recurrent Neural Network (RNN)
Recurrent Neural Network [ 29 , 30 ] is also a type of ANN used to make a Deep Learning where information can move in both directions between the deep layers and the first layers. This allows it to keep information from the near past in memory. For this reason, RNN is particularly suited to applications involving context, and more particularly to the processing of temporal sequences such as learning and signal generation. However, for applications involving long time differences (typically the classification of video sequences), this “short-term memory” is not sufficient because forgetting begins after about fifty iterations.
Generative Adversarial Network (GAN)
GAN [ 31 ] is a Deep Learning technique. It is based on the competition of two networks within a framework. These two networks are called “generator” and “discriminator”. The generator is a type of CNN whose role is to create new instances of an object which means that outputs are produced without it being possible to determine if they are false. On the other hand, the discriminator is a “deconvolutive” neural network that determines the authenticity of the object (whether or not it is part of a data set).
Reinforcement Learning
Reinforcement Learning [ 32 , 33 ] is a method of learning for machine learning models. Basically, this method lets the algorithm learn from its own mistakes. To learn how to make the right decisions, the AI program is directly confronted with choices. If it is wrong, it is “penalized”. On the contrary, if it makes the right decision, it is “rewarded”. In order to get more and more rewards, AI will therefore do its best to optimize its decision-making.
Overview of Machine Learning approaches used to combat COVID-19
Zhang et al. [ 34 ] applied Support Vector Machine (SVM) model for COVID-19 cases detection and classification. The clinical information and blood/urine test data were used in their work to validate SVM’s performance. Simulation results demonstrated the effectiveness of the SVM model by achieving an accuracy of 81.48%, sensitivity of 83.33%, and specificity of 100%.
Hassanien et al. [ 35 ] proposed a new approach based on the hybridization of SVM with Multi-Level Thresholding for detecting COVID-19 infected patients from X-ray images. The performance of the hybrid approach was evaluated using 40 contrast-enhanced lungs X-ray images (15 normal and 25 with COVID-19). A similar work was done by Sethy et al. [ 36 ], in which a combined approach based on the combination of SVM with 13 pre-trained CNN models for COVID-19 detection from chest X-ray images were proposed. Experimental results showed that ResNet50 combined with SVM outperforms other CCN models combined with SVM by achieving an average classification accuracy of 95.33%.
Sun et al. [ 37 ] used SVM model for predicting the COVID-19 patients with severe/critical symptoms. 220 clinical/laboratory observations records and 336 cases of patients infected COVID-19 divided into training and testing datasets were used to validate the performance of the SVM model. Simulation results showed that the SVM model achieves an Area Under Curve (AUC) of 0.9996 and 0.9757 in the training and testing dataset, respectively.
Singh et al. [ 38 ] used four machine learning approaches (SVM with Bagging Ensemble, CNN, Extreme Learning Machine (ELM), Online Sequential ELM (OS-ELM)) for automatic detection of COVID-19 cases. The performance of the proposed approaches was tested using datasets of 702 CT scan images (344with COVID-19 and 358 normal). Experimental results revealed the efficiency of SVM with Bagging Ensemble by obtaining an accuracy, precision, sensitivity, specificity, F1-score, and AUC of 95.70%, 95.50%, 96.30%, 94.80%, 95.90%, and 95.80%, respectively.
Singh et al. [ 39 ] proposed Least Square-SVM (LS-SVM) and Autoregressive Integrated Moving Average (ARIMA) for the prediction of COVID-19 cases. A dataset of COVID-19 confirmed cases collected from five the most affected countries 1 was used to validate the proposed models. It was demonstrated that the LS-SVM model outperforms the ARIMA model by obtaining an accuracy of 80%.
Nour et al. [ 40 ] applied machine learning approaches such as SVM, Decision tree (DT), and KNN for automatic detection of positive COVID-19 cases. The performance of the proposed approaches was validated on a public COVID-19 radiology database divided into training and test sets with 70% and 30% rates, respectively.
Tabrizchi et al. [ 41 ] used SVM with Naive Bayes (NB), Gradient boosting decision tree (GBDT), AdaBoost, CNN, and Multilayer perceptron (MLP) for rapid diagnosis of COVID-19. A dataset of 980 CT scan images (430 with COVID-19 and 550 normal) was used in the simulation and results showed that SVM outperforms other machine-learning approaches by achieving an average accuracy, precision, sensitivity, and F1-score of 99.20%, 98.19%, 100%, and 99.0%, respectively.
Regression Approaches
Yue et al. [ 42 ] used a linear regression model for the prediction of COVID-19 infected patients. CT images of 52 patients collected from five hospitals in Ankang, Lishui, Zhenjiang, Lanzhou, and Linxia were used to evaluate the performance of the regression model. Simulation results demonstrated that the linear regression model outperforms the Random Forest algorithm.
Another similar work was done by Shi et al. [ 43 ], in which a least absolute shrinkage and selection operator (LASSO) logistic regression model was proposed. The effectiveness of the proposed model was evaluated based on CT images taken from 196 patients (151 non-severe patients and 45 severe patients). Experimental results showed the high performance of the proposed model compared to quantitative CT parameters and PSI score by achieving an accuracy of 82.70%, sensitivity of 82.20%, specificity of 82.80%, and AUC of 89%
Yan et al. [ 44 ] proposed a supervised regression model, called XGBoost, for predicting COVID-19 patients. A database of blood samples of 485 infected patients in the region of Wuhan, China was used in simulations and results showed that XGBoost gives good performance by achieving an overall accuracy of 90% in the detection of patients with COVID-19.
Salama et al. [ 45 ] used the linear regression model with SVM and ANN for the prediction of COVID-19 infected patients. The effectiveness of the proposed models was assessed based on the Epidemiological dataset collected from many health reports of real-time cases. Simulation results demonstrated that SVM has the lowest mean absolute error with the value of 0.21, while the regression model has the lowest root mean squared error with a value of 0.46.
Gupta et al. [ 46 ] proposed a linear regression technique with mathematical SEIR (Susceptible, Exposed, Infectious, Recovered) model for COVID-19 outbreak predictions. It was tested using data collected from John Hopkins University repository taking into account the root mean squared log error (RMSLE) metric. Simulation results showed that SEIR model has the lowest RMSLE with the value of 1.52.
In the work of Chen and Liu [ 47 ], Logistic Regression with Random Forest, Partial Least Squares Regression (PLSR), Elastic Net, and Bagged Flexible Discriminant Analysis (BFDA) were proposed for predicting the severity of COVID-19 patients. The efficiency of the proposed models was evaluated using data of 183 severely infected COVID-19 patients and results showed that the logistic regression model outperforms other machine learning models by achieving a sensitivity of 89.20%, specificity of 68.70%, and AUC of 89.20%.
Another similar work was done by Ribeiro et al. [ 48 ], in which six machine learning approaches such as stacking-ensemble learning (SEL), support vector regression (SVR), cubist regression (CUBIST), auto-regressive integrated moving average (ARIMA), ridge regression (RIDGE), and random forest (RF) were employed for prediction purposes in COVID-19 datasets.
Yadav et al. [ 49 ] used three machine learning approaches (Linear Regression, Polynomial Regression, and SVR) for COVID-19 epidemic prediction and analysis. A dataset containing the total number of COVID19 positive cases was collected from different countries such as South Korea, China, US, India, and Italy. Results showed the superiority of SVR compared to Linear Regression and Polynomial Regression. The average accuracy for SVR, Linear Regression, and Polynomial Regression are 99.47%, 65.01%, and 98.82%, respectively.
Matos et al. [ 50 ] proposed four linear regression models (Penalized binomial regression (PBR, Conditional inference trees (CIR), Generalised linear (GL), and SVM with linear kernel) for COVID-19 diagnosis. CT images and Clinical data collected from 106 patients were used in the simulation and results showed that SVM with linear kernel gives better results compared to other models by providing an accuracy of 0.88, sensitivity of 0.90, specificity of 0.87, and AUC of 0.92.
Khanday et al. [ 51 ] proposed Logistic regression with six machine learning approaches (Adaboost, Stochastic Gradient Boosting, Decision Tree, SVM, Multinomial Naïve Bayes, and Random Forest) for COVID-19 detection and classification. It was evaluated using 212 clinical reports divided into four classes including COVID, ARDS, SARS, and Both (COVID, ARDS). Simulation results showed that logistic regression provides excellent performance by obtaining 94% of precision, 96% of sensitivity, accuracy of 96.20%, and 95% of F1-score.
Yang et al. [ 52 ] proposed Gradient Boosted Decision Tree (GBDT) with Decision Tree, Logistic Regression, and Random Forest for COVID-19 diagnosis. 27 routine laboratory tests collected from the New York Presbyterian Hospital/Weill Cornell Medicine (NYPH/WCM) were used to evaluate this technique. Experimental results revealed the efficiency of GBDT by achieving a sensitivity, specificity, and AUC of 76.10 %, 80.80%, and 85.40%, respectively.
Saqib [ 53 ] developed a novel model (PBRR) by combining Bayesian Ridge Regression (BRR) with n-degree Polynomial for forecasting COVID-19 outbreak progression. The performance of the PBRR model was validated using public datasets collected from John Hopkins University available until 11th May 2020. Experimental results revealed the good performance of PBRR with an average accuracy of 91%.
Random Forest Algorithm
Shi et al. [ 54 ] proposed an infection Size Aware Random Forest method (iSARF) for diagnosis of COVID-19. A dataset of 1020 CT images (1658 with COVID-19, and 1027 with pneumonia) was used to assess the performance of iSARF. Simulation results demonstrated that iSARF provides good performance by yielding the sensitivity of 90.7%, specificity of 83.30%, and accuracy of 87.90% under five-fold cross-validation.
Iwendi et al. [ 55 ] combined RF model with AdaBoost algorithm for COVID-19 disease severity prediction. The efficiency of the boosted RF model was evaluated based on COVID-19 patient’s geographical, travel, health, and demographic data. Boosted RF model gives an accuracy of 94% and F1-Score of 86% on the dataset used.
In the work of Brinati et al. [ 56 ], seven machine learning approaches (Random Forest, Logistic Regression, KNN, Decision Tree, Extremely Randomized Trees, Naïve Bayes, and SVM) were proposed for the identification of COVID-19 positive patients. Routine blood exams collected from 279 patients were used in the simulation and results demonstrated the feasibility and effectiveness of the Random Forest algorithm by achieving an accuracy, precision, sensitivity, specificity, and AUC of 82%, 83%, 92%, 65%, and 84%, respectively.
The main characteristics of the predefined Supervised Learning approaches are given in Table Table1 1 .
Summary of supervised learning approaches for detection, diagnosis, and prediction of COVID-19 cases
Deep Learning Approaches
The most applied method to detect, predict and diagnostic COVID-19 are based on Deep Learning with its different techniques. In the following, we summarize the found approaches in respect of the classification given in Fig. 8 . We gather in Tables Tables2, 2 , ,3, 3 , ,4, 4 , ,5 5 and and6 6 are their main features.
Summary of convolutional neural networks (CNN) approaches for detection, diagnosis, and prediction of COVID-19 cases
Summary of Recurrent Neural Networks (RNN) approaches for detection, diagnosis, and prediction of COVID-19 case
Summary of Specialized CNN approaches for detection, diagnosis, and prediction of COVID-19 cases
Summary of Generative Adversarial Network (GAN) approaches for detection, diagnosis, and prediction of COVID-19 cases
Summary of other deep learning approaches for detection, diagnosis, and prediction of COVID-19 cases
Wang et al. [ 60 ] proposed a deep CNN model, called Residual Network34 (ResNet34), for COVID-19 diagnosis in CT scan images. The effectiveness of ResNet34 was validated using CT scan images collected from 99 patients (55 patients with typical viral pneumonia and 44 patients with COVID-19). Simulation results showed that ResNet34 achieves an overall accuracy of 73.10%, specificity of 67%, and sensitivity of 74%.
Narin et al. [ 61 ] used three pre-trained techniques including ResNet50, InceptionV3, and InceptionResNetV2 for automatic diagnosis and detection of COVID-19. The case studies included four classes including normal, COVID-19, bacterial, and viral pneumonia patients. The authors demonstrated that ResNet50 gives the highest accuracy in three different datasets.
Maghdid et al. [ 62 ] proposed a CNN model with AlexNet for COVID-19 diagnosis. A dataset of 361 CT images and 170 X-ray images of COVID-19 disease collected from five different sources was used in the simulation. Quantitative results demonstrated that AlexNet achieves an accuracy of 98%, a sensitivity of 100%, and a specificity of 96% in X-ray images, while the modified CNN model achieves 94.10% of accuracy, 90% of sensitivity, and 100% of specificity in CT-images.
Wang et al. [ 63 ] employed eight deep learning (DL) models (fully convolutional network (FCN-8 s), UNet, VNet, 3D UNet++, dual-path network (DPN-92), Inceptionv3, ResNet50, and Attention ResNet50) for COVID-19 detection. The efficiency of the proposed models was evaluated using 1,136 CT images (723 with COVID-19 and 413 normal) collected from five hospitals. Simulation results demonstrated the superiority of 3D UNet++ compared to other CNN models.
In CT scan images, UNet++ was employed by Chen et al. [ 64 ] for COVID-19 detection. The performance of UNet++ was assessed based on a dataset of 106 CT scan images. Simulation results showed that UNet++ provides a per-patient accuracy of 95.24%, sensitivity of 100%, specificity of 93.55%. A per-image accuracy of 98.85%, sensitivity of 94.34%, specificity of 99.16% were also achieved.
Apostolopoulos et al. [ 65 ] proposed five deep CNN models (VGG19, MobileNetv2, Inception, Xception, and Inception ResNetv2) for COVID-19 detection cases. The proposed models were tested using two datasets of 1428 and 1442 images, respectively. In the first dataset (224 with COVID-19, 700 with bacterial pneumonia, and 504 normal), MobileNetv2 approach provided better results with a two-class problem accuracy, three-class problem accuracy, sensitivity, and specificity of 97.40%, 92.85%, 99.10%, and 97.09%, respectively. In the second dataset (224 with COVID-19, 714 with bacterial pneumonia, and 504 normal), MobileNetv2 approach also provided better performance by achieving a two-class problem accuracy, three-class problem accuracy, sensitivity, and specificity of 96.78%, 94.72%, 98.66%, and 96.46%, respectively.
Another deep CNN model was developed by Zhang et al. [ 66 ] which is composed of three components (a backbone network, a classification head, and an anomaly detection head). This technique was evaluated using 100 chest X-ray images of 70 patients taken from the Github repository. 1431 additional chest X-ray images of 1008 patients taken from the public Chest X-ray14 data were also used to facilitate deep learning. Simulation results showed that the proposed model is an effective diagnostic tool for low-cost and fast COVID-19 screening by achieving the accuracy of 96% for COVID-19 cases and 70.65% for non-COVID-19 cases.
Another intersting project was done by Ghoshal and Tucker [ 67 ], in which a Bayesian Convolutional Neural Networks (BCNN) was used in conjunction with Dropweights for COVID-19 diagnosis and classification.
Toraman et al. [ 68 ] proposed a CNN model, called CAPSNET, for fast and accurate diagnostics of COVID-19 cases. CAPSNET model was evaluated using two datasets of 2100 and 13,150 cases, respectively. In the first dataset (1050 with COVID-19 and 1050 no-findings), CAPSNET provided better results by achieving an accuracy, precision, sensitivity, specificity, F1-score of 97.23%, 97.08%, 97.42%, 97.04%, and 97.24% respectively. In the second dataset (1050 with COVID-19, 1050 no-findings, and 1050 pneumonia), CAPSNET provided better performance by achieving an accuracy, precision, sensitivity, specificity, and F1-score of 84.22%, 84.61%, 84.22%, 91.79%, and 84.21% respectively.
Hammoudi et al. [ 69 ] investigated six deep CNN models (ResNet34, ResNet50, DenseNet169, VGG19, InceptionResNetV2, and RNN-LSTM) for COVID-19 screening and detection. A dataset of 5,863 children’s X-Ray images (Normal and Pneumonia) was exploited to evaluate the techniques proposed. Simulation results showed that DenseNet169 outperforms other deep CNN models by obtaining an average accuracy of 95.72%.
Ardakani et al. [ 70 ] proposed ten deep CNN models (AlexNet, VGG16, VGG19, SqueezeNet, GoogleNet, MobileNetV2, ResNet18, ResNet50, ResNet101, and Xception) for COVID-19 diagnosis. A dataset of 1020 CT images (108 with COVID-19, and 86 with bacteria pneumonia) was used to benchmark the efficiency. Simulation results showed the high performance of ResNet101 compared to other deep CNN models by achieving an accuracy of 99.51%, sensitivity of 100%, AUC of 99.4%, and specificity of 99.02%. Xu et al. [ 71 ] proposed a hybrid deep learning model, called ResNet+, based on combining the traditional ResNet with location-attention mechanism for COVID-19 diagnosis. The effectiveness of ResNet+ was evaluated using 618 Computer Tomography (CT) images (175 normal, 219 with COVID-19, 224 with Influenza-A viral pneumonia) and results demonstrated that ResNet+ provides an overall accuracy of 86.70%, sensitivity of 81.50%, precision of 80.80%, and F1-score of 81.10%. It is also revealed that the proposed ResNet+ is a promising supplementary diagnostic technique for clinical doctors.
Cifci [ 72 ] proposed two deep CNN model (AlexNet and InceptionV4) for Diagnosis and prognosis analysis of COVID-19 cases. The effectiveness of the proposed models was evaluated using 5800 CT images divided into 80% training and 20% test. It was demonstrated that AlexNet outperforms InceptionV4 by achieving an overall accuracy of 94.74%, a sensitivity of 87.37%, and a specificity of 87.45%. Bai et al. [ 73 ] did a similar work by proposin an EfficientNet B4 CNN model with a fully connected neural network for the detection and classification of COVID-19 cases. CT scan images of 521 patients were used in the simulation.
Loey et al. [ 74 ] proposed three deep CNN approaches (Alexnet, Googlenet, and Restnet18) with GAN model for COVID-19 detection. The proposed approaches were evaluated using three scenarios: i) four classes (normal, viral pneumonia, bacteria pneumonia, and COVID-19 images); ii) three classes (COVID-19, Normal, and Pneumonia); and iii) two classes (COVID-19, Normal). Experimental results demonstrated that Googlenet gives better performance in the first and third scenario by achieving an accuracy of 80.60%, and 100%, respectively. Alexnet provides better results in the second scenario by achieving an accuracy of 85.20%.
Singh et al. [ 75 ] proposed a novel deep learning approach based on convolutional neural networks with multi-objective differential evolution (MODE) for the classification of COVID-19 patients. In addition, Mukherjee et al. [ 76 ] proposed a shallow light-weight CNN model for automatic detection of COVID-19 cases from Chest X-rays in a similar manner.
Ozkaya et al. [ 77 ] proposed an effective approach based on the combination of CNN model with the ranking method and SVM technique for COVID-19 detection. The case studies included two datasets generated from 150 CT images, each dataset contains 3000 normal images and 3000 with COVID-19. Simulation results showed the high performance and robustness of the proposed approach compared to VGG16, GoogleNet, and ResNet50 models in terms of accuracy, sensitivity, specificity, sensitivity, F1-score, and Matthews Correlation Coefficient (MCC) metrics.
Toğaçar et al. [ 78 ] proposed two CNN models (MobileNetV2, SqueezeNet) combined with SVM for COVID-19 detection. The efficiency of the proposed models was validated using a dataset of X-ray images divided into three classes: normal, with COVID-19, and with pneumonia. The accuracy obtained in their work is of 99.27%.
Pathak et al. [ 79 ] proposed a ResNet50 deep transfer learning technique for the detection and classification of COVID-19 infected patients. The effectiveness of ResNet50 was evaluated using 852 CT images collected from various datasets (413 COVID-19 (+) and 439 normal or pneumonia). Simulation results showed that ResNet50 model gives efficient performance by achieving a specificity, precision, sensitivity, accuracy of 94.78%, 95.19%, 91.48%, and 93.02%, respectively.
Elasnaoui et al. [ 80 ] proposed seven Deep CNN models including baseline CNN, VGG16, VGG19, DenseNet201, InceptionResNetV2, InceptionV3, Xception, Resnet50, and MobileNetV2 for automatic classification of pneumonia images. Chest X-Ray & CT datasets containing 5856 images (4273 pneumonia and 1583 normal) were used to validate the proposed models and results demonstrated that Resnet50, MobileNetV2, and InceptionResnetV2 provide high performance with an overall accuracy more than 96% against other CNN models with an accuracy around 84%. Another similar work was done by Zhang et al. [ 81 ], in which a diagnosis COVID-19 system based on 3D ResNet18 deep learning technique with five deep learning-based segmentation models (Unet, DRUNET, FCN, SegNet & DeepLabv3) for Diagnosis and prognosis prediction of COVID-19 cases.
Rajaraman and Antali [ 82 ] used five deep CNN models (VGG16, InceptionV3, Xception, DenseNet201, NasNetmobile) for COVID-19 screening. Six datasets of x-ray images including Pediatric CXR, RSNA CXR, CheXpert CXR, NIH CXR-14, Twitter COVID-19 CXR, and Montreal COVID-19 CXR were used to validate the effectiveness of the proposed models. The accuracy obtained was 99.26%.
Tsiknakis et al. [ 83 ] proposed a modified deep CNN model (Modified InceptionV3) for COVID-19 screening on chest X-rays. The Modified InceptionV3 was evaluated using two chest X-ray datasets, the first dataset was collected from [ 84 ], the second one was collected from the QUIBIM imagingcovid19 platform database and various public repositories. Experimental results showed that the modified InceptionV3 model gives an average accuracy, AUC, sensitivity, and specificity of 76%, 93%, 93%, and 91.80%, respectively.
Ahuja et al. [ 85 ] presented pre-trained transfer learning models (ResNet18, ResNet50, ResNet101, and SqueezeNet) for automatic detection of COVID-19 cases. Another similar work was done by Oh et al. [ 86 ], in which a patch-based convolutional neural network was proposed based on ResNet18.
Elasnaoui and Chawki [ 87 ] used seven pre-trained deep learning models (VGG16, VGG19, DenseNet201, InceptionResNetV2, InceptionV3, Resnet50, and MobileNetV2) for automated detection and diagnosis of COVID-19 disease. The effectiveness of the proposed models was assessed using chest X-ray & CT dataset of 6087 images. Simulation results showed the superiority of InceptionResNetV2 compared to other deep CNN models by achieving an accuracy, precision, sensitivity, specificity, and F1-score of 92.60%, 93.85%, 82.80%, 97.37%, and 87.98%, respectively.
Chowdhury et al. [ 88 ] introduced eight deep CNN (DenseNet201, RestNet18, MobileNetv2, InceptionV3, VGG19, ResNet101, CheXNet, and SqueezNet) for COVID-19 detection. A dataset of 3487 x-ray images (423 with COVID-19, 1485 with viral pneumonia, and 1579 normal) with and without image augmentation was used in the validation of the proposed models. Simulation results showed that CheXNet gives better results when image augmentation was not applied with an accuracy, precision, sensitivity, specificity, F1-score of 97.74%, 96.61 %, 96.61%, 98.31%, and 96.61% respectively. However, when image augmentation was used, DenseNet201 outperforms other deep CNN models by achieving an accuracy, precision, sensitivity, specificity, and F1-score of 97.94%, 97.95%, 97.94%, 98.80%, and 97.94%, respectively.
Apostolopoulos et al. [ 89 ] proposed a deep CNN model (MobileNetv2) for COVID-19 detection and classification. The efficiency of MobileNetv2 was assessed using a large-scale dataset of 3905 X-ray images and results showed its excellent performance by achieving an accuracy, sensitivity, specificity of 99.18%, 97.36%, and 99.42%, respectively in the detection of COVID-19.
Rahimzadeh and Attar [ 90 ] proposed a modified deep CNN model based on the combination of Xception and ReNet50V2 for detecting COVID-19 from chest X-ray images. The proposed model was tested using 11,302 chest X-ray images (31 with COVID-19, 4420 with pneumonia, and 6851 normal cases). Experimental results showed that the combined model gives an average accuracy, precision, sensitivity, and specificity of 91.4%, 72.8%, 87.3%, and 94.2%, respectively. In a similar work, Abbas et al. [ 91 ] adapted a Convolutional Neural Network model, called Decompose Transfer Compose (DeTraC). The effectiveness of the DeTraC model was validated using a dataset of X-ray images collected from several hospitals and institutions around the world. As the results 95.12% accuracy, 97.91% sensitivity, and 1.87% specificity were obtained.
Afshar et al. [ 92 ] developed a deep CNN model (COVID-CAPS) using on Capsule Networks for COVID-19 identification and diagnosis. The effectiveness of COVID-CAPS was tested using two publicly available chest X-ray datasets. [ 84 , 93 ] As the results 98.30% accuracy, 80% sensitivity, and 8.60% specificity were obtained.
Brunese et al. [ 94 ] adopted a deep CNN approach (VGG-16) for automatic and faster COVID-19 detection from chest X-ray images. The robustness of VGG-16 was evaluated using 6523 chest X-ray images (2753 with pneumonia disease, 250 with COVID-19, while 3520 healthy) and results showed that VGG-16 achieves an accuracy of 97% for the COVID-19 detection and diagnosis.
Jin et al. [ 95 ] proposed a deep learning-based AI system for diagnosis of COVID-19 in CT images. 10,250 CT scan images (COVID-19, viral pneumonia, influenza-A/B, normal) taken from three centers in China and three publicly available databases were used in the simulation and results showed that the proposed model achieves an AUC of 97.17%, a sensitivity of 90.19%, and a specificity of 95.76%.
Truncated Inception Net was proposed by Das et al. [ 96 ] as a Deep CNN model for COVID-19 cases detection. Six different datasets were used in the simulation considering healthy, with COVID-19, with Pneumonia, and with Tuberculosis cases. It was demonstrated that Truncated Inception Net provides accuracy, precision, sensitivity, specificity, and F1-score of 98.77%, 99%, 95%, 99%, and 97%, respectively.
Asif et al. [ 97 ] proposed a Deep CNN model (Inception V3) with transfer learning for automatic detection of COVID-19 patients cases. A dataset consists of 3550 chest x-ray images (864 with COVID-19, 1345 with viral pneumonia, and 1341 normal) was used to test Inception V3. Simulation results proved the efficiency of the Inception V3 by achieving an accuracy of 98%.
Punn and Agrawal [ 98 ] introduced five fine-tuned deep learning approaches (baseline ResNet, Inceptionv3, InceptionResNetv2, DenseNet169, and NASNetLarge) for automated diagnosis and classification of COVID-19. The performance of the proposed approaches was validated using three datasets of X-ray and CT images collected from Radiological Society of North America (RSNA), [ 99 ] U.S. national library of medicine (USNLM), [ 100 ] and COVID-19 image data collection. [ 84 ] Simulation results showed that NASNetLarge outperforms other CNN models by achieving 98% of accuracy, 88% of precision, 90% of sensitivity, 95% of specificity, and 89% of F1-score.
Shelke et al. [ 101 ] proposed three CNN models (VGG16, DenseNet161, and ResNet18) for COVID-19 diagnosis and analysis. The proposed models were tested using two datasets of 1191 and 1000 X-ray images, respectively. In the first dataset (303 with COVID-19, 500 with bacterial pneumonia, and 388 normal), VGG16 approach provided better results with an accuracy of 95.9%. In the second dataset (500 with COVID-19 and 500 normal), DenseNet161 approach provided better performance by achieving an accuracy of 98.9%.
Rajaraman et al. [ 102 ] proposed eight deep CNN models (VGG16, VGG19, InceptionV3, Xception, InceptionResNetV2, MobileNetV2, DenseNet201, NasNetmobile) for COVID-19 screening. Four datasets of x-ray images including Pediatric CXR, RSNA CXR, Twitter COVID-19 CXR, and Montreal COVID-19 CXR were used to validate the effectiveness of the proposed models. Experimental results demonstrated that the weighted average of the best-performing pruned models enhances performance by providing an accuracy, precision, sensitivity, AUC, F1-score of 99.01%, 99.01%, 99.01%, 99.72%, and 99.01%, respectively.
Another similar work was done by Luz et al. [ 103 ], which can be considered as an extension of EfficientNet for COVID-19 detection and diagnosis in X-Ray Chest images. It was compared with MobileNet, MobileNetV2, ResNet50, VGG16, and VGG19. Simulation results demonstrated the effectiveness of EfficientNet compared to other deep CNN models by achieving an overall accuracy of 93.9%, sensitivity of 96.8%, and a positive prediction rate of 100%.
Jaiswal et al. [ 104 ] employed DenseNet201 based transfer learning for COVID-19 detection and diagnosis. The performance of DenseNet201 was validated using 2492 chest CT-scan images (1262 with COVID-19 and 1230 healthy) taken into account precision, F1-measure, specificity, sensitivity, and accuracy metrics. Quantitative results showed the effectiveness of compared to VGG16, Resnet152V2, and InceptionResNet by providing a precision, F1-measure, specificity, sensitivity, and accuracy of 96.29%, 96.29%, 96.29% and 96.21%, and 96.25%, respectively.
Sharma [ 105 ] employed a ResNet50 CNN-based approach for COVID-19 detection. 2200 CT images (800 with COVID-19, 600 viral pneumonia, and 800 normal healthy) collected from various hospitals in Italy, China, Moscow, and India were used in the simulation and results showed that ResNet50 outperforms ResNet+ by giving a specificity, sensitivity, accuracy of 90.29%, 92.1%, and 91.0%, respectively. Pu et al. [ 106 ] conducted a similar work.
Alotaibi [ 107 ] used four pre-trained CNN models (RESNET50, VGG19, DENSENET121, and INCEPTIONV3) for the detection of COVID-19 cases. A dataset of X-ray images (219 with COVID-19, 1341 Normal, and 1345 with Viral Pneumonia) was used in the experimentation and results demonstrated the better performance of DENSENET121 compared to RESNET50, VGG19, and INCEPTIONV3 by achieving an accuracy, precision, sensitivity, and F1-score of 98.71%, 98%, 98%, and 97.66%, respectively.
Goyal and Arora [ 108 ] proposed three CNN models (VGG16, VGG19, and ResNet50) for COVID-19 detection. This technique was evaluated using 748 chest X-ray images (250 with COVID-19, 300 normal, and 198 with pneumonia bacteria) and results showed that VGG19 outperforms VGG16 and ResNet50 by achieving an accuracy of 98.79% and 98.12% in training and testing cases, respectively. A similar work was done by Das et al. [ 109 ], in which an extreme version of the Inception (Xception) model for the automatic detection of COVID-19 infection cases in X-ray images.
Rahaman et al. [ 110 ] used 15 different pre-trained CNN models for COVID-19 cases identification. 860 chest X-Ray images (260 with COVID-19, 300 healthy, and 300 pneumonia) were employed to investigate the effectiveness of the proposed models. Simulation results showed that the VGG19 model outperforms other deep CNN models by obtaining an accuracy of 89.3%, precision of 90%, sensitivity of 89%, and F1-score of 90%.
Altan and Karasu [ 111 ] proposed a hybrid approach based on CNN model (EfficientNet-B0), two-dimensional (2D) curvelet transformation, and chaotic salp swarm algorithm (CSSA) for COVID-19 detection. 2905 real raw chest X-ray images (219 with COVID-19, 1345 viral pneumonia, and 1341 normal) were used. Another similar work was done where a Confidence-aware anomaly detection (CAAD) was proposed based on EfficientNetB0
Ni et al. [ 112 ] proposed a CNN model, called MVPNet, for automatic detection of COVID-19 cases. 19,291 pulmonary CT scans images (3854 with COVID-19, 6871 with bacterial pneumonia, and 8566 healthy) were employed to validate the performance of the MVPNet model. Experimental results demonstrated that MVPNet achieves a sensitivity of 100%, specificity of 65%, accuracy of 98%, and F1-score of 97%.
Nguyen et al. [ 113 ] employed two deep CNN models (EfficientNet and MixNet) for the detection of COVID-19 infected patients from chest X-ray (CXR) images. The effectiveness of the proposed approach was validated using two real datasets consisting of: i) 13,511 training images and 1,489 testing images; ii) 14,324 training images and 3,581 testing images. Simulation results demonstrated that the proposed approach outperforms some well-established baselines by yielding an accuracy larger than 95%.
Islam et al. [ 114 ] proposed four CNN models( VGG19, DenseNet121, InceptionV3, and InceptionResNetV2) and recurrent neural network (RNN) for COVID-19 diagnosis. A similar work was done by Mei et al. [ 115 ] with proposing a combination of SVM, random forest, MLP, and CNN.
Khan and Aslam [ 116 ] presented four CNN models (DenseNet121, ResNet50, VGG16, and VGG19) for COVID-19 diagnosis. The superiority of the proposed models was evaluated using a dataset of 1057 X-ray images including 862 normal and 195 with COVID-19. Experimental results demonstrated that VGG-19 model achieves better performance than DenseNet121, ResNet50, and VGG16 by achieving an accuracy, sensitivity, specificity, F1-score of 99.33%, 100%, 98.77%, and 99.27%, respectively.
Perumal et al. [ 117 ] used deep CNN models (VGG16, Resnet50, and InceptionV3) and Haralick features for the detection of COVID-19 cases. A dataset of X-ray and CT images collected from various resources available in Github open repository, RSNA, and Google images was used in the simulation and results showed that the proposed models outperform other existing models with an average accuracy of 93%, precision of 91%, and sensitivity of 90%.
Kumar et al. [ 118 ] used various deep learning models (VGG, DenseNet, AlexNet, MobileNet, ResNet, and Capsule Network) with blockchain and federated-learning technology for COVID-19 detection from CT images. These techniques were evaluated using a dataset of 34,006 CT scan images taken from the GitHub repository ( https://github.com/abdkhanstd/COVID-19 ). Simulation results revealed that the Capsule Network model outperforms other deep learning models by achieving an accuracy of 0.83 and sensitivity of 0.967 and precision of 0.83.
Zebin et al. [ 119 ] proposed three Deep CNN models (modified VGG16, ResNet50, and EfficientNetB0) for COVID-19 detection. A dataset of X-ray images (normal, non-COVID-19 pneumonia, and COVID-19) taken from COVID-19 image Data Collection was used to evaluate them. The overall accuracy of 90%, 94.30%, and 96.80% for the VGG16, ResNet50, and EfficientNetB0 were obtained.
Abraham and Nair [ 120 ] proposed a combined approach based on the combination of five multi-CNN models (Squeezenet, Darknet-53, MobilenetV2, Xception, and Shufflenet) for the automated detection of COVID-19 cases from X-ray images.
Ismael and Şengür [ 121 ] proposed three deep learning techniques for COVID-19 detection from chest X-ray images. The first technique was proposed based on five pre-trained deep CNN models (ResNet18, ResNet50, ResNet101, VGG16, and VGG19), the second deep learning model was proposed using CNN model with end-to-end training, the third and the last technique was proposed using pre-trained CNN models and SVM classifiers with various kernel functions. A dataset of 380 chest X-ray images (180 with COVID-19 and 200 normal (healthy)) was used for validation experimentation and results showed the efficiency of CNN techniques compared to various local texture descriptors.
Goel et al. [ 122 ] proposed an optimized convolutional neural network model, called OptCoNet, for COVID-19 diagnosis. A dataset of 2700 X-ray images (900 with COVID-19, 900 normal, and 900 with pneumonia) was employed to assess the performance of OptCoNet and results showed is effectiveness by providing accuracy, precision, sensitivity, specificity, and F1-score values of 97.78%, 92.88%, 97.75%, 96.25%, and 95.25%, respectively.
Bahel and Pillali [ 123 ] proposed five deep CNN models (InceptionV4, VGG 19, ResNetV2-152, and DenseNet) for detecting COVID-19 from chest X-Ray images. These techniques were evaluated based on a dataset of 300 chest x-ray images of infected and uninfected patients. Heat map filter was used on the images for helping the CNN models to perform better. Simulation results showed that DenseNet outperforms other deep CNN models such as InceptionV4, VGG19, and ResNetV2-152.
Sitaula and Hossain [ 124 ] proposed a novel deep learning model based on VGG-16 with the attention module for COVID-19 detection and classification. Authors conducted extensive experiments based on three X-ray image datasets D1 (Covid-19, No findings, and Pneumonia), D2 (Covid, Normal, Pneumonia Bacteria, Pneumonia Viral), and D3 (Covid, Normal, No findings, Pneumonia Bacteria, and Pneumonia Viral) to test this technique. Experimental results revealed the stable and promising performance compared to the state-of-the-art models by obtaining an accuracy of 79.58%, 85.43%, and 87.49% in D1, D2, and D3, respectively.
Jain et al. [ 125 ] proposed three CNN models (Inception V3, Xception, and ResNeXt) for COVID-19 detection and analysis. 6432 chest x-ray images divided into two classes including training set (5467) and validation set (965) were used to analyze the approaches performance. Simulation results showed that Xception model gives the highest accuracy with 97.97% as compared to other existing models.
Yasar and Ceylan [ 126 ] proposed a novel model based on CNN model with local binary pattern and dual-tree complex wavelet transform for COVID-19 detection on chest X-ray images. This approach was validated using two datasets of X-ray images: i) dataset of 230 images (150 with Covid-19 and 80 normal) and ii) dataset of 476 images (150 with Covid-19 and 326 normal). Experimental results showed that the proposed model gives good performance by achieving an accuracy, sensitivity, specificity, F1-score, and AUC of 98.43%, 99.47%, 98%, 98.81%, and 99.90%, respectively for the first dataset. For the second dataset, the proposed model achieves an accuracy, sensitivity, specificity, F1-score and, AUC of 98.91%, 99.20%, 99.39%, 98.28%, and 99.91%, respectively.
Khalifa et al. [ 127 ] proposed a new approach based on three deep learning models (Resnet50, Shufflenet, and Mobilenet) and GAN for detecting COVID-19 in CT chest Medical Images. In a similar work, Mukherjee et al. [ 128 ] proposed a lightweight (9 layered) CNN-tailored deep neural network model. It was demonstrated that the proposed model outperforms InceptionV3.
Hira et al. [ 142 ] used nine CNN models (AlexNet, GoogleNet, ResNet50, SeResNet50, DenseNet121, InceptionV4, InceptionResNetV2, ResNeXt50, and SeResNeXt50) for the detection of COVID–19 disease. The efficiency of the proposed models was validated using four scenarios: (i) two classes (224 with COVID–19 and 504 Normal); (ii) three classes (224 with COVID–19, 504 Normal, and 700 with bacterial Pneumonia); (iii) three classes (224 with COVID-19, 504 Normal, and 714 with bacterial and viral Pneumonia) and (iv) four classes (1346 normal, 1345 viral pneumonia, 2358 bacteria pneumonia, and with 183 COVID-19). Experimental results demonstrated that SeResNeXt50 outperforms other methods in terms of accuracy, precision, sensitivity, specificity, and F1-score.
Jelodar et al. [ 147 ] proposed a novel model based on LSTM with natural language process (NLP) for COVID-19 cases classification. The effectiveness of the proposed model was validated using a dataset of 563,079 COVID-19-related comments collected from the Kaggle website (between January 20, 2020 and March 19, 2020) and results showed its efficiency and robustness on this problem area to guide related decision-making.
Chimmula et al. [ 148 ] used LSTM model for forecasting of COVID-19 cases in Canada. The performance of LSTM was validated using data collected from Johns Hopkins University and Canadian Health Authority with several confirmed cases and results showed that the LSTM model achieves better performance when compared with other forecasting models.
Jiang et al. [ 149 ] developed a novel model, called BiGRU-AT, based on bidirectional GRU with an attention mechanism for COVID-19 detection and diagnosis. The performance of BiGRU-AT was assessed using breathing and thermal data extracted from people wearing masks. Simulation results showed that BiGRU-AT achieves an accuracy, sensitivity, specificity, and F1-score of 83.69%, 90.23%, 76.31%, and 84.61%, respectively.
Mohammed et al. [ 150 ] proposed LSTM with ResNext+ and slice attention module for COVID-19 detection. A total of of 302 CT volumes (20 with confirmed COVID19 and 282 normal) was used for testing and training the proposed model. According to the results, the proposed model provides an accuracy of 77.60%, precision of 81.90%, sensitivity of 85.50%, specificity of 79.30%, and F1-score of f 81.40%.
Islam et al. [ 151 ] introduced a novel model based on the hybridization of LSTM with CNN for automatic diagnosis of COVID-19 cases. The effectiveness of the hybrid model was validated using a dataset of 4575 X-ray images (1525 images with COVID-19, 1525 with viral pneumonia, and 1525 normal). Simulation results showed that the hybrid model outperforms other existing models by achieving an accuracy, sensitivity, specificity, and F1-score of 99.20%, 99.30%, 99.20%, and 98.90%, respectively.
Aslan et al. [ 152 ] proposed a hybrid approach based on the hybridization of Bidirectional LSTM (BiLSTM) with CNN Transfer Learning (mAlexNet) for COVID-19 detection. A dataset of 2905 X-ray images (219 with COVID-19, 1345 with viral pneumonia, and 1341 normal) was used in the simulation and results showed that the hybrid approach outperforms mAlexNet model by giving an accuracy, precision, sensitivity, specificity, F1-score, and AUC of 98.70%, 98.77%, 98.76%, 99.33%, 98.76%, and 99%, respectively (Tables (Tables2, 2 , ,3, 3 , ,4, 4 , ,5, 5 , ,6 6 ).
Specialized CNN Approaches for COVID–19
Song et al. [ 155 ] developed a deep-learning model, called Details Relation Extraction neural Network (DRE-Net), for accurate identification of COVID-19-infected patients. 275 chest scan images (86 normal, 88 with COVID-19, and 101 with bacteria pneumonia) were used to validate the performance of DRE-Net. Simulation results showed that DRE-Net can identify COVID-19 infected patients with an average accuracy of 94%, AUC of 99%, and sensitivity of 93%.
Li et al. [ 156 ] proposed a deep learning method, called COVNet, for COVID-19 diagnosis from CT scan images. A dataset of 4356 chest CT images from 3222 patients collected from six hospitals between August 2016 and February 2020 was used in the simulation and results showed that the proposed COVNet achieves an AUC, sensitivity, and specificity of 96%, 90%, and 96%, respectively. Zheng et al. conducted a similar study [ 157 ] by proposing a 3D deep CNN model, called DeCoVNet, for detecting COVID-19 from 3D CT images.
Ucar and Korkmaz [ 158 ] proposed a novel and efficient Deep Bayes-SqueezeNet-based system (COVIDiagnosis-Net) for COVID-19 Diagnosis. A dataset of 5949 chest X-ray images including 1583 normal, 4290 pneumonia, and 76 COVID-19 infection cases was employed in the simulation and results showed that COVIDiagnosis-Net outperforms existing network models by achieving 98.26% of accuracy, 99.13% of specificity, and 98.25% of F1-score.
DarkCovidNet was proposed by Ozturk et al. [ 159 ] for automated detection of COVID-19. The efficiency of DarkCovidNet was evaluated using two datasets: i) A COVID-19 X-ray image database developed by Cohen JP [ 84 ] and ii) ChestX-ray8 database provided by Wang et al. [ 160 ]. Simulation results showed that DarkCovidNet gives accurate diagnostics of 98.08% and 87.02% for binary classification (COVID vs. No-Findings) and multi-class classification (COVID vs. No-Findings vs. Pneumonia), respectively.
Wang and Wong [ 161 ] proposed a deep learning model, called Covid-Net, for detecting COVID-19 Cases from Chest X-Ray Images. Quantitative and qualitative results showed the efficiency and superiority of the proposed Covid-Net model compared to VGG-19 and ResNet-50 techniques.
In [ 162 ], Born et al. proposed POCOVID-Net for the automatic detection of COVID-19 cases. A lung ultrasound (POCUS) dataset consisting of 1103 images (654 COVID-19, 277 bacterial pneumonia, and 172 normal) sampled from 64 videos was used for evaluating the effectiveness of POCOVID-Net model. According to the results, POCOVID-Net model provides good performance with 0.89 accuracy, 0.88 precision, 0.96 sensitivity, 0.79 specificity, and 0.92 F1-score.
COVID-19Net was proposed by Wang et al. [ 163 ] for the diagnostic and prognostic analysis of COVID-19 cases in CT images. A dataset of chest CT images collected from six cities or provinces including Wuhan city in China was used for the simulation and results showed the good performance of COVID-19Net by achieving an AUC of 87%, an accuracy of 78.32%, a sensitivity of 80.39%, F1-score of 77%, and a specificity of 76.61%.
Khan et al. [ 164 ] proposed a new model (CoroNet) for COVID-19 detection and diagnosis. CoroNet was validated using three scenarios: i) 4-class CoroNet (normal, viral pneumonia, bacteria pneumonia, and COVID-19 images); ii) 3-class CoroNet (COVID-19, Normal and Pneumonia); and iii) binary 2-class CoroNet (COVID-19, Normal and Pneumonia). Experimental results demonstrated the superiority of CoroNet compared to some studies in the literature by achieving an accuracy of 89.5%, 94.59%, and 99% for 4-class, 3-class, and binary 2-class scenarios, respectively.
Mahmud et al. [ 165 ] proposed a novel multi-dilation deep CNN model (CovXNeT) based on depthwise dilated convolutions for automatic COVID-19 detection. Three datasets of 5856, 610, and 610 x-ray images were used for evaluating the effectiveness of CovXNeT. Experimental results revealed the performance of CovXNeT compared to other approaches in the literature by providing an accuracy of 98.1%, 95.1%, and 91.70% for the dataset of 5856 images, dataset of 610 images, and dataset of 610 images, respectively.
siddhartha and Santra [ 166 ] proposed a novel model, called COVIDLite, based on a depth-wise separable deep neural network (DSCNN) with white balance and CLAHE for the detection of COVID-19 cases. Two datasets of X-ray images: i)1458 images (429 COVID-19, 495 viral pneumonia, and 534 normal) and ii) 365 images (107 COVID-19, 124 viral pneumonia, and 134 normal) were used for testing the effectiveness of COVIDLite. Simulation results revealed that COVIDLite performs for both 2-class and 3-class scenario by achieving an accuracy of 99.58% and 96.43%, respectively.
Ahmed et al. [ 167 ] proposed a novel CNN model, called ReCoNet, for COVID-19 detection. The effectiveness of ReCoNet was evaluated based on COVIDx [ 161 ] and CheXpert [ 168 ] datasets containing 15.134 and 224.316 CXR images, respectively. Experimental results demonstrated that ReCoNet outperforms COVID-Net and other state-of-the-art techniques by yielding an accuracy, sensitivity, and specificity of 97.48%, 96.39%, and 97.53%, respectively.
Haghanifar et al. [ 169 ] developed a novel approach, called COVID-CXNET, based on the well-known CheXNet model for automatic detection of COVID-19 cases. The effectiveness of COVID-CXNET was tested using a dataset of 3,628 chest X-ray images (3,200 normal and 428 with COVID-19) divided into two classes including training set (80%)and validation set (20%). Experimental results showed that COVID-CXNET gives an accuracy of 99.04% and F1-score of 96%.
Turkoglu [ 170 ] proposed a COVIDetectioNet model with AlexNet and SVM for COVID-19 diagnosis and classification. A dataset of 6092 X-ray images (1583 Normal, 219 with COVID19, and 4290 with Pneumonia) collected from the Github and Kaggle databases was used in the experimentation. Simulation results demonstrated the better performance of COVIDetectioNet compared to other deep learning approaches by achieving an accuracy of 99.18%.
Tammina [ 171 ] proposed a novel deep learning approach, called CovidSORT for COVID-19 detection. 5910 Chest X-ray images collected from retrospective cohorts of pediatric Women patients and Children’s Medical Center of Guangzhou, China were used to validate the CovidSORT performance. Simulation results demonstrated that the CovidSORT model provides an accuracy of 96.83%, precision of 98.75%, sensitivity of 96.57%, and F1-score of 97.65%.
Al-Bawi et al. [ 172 ] developed an efficient model based on VGG with the convolutional COVID block (CCBlock) for the automatic diagnosis of COVID-19. To evaluate It, 1,828 x-ray images were used including 310 with COVID-19 cases, 864 with pneumonia, and 654 normal images. According to the results, the proposed model gives the highest diagnosis performance by achieving an accuracy of 98.52% and 95.34% for two and three classes, respectively.
Jamshidi et al. [ 181 ] used Generative Adversarial Network (GAN), Extreme Learning Machine (ELM), RNN, and LSTM for COVID–19 diagnosis and treatment. Sedik et al. [ 182 ] proposed a combined model based on GAN with CNN and ConvLSTM for COVID–19 infection detection. Two datasets of X-ray and CT images were used in the simulation and results showed the effectiveness and performance of the combined model by achieving 99% of accuracy, 97.70% of precision, 100% of sensitivity, 97.80% of specificity, and 99% of F1-score.
Other Deep Learning Approaches
Farid et al. [ 184 ] proposed a Stack Hybrid Model, called Composite Hybrid Feature Selection Model (CHFS), based on the hybridization of CNN and machine learning approaches for early diagnosis of covid19. The performance of CHFS was evaluated based on a dataset containing 51 CT images divided into training and testing sets. Simulation results showed that CHFS achieves an F1-score, precision, sensitivity, accuracy of 96.10%, 96.10%, 96.10%, and 96.07%, respectively.
Hwang et al. [ 185 ] implemented a Deep Learning-Based Computer-Aided Detection (CAD) System for the identification of COVID-19 infected patients. CAD system was trained based on chest X-ray and CT images and results showed that CAD system achieves 68.80% of sensitivity, 66.70% of specificity with chest X-ray images and 81.5% of sensitivity, 72.3% of specificity with CT images.
Amyar et al. [ 186 ] proposed a multi-task deep learning approach for COVID-19 detection and classification from CT images. A dataset of images collected from 1369 patients (449 with COVID-19, 425 normal, 98 with lung cancer, and 397 of different kinds of pathology) was used to evaluate the performance of the proposed approach. Results showed that the proposed approach achieves an AUC of 0.97, an accuracy of 94.67, a sensitivity of 0.96, and a specificity of 0.92.
For COVID-19 pneumonia diagnosis, Ko et al. [ 187 ] proposed fast-track COVID-19 classification network (FCONet), which uses as backbone one of the pre-trained deep learning models (VGG16, ResNet50, Inceptionv3, or Xception). A set of 3993 chest CT images divided into training and test classes were used to evaluate the performance of the proposed FCONet. Experimental results demonstrated that FCONet with ResNet50 gives excellent diagnostic performance by achieving a sensitivity of 99.58%, specificity 100%, accuracy 99.87%, and AUC of 100%.
Basu and Mitra [ 188 ] proposed a domain extension transfer learning (DETL) with three pre-trained deep CNN models (AlexNet, VGGNet, and ResNet) for COVID-19 screening. 1207 X-ray images (350 normal, 322 with pneumonia, 305 with COVID-19, and 300 other diseases) were employed to validate the proposed model. Experimental results showed that DETL with VGGNet gives a better accuracy of 90.13%.
Elghamrawy [ 189 ] developed a new approach (DLBD-COV) based on H2O’s Deep-Learning-inspired model with Big Data analytic for COVID-19 detection. The efficiency of DLBD-COV was validated based on CT images collected from [ 84 ] and X-ray images collected from [ 190 ] taking into account five metrics such as accuracy, precision, Sensitivity, and computational time. Simulation results showed that DLBD-COV provides a superior accuracy compared to other CNN models such as DeConNet and ResNet+.
Sharma et al. [ 191 ] proposed an deep learning model for rapid identifying and screening of COVID-19 patients. The efficiency of the proposed model was validated using chest X-ray images of adult COVID-19 patients (COVID-19, non-COVID-19, pneumonia, and tuberculosis images) and results showed its efficiency compared to previously published methods.
Hammam et al. [ 192 ] proposed a stacked ensemble deep learning model for COVID-19 vision diagnosis. The efficiency of the proposed model was validated using a dataset of 500 X-ray images divided into three classes including the training set (80%), validation set (10%), and testing set (10%). Simulation results showed the superior performance of the proposed model compared to any other single model by achieving 98.60% test accuracy. A similar work was done by Mohammed et al. [ 193 ], in which a Corner-based Weber Local Descriptor (CWLD) was prpoposed for diagnosis of COVID-19 from chest X-Ray images.
Li et al. [ 194 ] proposed a stacked auto-encoder detector model for the diagnosis of COVID-19 Cases on CT scan images. Authors used in their experimentation a dataset of 470 CT images (275 with COVID-19 and 195 normal) collected from UC San Diego. According to the results, the proposed model performs well and achieves an average accuracy of 94.70%, precision of 96.54%, sensitivity of 94.10%, and F1-score of 94.80%. Al-antari et al. [ 195 ] introduced a novel model (CAD-based YOLO Predictor) based on fast deep learning computer-aided diagnosis system with YOLO predictor for automatic diagnosis of COVID-19 cases from digital X-ray images. The proposed system was trained using two different digital X-ray datasets: COVID-19 images [ 84 , 88 ] and ChestX-ray8 images [ 196 ]. According to the experimentation, CAD-based YOLO Predictor achieves an accuracy of 97.40%, sensitivity of 85.15%, specificity of 99.06%, and F1-score of 84.81%.
Gianchandani et al. [ 197 ] proposed two ensemble deep transfer learning models for Rapid COVID-19 diagnosis. The proposed models were validated using two datasets of X-ray images obtained from Kaggle datasets resource [ 198 ] and the University of Dhaka and Qatar University. [ 88 ]
Other Machine Learning Approaches
Chakraborty and Ghosh [ 204 ] developed a hybrid method (ARIMA–WBF) based on the hybridization of ARIMA model and Wavelet-based forecasting (WBF) model for predicting the number of daily confirmed COVID-19 cases. The effectiveness of ARIMA-WBF was validated using datasets of 346 cases taken from five countries (70: Canada, 71: France, 64: India, 76: South Korea, and 65: UK). Simulation results showed the performance and robustness of ARIMA-WBF in the prediction of COVID-19 cases.
Tuncer et al. [ 205 ] proposed a feature generation technique, called Residual Exemplar Local Binary Pattern (ResExLBP) with iterative ReliefF (IRF) and five machine learning methods (Decision tree, linear discriminant, SVM, kNN, and subspace discriminant) for automatic COVID-19 detection. The efficiency of the proposed model was validated using datasets of X-ray images collected from the GitHub website and Kaggle site. Simulation results showed that ResExLBP with IRF and SVM gives better performance compared to other models by providing 99.69% accuracy, 98.85% sensitivity, and 100% specificity.
Tuli et al. [ 206 ] developed a novel model based on machine learning and Cloud Computing for real-time prediction of COVID-19. The effectiveness of the proposed model was validated using 2Our World In Data (COVID-19 Dataset) taken from the Github repository ( https://github.com/owid/covid-19-data/tree/master/public/data/ ). Simulation results showed that the proposed model gives good performance on this problem area.
Pereira et al. [ 207 ]used MLP with KNN, SVM, Decision Trees, and Random Forest for COVID-19 identification in chest X-ray images. The efficiency of the proposed models was evaluated based on RYDLS-20 database of 1144 chest X-ray images divided into training and test sets with 70% and 30% rates. Experimental results showed the superiority of MLP compared to other machine learning approaches by providing an F1-Score of 89%.
Albahri et al. [ 208 ] used a machine learning model combined with a novel Multi-criteria-decision-method (MCDM) for the identification of COVID-19 infected patients. The effectiveness of the proposed model was evaluated based on Blood sample images. Simulation results revealed that the proposed model is a good tool for identifying infected COVID-19 cases.
Wang et al. [ 209 ] developed a hybrid model based on FbProphet technique and Logistic Model for COVID-19 epidemic trend prediction. The hybrid model was validated using COVID-19 epidemiological time-series data and results revealed the effectiveness of the hybrid model for the prediction of the turning point and epidemic size of COVID-19.
Ardakani et al. [ 210 ] proposed a machine learning-based Computer-Aided Detection (CAD) System (COVIDiag) for COVID-19 diagnosis. The performance of COVIDiag was evaluated using CT images of 612 patients (306 with COVID-19 and 306 normal). Experimental results demonstrated the effectiveness of COVIDiag compared to SVM, KNN, NB, and DT by achieving the sensitivity, specificity, and accuracy of 93.54%, 90.32%, and 91.94%, respectively.
The summary of other Machine Learning approaches is given in Table Table7 7 .
Summary of other Machine Learning approaches for detection, diagnosis, and prediction of COVID-19 cases
Machine Learning is the field of AI that has been applied to deal with COVID-19. The finding from this study reveals that:

Approaches of machine learning used to deal with COVID-19
- Techniques basically known in the field of Unsupervised Learning did not appear in the reviewed papers. However, in case of unlabeled data, deep Learning makes an automatic learning which is a form of an unsupervised learning;
- Similarly, techniques of Reinforcement Learning are not explored in the summarized approaches;

Deep learning approaches used to deal with COVID-19

Supervised learning techniques used to deal with COVID-19

Metrics used in the evaluation of COVID-19 related approaches
Despite all these contributions, there are still some remaining challenges in applying ML to deal with COVID-19. Actually, handling new datasets generated in real time is facing several issues limiting the efficiency of results. In fact, many of the proposed approaches are based on small datasets. They are, in most cases, incomplete, noisy, ambiguous and with a significant ratio of missing patterns. Consequently, the training is not efficient and the risk of over-fitting is high because of the high variance and errors on the test set. Therefore, the need to build large datasets becomes unavoidable. However, it is not sufficient. In fact, without a complete and standard dataset, it is difficult to conclude which method provides the best results. To overcome that, a deep work of merging existing datasets and cleaning them up, by removing / imputing missing data and removing redundancy, is required.
The COVID-19 pandemic has deeply marked the year 2020 and has made the researchers community in different fields react. This paper demonstrated the interest attached by data scientists to this particular situation. It provided a survey of Machine Learning based research classified into two categories (Supervised Learning approaches and Deep Learning approaches) to make detection, diagnosis, or prediction of the COVID-19. Moreover, it gave an analysis and statistics on published works. The review included more than 160 publications coming from more than 6 famous scientific publishers. The learning is based on various data supports such as X-Ray images, CT images, Text data, Time series, Sounds, Coughing/Breathing videos, and Blood Samples. Our study presented a synthesis with accurate ratios of use of each of the ML techniques. Also, it summarized the metrics employed to validate the different models. The statistical study showed that 6 metrics are frequently used with favor to accuracy, sensitivity, and specificity which are evaluated in almost equal proportions. Among the ML techniques, it is shown that 79% of them are based on Deep Learning. In 65% of cases, CNN architecture was used. However, 17% of the reviewed papers proposed a Specialize CNN architecture adapted to COVID-19. Supervised Learning is also present in 16% of cases either to make classification by using mainly SVM or to make regression where Random Forest Algorithms and Linear regression are the most dominant techniques. In addition of them, hybrid approaches are also explored to address the topic of COVID-19. They represent 5% of the reviewed methods in this paper. Most of them mix CNN with other techniques and/or meta-heuristics in order to outperform the classical ones. They demonstrated good performance in terms of accuracy and F1-Score, thus, it would be worth investigating them further. Given this state of the art and the number of techniques proposed, research must now focus on the quality of the data used and their harmonization. Indeed, until now, the studies carried out have been based on different types of datasets and different volumes of datasets. The data considered are overall those present in each country where the disease of COVID-19 has not necessarily evolved in the same way. Thus, it is essential to create benchmarks with real-world datasets to train future models on them.
Declaration
The authors declare that there is no conflict of interest with any person(s) or Organization(s).
1 https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports .
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yassine Meraihi, Email: [email protected] .
Asma Benmessaoud Gabis, Email: zd.ise@duoassemneb_a .
Seyedali Mirjalili, Email: [email protected] .
Amar Ramdane-Cherif, Email: rf.qsvu.vsil@acr .
Fawaz E. Alsaadi, Email: moc.liamg@uakzawaf .
Help | Advanced Search
Computer Science > Machine Learning
Title: application of machine learning in agriculture: recent trends and future research avenues.
Abstract: Food production is a vital global concern and the potential for an agritech revolution through artificial intelligence (AI) remains largely unexplored. This paper presents a comprehensive review focused on the application of machine learning (ML) in agriculture, aiming to explore its transformative potential in farming practices and efficiency enhancement. To understand the extent of research activity in this field, statistical data have been gathered, revealing a substantial growth trend in recent years. This indicates that it stands out as one of the most dynamic and vibrant research domains. By introducing the concept of ML and delving into the realm of smart agriculture, including Precision Agriculture, Smart Farming, Digital Agriculture, and Agriculture 4.0, we investigate how AI can optimize crop output and minimize environmental impact. We highlight the capacity of ML to analyze and classify agricultural data, providing examples of improved productivity and profitability on farms. Furthermore, we discuss prominent ML models and their unique features that have shown promising results in agricultural applications. Through a systematic review of the literature, this paper addresses the existing literature gap on AI in agriculture and offers valuable information to newcomers and researchers. By shedding light on unexplored areas within this emerging field, our objective is to facilitate a deeper understanding of the significant contributions and potential of AI in agriculture, ultimately benefiting the research community.
Submission history
Access paper:.
- HTML (experimental)
- Other Formats
References & Citations
- Google Scholar
- Semantic Scholar
BibTeX formatted citation
Bibliographic and Citation Tools
Code, data and media associated with this article, recommenders and search tools.
- Institution
arXivLabs: experimental projects with community collaborators
arXivLabs is a framework that allows collaborators to develop and share new arXiv features directly on our website.
Both individuals and organizations that work with arXivLabs have embraced and accepted our values of openness, community, excellence, and user data privacy. arXiv is committed to these values and only works with partners that adhere to them.
Have an idea for a project that will add value for arXiv's community? Learn more about arXivLabs .

Frequently Asked Questions
JMLR Papers
Select a volume number to see its table of contents with links to the papers.
Volume 25 (January 2024 - Present)
Volume 24 (January 2023 - December 2023)
Volume 23 (January 2022 - December 2022)
Volume 22 (January 2021 - December 2021)
Volume 21 (January 2020 - December 2020)
Volume 20 (January 2019 - December 2019)
Volume 19 (August 2018 - December 2018)
Volume 18 (February 2017 - August 2018)
Volume 17 (January 2016 - January 2017)
Volume 16 (January 2015 - December 2015)
Volume 15 (January 2014 - December 2014)
Volume 14 (January 2013 - December 2013)
Volume 13 (January 2012 - December 2012)
Volume 12 (January 2011 - December 2011)
Volume 11 (January 2010 - December 2010)
Volume 10 (January 2009 - December 2009)
Volume 9 (January 2008 - December 2008)
Volume 8 (January 2007 - December 2007)
Volume 7 (January 2006 - December 2006)
Volume 6 (January 2005 - December 2005)
Volume 5 (December 2003 - December 2004)
Volume 4 (Apr 2003 - December 2003)
Volume 3 (Jul 2002 - Mar 2003)
Volume 2 (Oct 2001 - Mar 2002)
Volume 1 (Oct 2000 - Sep 2001)
Special Topics
Bayesian Optimization
Learning from Electronic Health Data (December 2016)
Gesture Recognition (May 2012 - present)
Large Scale Learning (Jul 2009 - present)
Mining and Learning with Graphs and Relations (February 2009 - present)
Grammar Induction, Representation of Language and Language Learning (Nov 2010 - Apr 2011)
Causality (Sep 2007 - May 2010)
Model Selection (Apr 2007 - Jul 2010)
Conference on Learning Theory 2005 (February 2007 - Jul 2007)
Machine Learning for Computer Security (December 2006)
Machine Learning and Large Scale Optimization (Jul 2006 - Oct 2006)
Approaches and Applications of Inductive Programming (February 2006 - Mar 2006)
Learning Theory (Jun 2004 - Aug 2004)
Special Issues
In Memory of Alexey Chervonenkis (Sep 2015)
Independent Components Analysis (December 2003)
Learning Theory (Oct 2003)
Inductive Logic Programming (Aug 2003)
Fusion of Domain Knowledge with Data for Decision Support (Jul 2003)
Variable and Feature Selection (Mar 2003)
Machine Learning Methods for Text and Images (February 2003)
Eighteenth International Conference on Machine Learning (ICML2001) (December 2002)
Computational Learning Theory (Nov 2002)
Shallow Parsing (Mar 2002)
Kernel Methods (December 2001)
Federated learning in Emotion Recognition Systems based on physiological signals for privacy preservation: a review
- Published: 03 June 2024
Cite this article

- Neha Gahlan ORCID: orcid.org/0000-0002-2694-7468 1 &
- Divyashikha Sethia 1
Automated Emotion Recognition Systems (ERS) with physiological signals help improve health and decision-making in everyday life. It uses traditional Machine Learning (ML) methods, requiring high-quality learning models for physiological data (sensitive information). However, automated ERS enables data attacks and leaks, significantly losing user privacy and integrity. This privacy problem can be solved using a novel Federated Learning (FL) approach, which enables distributed machine learning model training. This review examines 192 papers focusing on emotion recognition via physiological signals and FL. It is the first review article concerning the privacy of sensitive physiological data for an ERS. The paper reviews the different emotions, benchmark datasets, machine learning, and federated learning approaches for classifying emotions. It proposes a novel multi-modal Federated Learning for Physiological signals based on Emotion Recognition Systems (Fed-PhyERS) architecture, experimenting with the AMIGOS dataset and its applications for a next-generation automated ERS. Based on critical analysis, this paper provides the key takeaways, identifies the limitations, and proposes future research directions to address gaps in previous studies. Moreover, it reviews ethical considerations related to implementing the proposed architecture. This review paper aims to provide readers with a comprehensive insight into the current trends, architectures, and techniques utilized within the field.
This is a preview of subscription content, log in via an institution to check access.
Access this article
Price includes VAT (Russian Federation)
Instant access to the full article PDF.
Rent this article via DeepDyve
Institutional subscriptions

Data Availability
Data sharing does not apply to this article as no datasets were generated or analyzed during the current study. The datasets mentioned in the current study are cited with their base papers.
https://tinyurl.com/4dcp62es
https://www.tensorflow.org/federated
http://www.eecs.qmul.ac.uk/mmv/datasets/amigos/index.html
https://living.ai/emo/
https://uxplanet.org/designing-emotional-ui-b11fa0fda5c
Schmidt et al (2018) Wearable Affect and Stress Recognition: A Review. arXiv preprint arXiv:1811.08854 . https://doi.org/10.48550/arXiv.1811.08854
Peng et al (2015) A mixed bag of emotions: Model, predict, and transfer emotion distributions supplementary material. https://doi.org/https://shorturl.at/mI146
Shikha et al (2022) HRV and GSR as Viable Physiological Markers for Mental Health Recognition. In: 2022 14th International conference on communication systems & NETworkS (COMSNETS), pp 37–42. https://doi.org/10.1109/COMSNETS53615.2022.9668439 . IEEE
Shikha et al (2021) Stacked Sparse Autoencoder and Machine Learning Based Anxiety Classification using EEG Signals. In: The first international conference on AI-ML-Systems, pp 1–7. https://doi.org/10.1145/3486001.3486227
De Nadai et al (2016) Enhancing Safety of Transport by Road by On-line Monitoring of Driver Emotions. In: 2016 11th System of systems engineering conference (SoSE), pp 1–4. https://doi.org/10.1109/SYSOSE.2016.7542941 . Ieee
Alipour-Vaezi M, Aghsami A, Rabbani M (2022) Introducing a novel revenue-sharing contract in media supply chain management using data mining and multi-criteria decision-making methods. Soft Comput 26(6):2883–2900. https://doi.org/10.1007/s00500-021-06609-0
Article Google Scholar
Verschuere et al (2006) Psychopathy and Physiological Detection of Concealed Information: A Review. Psychologica Belgica 46(1–2)
Guo et al (2013) Pervasive and Unobtrusive Emotion Sensing for Human Mental Health. In: 2013 7th International conference on pervasive computing technologies for healthcare and workshops, pp 436–439. IEEE
Mao et al (2014) Learning Salient Features for Speech Emotion Recognition Using Convolutional Neural Networks. IEEE Trans Multimed 16(8):2203–2213. https://doi.org/10.1109/TMM.2014.2360798
Zhang et al (2016) Facial Emotion Recognition Based on Biorthogonal Wavelet Entropy, Fuzzy Support Vector Machine, and stratified cross validation. IEEE Access 4:8375–8385. https://doi.org/10.1109/ACCESS.2016.2628407
Li et al (2017) Emotion Recognition from EEG using RASM and LSTM. In: International conference on internet multimedia computing and service, pp 310–318. https://doi.org/10.1007/978-981-10-8530-7_30 . Springer
Valenza et al (2014) Revealing Real-Time Emotional Responses: A Personalized Assessment Based on Heartbeat Dynamics. Sci Rep 4(1):1–13. https://doi.org/10.1038/srep04998
Benedek M, Kaernbach C (2010) A Continuous Measure of Phasic Electrodermal Activity. J Neurosci Methods 190(1):80–91. https://doi.org/10.1016/j.jneumeth.2010.04.028
Peter et al (2005) A Wearable Multi-Sensor System for Mobile Acquisition of Emotion-Related Physiological Data. In: International conference on affective computing and intelligent interaction, pp 691–698. https://doi.org/10.1007/11573548_89 . Springer
Krumova et al (2008) Long-Term Skin Temperature Measurements-A Practical Diagnostic Tool in Complex Regional Pain Syndrome. Pain 140(1):8–22. https://doi.org/10.1016/j.pain.2008.07.003
Sharma et al (2021) A computerized approach for automatic human emotion recognition using sliding mode singular spectrum analysis. IEEE Sensors J 21(23):26931–26940. https://doi.org/10.1109/JSEN.2021.3120787
Hasnul MA, Ab Aziz NA, Abd Aziz A (2023) Augmenting ECG data with multiple filters for a better emotion recognition system. Arab J Sci Eng 1–22. https://doi.org/10.1007/s13369-022-07585-9
Younis EM, Zaki SM, Kanjo E, Houssein EH (2022) Evaluating ensemble learning methods for multi-modal emotion recognition using sensor data fusion. Sensors 22(15):5611. https://doi.org/10.3390/s22155611
Dar et al (2020) CNN and LSTM-Based Emotion Charting using Physiological Signals. Sensors 20(16):4551. https://doi.org/10.3390/s20164551
Yang et al (2018) Emotion Recognition from Multi-Channel EEG Through Parallel Convolutional Recurrent Neural Network. In: 2018 International joint conference on neural networks (IJCNN), pp 1–7. https://doi.org/10.1109/IJCNN.2018.8489331 . IEEE
Tang et al (2017) Multimodal Emotion Recognition Using Deep Neural Networks. In: International conference on neural information processing, pp 811–819. https://doi.org/10.1007/978-3-319-70093-9_86 . Springer
Khateeb et al (2021) Multi-Domain Feature Fusion for Emotion Classification Using DEAP Dataset. IEEE Access 9:12134–12142. https://doi.org/10.1109/ACCESS.2021.3051281
Galvão et al (2021) Predicting Exact Valence and Arousal Values from EEG. Sensors 21(10):3414. https://doi.org/10.3390/s21103414
Liakopoulos et al (2021) CNN-Based Stress and Emotion Recognition in Ambulatory Settings. In: 2021 12th International conference on information, intelligence, systems & applications (IISA), pp 1–8. https://doi.org/10.1109/IISA52424.2021.9555508 . IEEE
Bhatti A, Behinaein B, Hungler P, Etemad A (2022) Attx: Attentive cross-connections for fusion of wearable signals in emotion recognition. arXiv preprint arXiv:2206.04625 . https://doi.org/10.48550/arXiv.2206.04625
Deng et al (2021) SFE-Net: EEG-Based Emotion Recognition with Symmetrical Spatial Feature Extraction. In: Proceedings of the 29th ACM international conference on multimedia, pp 2391–2400. https://doi.org/10.1145/3474085.3475403
Tan C, Šarlija M, Kasabov N (2021) Neurosense: Short-term emotion recognition and understanding based on spiking neural network modelling of spatio-temporal eeg patterns. Neurocomputing 434:137–148. https://doi.org/10.1016/j.neucom.2020.12.098
Shu et al (2018) A Review of Emotion Recognition using Physiological Signals. Sensors 18(7):2074. https://doi.org/10.3390/s18072074
Egger et al (2019) Emotion Recognition from Physiological Signal Analysis: A Review. Electron Notes Theo Comput Sci 343:35–55. https://doi.org/10.1016/j.entcs.2019.04.009
Dzedzickis et al (2020) Human Emotion Recognition: Review of Sensors and Methods. Sensors 20(3):592. https://doi.org/10.3390/s20030592
Saganowski et al (2020) Emotion Recognition using Wearables: A Systematic Literature Review-Work-in-Progress. In: 2020 IEEE International conference on pervasive computing and communications workshops (PerCom workshops), pp 1–6. https://doi.org/10.1109/PerComWorkshops48775.2020.9156096 . IEEE
Saxena et al (2020) Emotion Recognition and Detection Methods: A Comprehensive Survey. J Artif Intell Syst 2(1):53–79. https://doi.org/10.33969/AIS.2020.21005
Saganowski S, Perz B, Polak A, Kazienko P (2022) Emotion recognition for everyday life using physiological signals from wearables: A systematic literature review. IEEE Trans Affect Comput. https://doi.org/10.1109/TAFFC.2022.3176135
Lin W, Li C (2023) Review of Studies on Emotion Recognition and Judgment Based on Physiological Signals. Appl Sci 13(4):2573. https://doi.org/10.3390/app13042573
Miranda-Correa et al (2018) Amigos: A Dataset for Affect, Personality and Mood Research on Individuals and Groups. IEEE Trans Affect Comput 12(2):479–493. https://doi.org/10.1109/TAFFC.2018.2884461
Wang et al (2020) A Review of Emotion Sensing: Categorization Models and Algorithms. Multimed Tools App 79(47):35553–35582. https://doi.org/10.1007/s11042-019-08328-z
Graver MR (2002) Cicero on the Emotions: Tusculan Disputations 3 and 4. https://doi.org/10.1016/j.pain.2008.07.003
Ekman P (1992) An Argument for Basic Emotions. Cognit Emot 6(3–4):169–200. https://doi.org/10.1080/02699939208411068
Plutchik R (1982) A Psychoevolutionary Theory of Emotions. Sage Publications. https://doi.org/10.1177/053901882021004003
Lang PJ (1995) The Emotion Probe: Studies of Motivation and Attention. Am Psycho 50(5):372. https://doi.org/10.1037/0003-066X.50.5.372
Russell JA (1979) Affective space is bipolar. J Personality Soc Psycho 37(3):345. https://doi.org/10.1037/0022-3514.37.3.345
Article MathSciNet Google Scholar
Yang Y-H, Chen HH (2012) Machine Recognition of Music Emotion: A Review. ACM Trans Intell Syst Techno (TIST) 3(3):1–30. https://doi.org/10.1145/2168752.2168754
Russell JA, Mehrabian A (1977) Evidence for a three-factor theory of emotions. J Res Personality 11(3):273–294. https://doi.org/10.1016/0092-6566(77)90037-X
Mehrabian A, Russell JA (1974) An approach to environmental psychology
Bălan et al (2019) Emotion Classification Based on Biophysical Signals and Machine Learning Techniques. Symmetry 12(1):21. https://doi.org/10.3390/sym12010021
Abo-Zahhad et al (2015) A New EEG Acquisition Protocol for Biometric Identification Using Eye Blinking Signals. Int J Intell Syst App 7(6):48. https://doi.org/10.5815/ijisa.2015.06.05
Brain Lobes (2022). https://qbi.uq.edu.au/brain/brain-anatomy/lobes-brain . Accessed 7 Aug 2022
Sandler et al (2016) Positive Emotional Experience: Induced by Vibroacoustic Stimulation Using a Body Monochord in Patients with Psychosomatic Disorders: Is Associated with an Increase in EEG-theta and a Decrease in EEG-Alpha Power. Brain Topograph 29(4):524–538. https://doi.org/10.1007/s10548-016-0480-8
Wang J, Song Y, Gao Q, Mao Z (2023) Functional brain network based multi-domain feature fusion of hearing-Impaired EEG emotion identification. Biomed Signal Process Control 85:105013. https://doi.org/10.1016/j.bspc.2023.105013
Guo W, Xu G, Wang Y (2023) Multi-source domain adaptation with spatio-temporal feature extractor for EEG emotion recognition. Biomed Signal Process Control 84:104998. https://doi.org/10.1016/j.bspc.2023.104998
Purnomo et al (2009) Analysis the Dominant Location of Brain Activity in Frontal Lobe using K-Means method. In: International conference on instrumentation, communication, information technology, and biomedical engineering 2009, pp 1–3. IEEE
Lin et al (2007) Multilayer Perceptron for EEG Signal Classification during Listening to Emotional Music. In: TENCON 2007-2007 IEEE Region 10 Conference, pp 1–3. https://doi.org/10.1109/TENCON.2007.4428831 . IEEE
Guo W, Xu G, Wang Y (2022) Horizontal and vertical features fusion network based on different brain regions for emotion recognition. Knowl-Based Syst 247:108819. https://doi.org/10.1016/j.knosys.2022.108819
Saganowski et al (2020) Consumer Wearables and Affective Computing for Wellbeing Support. In: MobiQuitous 2020-17th EAI international conference on mobile and ubiquitous systems: computing, networking and services, pp 482–487. https://doi.org/10.1145/3448891.3450332
EMOTIVE EPOC+ (2022). https://www.emotiv.com/epoc/ . Accessed 7 Aug 2022
LaRocco et al (2020) A Systemic Review of Available Low-Cost EEG Headsets Used for Drowsiness Detection. Front Neuroinformatics 42. https://doi.org/10.3389/fninf.2020.553352
Berka et al (2004) Real-time Analysis of EEG Indexes of Alertness, Cognition, and Memory Acquired with a Wireless EEG Headset. Int J Human-Comput Interact 17(2):151–170. https://doi.org/10.1207/s15327590ijhc1702_3
VITALCONNECT (2022). https://vitalconnect.com/ . Accessed 7 Aug 2022
Polar H10 (2022). http://www.muscleoxygentraining.com/2021/03/polar-h10-ecg-tracing-short-how-to-guide.html . Accessed 7 Aug 2022
Garmin HRM-DUAL (2022). https://www.garmin.com/en-US/p/649059 . Accessed 7 Aug 2022
EMPATIC E4 (2022). https://www.empatica.com/en-gb/ . Accessed 7 Aug 2022
RING (2022). https://www.bitbrain.com/neurotechnology-products/biosignals-amplifier/ring . Accessed 7 Aug 2022
SHIMMER3 (2022). https://imotions.com/hardware/shimmer3-gsr/ . Accessed 7 Aug 2022
Samsung Gear live (2022). https://www.samsung.com/us/support/mobile/wearables/smartwatches/gear-live . Accessed 8 Aug 2022
Fitbit Charge 3 (2022). https://www.fitbit.com/global/us/technology/irregular-rhythm . Accessed 8 Aug 2022
Zheng et al (2017) Identifying Stable Patterns Over Time for Emotion Recognition From EEG. IEEE Trans Affect Comput 10(3):417–429. https://doi.org/10.1109/TAFFC.2017.2712143
Li et al (2018) Exploring EEG Features in Cross-Subject Emotion Recognition. Front Neurosci 12:162. https://doi.org/10.3389/fnins.2018.00162
Gao Y, Fu X, Ouyang T, Wang Y (2022) EEG-GCN: spatio-temporal and self-adaptive graph convolutional networks for single and multi-view EEG-based emotion recognition. IEEE Signal Process Lett 29:1574–1578. https://doi.org/10.1109/LSP.2022.3179946
Awan AW, Usman SM, Khalid S, Anwar A, Alroobaea R, Hussain S, Almotiri J, Ullah SS, Akram MU (2022) An Ensemble Learning Method for Emotion Charting Using Multimodal Physiological Signals. Sensors 22(23):9480. https://doi.org/10.3390/s22239480
Iyer A, Das SS, Teotia R, Maheshwari S, Sharma RR (2023) CNN and LSTM based ensemble learning for human emotion recognition using EEG recordings. Multimed Tools App 82(4):4883–4896. https://doi.org/10.1007/s11042-022-12310-7
Bagherzadeh S, Norouzi MR, Hampa SB, Ghasri A, Kouroshi PT, Hosseininasab S, Zadeh MAG, Nasrabadi AM (2024) A subject-independent portable emotion recognition system using synchrosqueezing wavelet transform maps of EEG signals and ResNet-18. Biomed Signal Process Control 90:105875. https://doi.org/10.1016/j.bspc.2023.105875
Bota et al (2019) A Review, Current Challenges, and Future Possibilities on Emotion Recognition Using Machine Learning and Physiological Signals. IEEE Access 7:140990–141020. https://doi.org/10.1109/ACCESS.2019.2944001
Dinde et al (2004) Human Emotion Recognition using Electrocardiogram Signals. Int J Recent Innov Trends Comput Commu 2(2):194–197
Google Scholar
Agrafioti et al (2011) ECG Pattern Analysis for Emotion Detection. IEEE Trans Affect Comput 3(1):102–115. https://doi.org/10.1109/T-AFFC.2011.28
Cheng et al (2017) A Novel ECG-Based Real-Time Detection Method of Negative Emotions in Wearable Applications. In: 2017 International conference on security, pattern analysis, and cybernetics (SPAC), pp 296–301. https://doi.org/10.1109/SPAC.2017.8304293 . IEEE
Tian et al (2018) RF-Based Fall Monitoring using Convolutional Neural Networks. Proc ACM Interact Mobile Wear Ubiquitous Techno 2(3):1–24. https://doi.org/10.1145/3264947
Ali et al (2018) A globally generalized emotion recognition system involving different physiological signals. Sensors 18(6):1905. https://doi.org/10.3390/s18061905
Hassani T (2021) Federated Emotion Recognition with Physiological Signals-GSR
Sharma et al (2019) A Dataset of Continuous Affect Annotations and Physiological Signals for Emotion Analysis. Sci Data 6(1):1–13. https://doi.org/10.1038/s41597-019-0209-0
Perry Fordson H, Xing X, Guo K, Xu X (2022) Emotion Recognition With Knowledge Graph Based on Electrodermal Activity. Front Neurosci 16:911767. https://doi.org/10.3389/fnins.2022.911767
Shukla J, Barreda-Angeles M, Oliver J, Nandi GC, Puig D (2019) Feature extraction and selection for emotion recognition from electrodermal activity. IEEE Trans Affect Comput 12(4):857–869. https://doi.org/10.1109/TAFFC.2019.2901673
Lisetti et al (2004) Using Noninvasive Wearable Computers to Recognize Human Emotions from Physiological Signals. EURASIP J Adv Signal Process 2004(11):1–16. https://doi.org/10.1155/S1110865704406192
Tarvainen et al (2014) Kubios HRV–Heart Rate Variability Analysis Software. 113(1):210–220. https://doi.org/10.1016/j.cmpb.2013.07.024
Guo et al (2016) Heart Rate Variability Signal Features for Emotion Recognition by using Principal Component Analysis and Support Vectors Machine. In: 2016 IEEE 16th International conference on bioinformatics and bioengineering (BIBE), pp 274–277. https://doi.org/10.1109/BIBE.2016.40 . IEEE
Costa et al (2017) EmotionCheck: A Wearable Device to Regulate Anxiety Through False Heart Rate Feedback. GetMobile: Mobile Comput Commu 21(2):22–25. https://doi.org/10.1145/3131214.3131222
Bota et al (2020) Emotion assessment using feature fusion and decision fusion classification based on physiological data: Are we there yet? Sensors 20(17):4723. https://doi.org/10.3390/s20174723
Castaneda et al (2018) A Review on Wearable Photoplethysmography Sensors and their Potential Future Applications in Health Care. Int J Biosens Bioelectron 4(4):195. https://doi.org/10.15406/ijbsbe.2018.04.00125
Bolanos et al (2006) Comparison of Heart Rate Variability Signal Features Derived from Electrocardiography and Photoplethysmography in Healthy Individuals. In: 2006 International conference of the IEEE engineering in medicine and biology society, pp 4289–4294. https://doi.org/10.1109/IEMBS.2006.260607 . IEEE
Cheang et al (2003) An Overview of Non-Contact Photoplethysmography. Dept. of Electron Electrical Engineering, Loughborough University, LE. 1(1)
Sonoda H, Tanaka, H (2021) Emotion estimation by acceleration pulse wave analysis. In: International symposium on affective science and engineering ISASE2021, pp 1–4. https://doi.org/10.5057/isase.2021-C000029 . Japan Society of Kansei Engineering
Fordson HP, Gardhouse K, Cicero N, Chikazoe J, Anderson A, Derosa E (2022) A Novel Deep Learning Based Emotion Recognition Approach to well Being from Fingertip Blood Volume Pulse. In: 2022 International conference on machine learning and cybernetics (ICMLC), pp 130–137. https://doi.org/10.1109/ICMLC56445.2022.9941301 . IEEE
Lebaka LN, Govarthan PK, Rani P, Ganapathy N, Ronickom A, Fredo J et al (2023) Automated Emotion Recognition System Using Blood Volume Pulse and XGBoost Learning. In: Healthcare transformation with informatics and artificial intelligence, pp 52–55. IOS Press, ???. https://doi.org/10.3233/SHTI230422
Folschweiller S, Sauer J-F (2021) Respiration-driven brain oscillations in emotional cognition. Front Neural Circ 15:761812. https://doi.org/10.3389/fncir.2021.761812
Siddiqui HUR, Zafar K, Saleem AA, Raza MA, Dudley S, Rustam F, Ashraf I (2023) Emotion classification using temporal and spectral features from IR-UWB-based respiration data. Multimed Tools App 82(12):18565–18583. https://doi.org/10.1007/s11042-022-14091-5
Rawn KP, Keller PS (2023) Child emotion lability is associated with within-task changes of autonomic activity during a mirror-tracing task. Psychophysiology 14354. https://doi.org/10.1111/psyp.14354
Vos et al (2012) The tell-tale: What do Heart Rate; Skin Temperature and Skin Conductance reveal about emotions of people with severe and profound intellectual disabilities? Res Dev Disabil 33(4):1117–1127. https://doi.org/10.1016/j.ridd.2012.02.006
Storey BD (2002) Computing Fourier Series and Power Spectrum with Matlab. TEX paper 660:661
Bos et al (2006) Eeg-based emotion recognition. Influ Vis Audit Stimuli 56(3):1–17
Rigas et al (2007) A User Independent, Biosignal Based, Emotion Recognition Method. In: International conference on user modeling, pp 314–318. Springer
Izard CE (2009) Emotion Theory and Research: Highlights, Unanswered Questions, and Emerging Issues. Ann Rev Psycho 60:1–25. https://doi.org/10.1146/annurev.psych.60.110707.163539
Xiefeng C, Wang Y, Dai S, Zhao P, Liu Q (2019) Heart sound signals can be used for emotion recognition. Sci Rep 9(1):6486. https://doi.org/10.1038/s41598-019-42826-2
Liu Z, Kong J, Qu M, Zhao G, Zhang C (2022) Progress in Data Acquisition of Wearable Sensors. Biosensors 12(10):889. https://doi.org/10.3390/bios12100889
Basu S, Bag A, Aftabuddin M, Mahadevappa M, Mukherjee J, Guha R (2016) Effects of emotion on physiological signals. In: 2016 IEEE annual india conference (INDICON), pp 1–6. https://doi.org/10.1109/INDICON.2016.7839091 . IEEE
Chatterjee A, Roy UK (2018) Non-Invasive Heart State Monitoring an Article on Latest PPG Processing. Biomed Pharmacol J 11(4):1885–1893. https://doi.org/10.13005/bpj/1561
Qi P, Chiaro D, Giampaolo F, Piccialli F (2023) A blockchain-based secure Internet of medical things framework for stress detection. Inf Sci 628:377–390. https://doi.org/10.1016/j.ins.2023.01.123
Valenza et al (2011) The Role of Nonlinear Dynamics in Affective Valence and Arousal Recognition. IEEE Trans Affect Comput 3(2):237–249. https://doi.org/10.1109/T-AFFC.2011.30
Alickovic et al (2015) The Effect of Denoising on Classification of ECG Signals. In: 2015 XXV International conference on information, communication and automation technologies (ICAT), pp 1–6. https://doi.org/10.1109/ICAT.2015.7340540 . IEEE
Bigirimana et al (2016) A Hybrid ICA-Wavelet Transform for Automated Artefact Removal in EEG-Based Emotion Recognition. In: 2016 IEEE International conference on systems, man, and cybernetics (SMC), pp 004429–004434. https://doi.org/10.1109/SMC.2016.7844928 . IEEE
Patel et al (2016) Suppression of Eye-Blink Associated Artifact Using Single Channel EEG Data by Combining Cross-Correlation with Empirical Mode Decomposition. IEEE Sensors J 16(18):6947–6954. https://doi.org/10.1109/JSEN.2016.2591580
Gao Q, Wang C-h, Wang Z, Song X-l, Dong E-z, Song Y (2020) EEG based emotion recognition using fusion feature extraction method. Multimed Tools App 79:27057–27074. https://doi.org/10.1007/s11042-020-09354-y
Rajpoot AS, Panicker MR et al (2022) Subject independent emotion recognition using EEG signals employing attention driven neural networks. Biomed Signal Process Control 75:103547. https://doi.org/10.1016/j.bspc.2022.103547
Gahlan N, Sethia D (2023) Three Dimensional Emotion State Classification based on EEG via Empirical Mode Decomposition. In: 2023 International conference on artificial intelligence and applications (ICAIA) alliance technology conference (ATCON-1), pp 1–6. https://doi.org/10.1109/ICAIA57370.2023.10169633 . IEEE
Jahankhani et al (2006) EEG Signal Classification using Wavelet Feature Extraction and Neural Networks. In: IEEE John Vincent Atanasoff 2006 international symposium on modern computing (JVA’06), pp 120–124. https://doi.org/10.1109/JVA.2006.17 . IEEE
Zhang et al (2008) Feature Extraction of EEG Signals using Power Spectral Entropy. In: 2008 International conference on biomedical engineering and informatics, vol 2, pp 435–439. https://doi.org/10.1109/BMEI.2008.254 . IEEE
Rubin et al (2016) Time, Frequency & Complexity Analysis for Recognizing Panic States from Physiologic Time-Series. In: PervasiveHealth, pp 81–88
Li et al (2018) Emotion Recognition of Human Physiological Signals Based on Recursive Quantitative Analysis. In: 2018 Tenth international conference on advanced computational intelligence (ICACI), pp 217–223. https://doi.org/10.1109/ICACI.2018.8377609 . IEEE
Xiong et al (2020) A Parallel Algorithm Framework for Feature Extraction of EEG Signals on MPI. Comput Math Methods Med 2020. https://doi.org/10.1155/2020/9812019
Nandi A, Xhafa F (2022) A Federated Learning Method for Real-Time Emotion State Classification from Multi-Modal Streaming. Methods. https://doi.org/10.1016/j.ymeth.2022.03.005
Nandi A, Xhafa F, Kumar R (2023) A Docker-based federated learning framework design and deployment for multi-modal data stream classification. Computing 1–35. https://doi.org/10.1007/s00607-023-01179-5
Topic et al (2021) Emotion Recognition Based on EEG Feature Maps Through Deep Learning Network. Eng Sci Tech Int J 24(6):1442–1454. https://doi.org/10.1016/j.jestch.2021.03.012
Sarkar et al (2020) Self-Supervised ECG Representation Learning for Emotion Recognition. IEEE Trans Affect Comput. https://doi.org/10.1109/TAFFC.2020.3014842
Martinez et al (2013) Learning Deep Physiological Models of Affect. IEEE Comput Intell Mag 8(2):20–33. https://doi.org/10.1109/MCI.2013.2247823
Zhang et al (2016) “BioVid Emo DB”: A Multimodal Database for Emotion Analyses Validated by Subjective Ratings. In: 2016 IEEE Symposium series on computational intelligence (SSCI), pp 1–6. https://doi.org/10.1109/SSCI.2016.7849931 . IEEE
Koelstra et al (2011) DEAP: A Database for Emotion Analysis; using Physiological Signals. IEEE Trans Affect Comput 3(1):18–31. https://doi.org/10.1109/T-AFFC.2011.15
Katsigiannis et al (2017) DREAMER: A Database for Emotion Recognition Through EEG and ECG Signals From Wireless Low-cost Off-the-Shelf Devices. IEEE J Biomed Health Informat 22(1):98–107. https://doi.org/10.1109/JBHI.2017.2688239
Gao et al (2019) HHHFL: Hierarchical Heterogeneous Horizontal Federated Learning for electroencephalography. arXiv preprint arXiv:1909.05784 . https://doi.org/10.48550/arXiv.1909.05784
Vyzas et al (1999) O ine and Online Recognition of Emotion Expression From Physiological Data. In: Workshop on emotion-based archetectures, at the third international conference on autonomous agents, Seattle, WA
Schneegass et al (2013) A Data Set of Real World Driving to Assess Driver Workload. In: Proceedings of the 5th international conference on automotive user interfaces and interactive vehicular applications, pp 150–157. https://doi.org/10.1145/2516540.2516561
Zheng et al (2015) Investigating critical frequency bands and channels for EEG-based emotion recognition with deep neural networks. IEEE Trans Auton Mental Dev 7(3):162–175. https://doi.org/10.1109/TAMD.2015.2431497
Schmidt et al (2018) Introducing WESAD, A Multimodal Dataset for Wearable Stress and Affect Detection. In: Proceedings of the 20th ACM international conference on multimodal interaction, pp 400–408. https://doi.org/10.1145/3242969.3242985
Healey et al (2005) Detecting Stress During Real-World Driving Tasks Using Physiological Sensors. IEEE Trans Intell Transp Syst 6(2):156–166. https://doi.org/10.1109/TITS.2005.848368
Soleymani et al (2011) A Multimodal Database for Affect Recognition and Implicit Tagging. IEEE Trans Affect Comput 3(1):42–55. https://doi.org/10.1109/T-AFFC.2011.25
Saganowski et al (2022) Emognition dataset: emotion recognition with self-reports, facial expressions, and physiology using wearables. Sci Data 9(1):1–11
Subramanian et al (2016) Ascertain: Emotion and Personality Recognition using Commercial Sensors. IEEE Trans Affect Comput 9(2):147–160. https://doi.org/10.1109/TAFFC.2016.2625250
Gao Z, Cui X, Wan W, Zheng W, Gu Z (2021) ECSMP: A dataset on emotion, cognition, sleep, and multi-model physiological signals. Data in Brief 39:107660. https://doi.org/10.1016/j.dib.2021.107660
Chilimbi et al (2014) Project adam: Building an efficient and scalable deep learning training system. In: 11th USENIX Symposium on operating systems design and implementation (OSDI 14), pp 571–582
Horvitz et al (2015) Data, privacy, and the greater good. Science 349(6245):253–255
Tuncer et al (2021) LEDPatNet19: Automated Emotion Recognition Model Based on Nonlinear LED Pattern Feature Extraction Function using EEG Signals. Cognit Neurodyn 1–12. https://doi.org/10.1007/s11571-021-09748-0
Nasoz et al (2004) Emotion recognition from physiological signals using wireless sensors for presence technologies. Cognit Tech Work 6(1):4–14. https://doi.org/10.1007/s10111-003-0143-x
Cheng et al (2020) Emotion Recognition From Multi-Channel EEG Via Deep Forest. IEEE J Biomed Health Informat 25(2):453–464. https://doi.org/10.1109/JBHI.2020.2995767
Sarma et al (2021) Emotion Recognition by Distinguishing Appropriate EEG Segments Based on Random Matrix Theory 70:102991. https://doi.org/10.1016/j.bspc.2021.102991
Gao et al (2020) EEG Based Emotion Recognition Using Fusion Feature Extraction Method. Multimed Tools App 79(37):27057–27074. https://doi.org/10.1007/s11042-020-09354-y
Adams et al (2014) Towards Personal Stress Informatics: Comparing Minimally Invasive Techniques for Measuring Daily Stress in the Wild. In: Proceedings of the 8th International conference on pervasive computing technologies for healthcare, pp 72–79. https://doi.org/10.4108/icst.pervasivehealth.2014.254959
Birjandtalab et al (2016) A Non-EEG Biosignals Dataset for Assessment and Visualization of Neurological Status. In: 2016 IEEE International workshop on signal processing systems (SiPS), pp 110–114. https://doi.org/10.1109/SiPS.2016.27 . IEEE
Jia et al (2014) A Novel Semi-Supervised Deep Learning Framework for Affective State Recognition on EEG Signals. In: 2014 IEEE International conference on bioinformatics and bioengineering, pp 30–37. https://doi.org/10.1109/BIBE.2014.26 . IEEE
Maaoui C (2010) Pruski A (2010) Emotion recognition through physiological signals for human-machine communication. Cut Edge Robot 317–332:11
Ferdinando et al (2016) Comparing Features From ECG Pattern and HRV Analysis for Emotion Recognition System. In: 2016 IEEE Conference on computational intelligence in bioinformatics and computational biology (CIBCB), pp 1–6. https://doi.org/10.1109/CIBCB.2016.7758108 . IEEE
Ragot et al (2017) Emotion Recognition Using Physiological Signals: Laboratory vs. Wearable Sensors. In: International conference on applied human factors and ergonomics, pp 15–22. https://doi.org/10.1007/978-3-319-60639-2_2 . Springer
Goran et al (2017) Wearable emotion recognition system based on GSR and PPG signals. In: Proceedings of the 2nd international workshop on multimedia for personal health and health care, pp 53–59. https://doi.org/10.1145/3132635.3132641
et al, T-VC (2017) Svm-based feature selection methods for emotion recognition from multimodal data. J Multimodal User Interfaces 11(1):9–23. https://doi.org/10.1007/s12193-016-0222-y
Wiem et al (2017) Emotion Classification in Arousal Valence Model using MAHNOB-HCI Database. Int J Adv Comput Sci App 8(3)
Agarwal R, Andujar M, Canavan S (2022) Classification of emotions using eeg activity associated with different areas of the brain. Pattern Recognit Lett 162:71–80. https://doi.org/10.1016/j.patrec.2022.08.018
Anuragi A, Sisodia DS, Pachori RB (2022) EEG-based cross-subject emotion recognition using Fourier-Bessel series expansion based empirical wavelet transform and NCA feature selection method. Inf Sci 610:508–524. https://doi.org/10.1016/j.ins.2022.07.121
Palo et al (2015) Use of Different Features for Emotion Recognition Using MLP Network. Comput Vis Robot 7–15. https://doi.org/10.1007/978-81-322-2196-8_2
Song et al (2018) (2020) EEG Emotion Recognition using Dynamical Graph Convolutional Neural Networks. IEEE Trans Affect Comput 11(3):532–541. https://doi.org/10.1109/TAFFC.2018.2817622
Tao et al (2020) EEG-Based Emotion Recognition Via Channel-Wise Attention and Self Attention. IEEE Trans Affect Comput. https://doi.org/10.1109/TAFFC.2020.3025777
Santamaria-Granados et al (2018) Using Deep Convolutional Neural Network for Emotion Detection on a Physiological Signals Dataset (AMIGOS). IEEE Access 7:57–67. https://doi.org/10.1109/ACCESS.2018.2883213
Iyer et al (2022) CNN and LSTM Based Ensemble Learning for Human Emotion Recognition Using EEG Recordings. Multimed Tools App 1–14. https://doi.org/10.1007/s11042-022-12310-7
Chakravarthi B, Ng S-C, Ezilarasan M, Leung M-F (2022) EEG-based emotion recognition using hybrid CNN and LSTM classification. Front Comput Neurosci 16:1019776. https://doi.org/10.3389/fncom.2022.1019776
Zali-Vargahan B, Charmin A, Kalbkhani H, Barghandan S (2023) Deep time-frequency features and semi-supervised dimension reduction for subject-independent emotion recognition from multi-channel EEG signals. Biomed Signal Process Control 85:104806. https://doi.org/10.1016/j.bspc.2023.104806
Singh MK, Singh M et al (2023) A deep learning approach for subject-dependent & subject-independent emotion recognition using brain signals with dimensional emotion model. Biomed Signal Process Control 84:104928. https://doi.org/10.1016/j.bspc.2023.104928
Li W, Tian Y, Hou B, Dong J, Shao S, Song A (2023) A Bi-Stream hybrid model with MLPBlocks and self-attention mechanism for EEG-based emotion recognition. Biomed Signal Process Control 86:105223. https://doi.org/10.1016/j.bspc.2023.105223
Liu et al (2019) Multimodal Emotion Recognition using Deep Canonical Correlation Analysis. arXiv preprint arXiv:1908.05349 . https://doi.org/10.48550/arXiv.1908.05349
Bhattacharyya et al (2020) A Novel Multivariate-Multiscale Approach for Computing EEG Spectral and Temporal Complexity for Human Emotion Recognition. IEEE Sensors J 21(3):3579–3591. https://doi.org/10.1109/JSEN.2020.3027181
Nath et al (2020) An Efficient Approach to EEG-Based Emotion Recognition using LSTM Network. In: 2020 16th IEEE International colloquium on signal processing & its applications (CSPA), pp 88–92. https://doi.org/10.1109/CSPA48992.2020.9068691 . IEEE
Zhang et al (2020) Corrnet: Fine-Grained Emotion Recognition for Video Watching using Wearable Physiological Sensors. Sensors 21(1):52. https://doi.org/10.3390/s21010052
Yang et al (2020) A Convolution Neural Network Based Emotion Recognition System Using Multimodal Physiological Signals. In: 2020 IEEE International conference on consumer electronics-taiwan (ICCE-Taiwan), pp 1–2. https://doi.org/10.1109/ICCE-Taiwan49838.2020.9258341 . IEEE
Zhang et al (2021) Multimodal Emotion Recognition Using A Hierarchical Fusion Convolutional Neural Network. IEEE Access 9:7943–7951. https://doi.org/10.1109/ACCESS.2021.3049516
Kumari N, Anwar S, Bhattacharjee V (2022) Time series-dependent feature of EEG signals for improved visually evoked emotion classification using EmotionCapsNet. Neural Comput App 34(16):13291–13303. https://doi.org/10.1007/s00521-022-06942-x
McMahan et al (2016) Communication-E Client Learning of Deep Networks from Decentralized Data. In: Proceedings of the 20th international conference on artificial intelligence and statistics (AISTATS). arXiv:1602.05629
Liu et al (2020) A Secure Federated Transfer Learning Framework. IEEE Intell Syst 35(4):70–82. https://doi.org/10.1109/MIS.2020.2988525
Chen et al (2020) Asynchronous Online Federated Learning for Edge Devices with Non-IID Data. In: 2020 IEEE International conference on big data (Big Data), pp 15–24. https://doi.org/10.1109/BigData50022.2020.9378161 . IEEE
Chen et al (2019) Communication-efficient federated deep learning with layerwise asynchronous model update and temporally weighted aggregation. IEEE Trans Neural Netw Learn Syst 31(10):4229–4238. https://doi.org/10.1109/TNNLS.2019.2953131
Rahman et al (2021) Challenges, Applications and Design Aspects of Federated Learning: A Survey. IEEE Access 9:124682–124700. https://doi.org/10.1109/ACCESS.2021.3111118
Vanhaesebrouck et al (2017) Decentralized Collaborative Learning of Personalized Models Over Networks. In: Artificial intelligence and statistics, pp 509–517. PMLR
Jiang et al (2017) Collaborative Deep Learning in Fixed Topology Networks. Adv Neural Inf Process Syst 30
Anwar MA, Agrawal M, Gahlan N, Sethia D, Singh GK, Chaurasia R (2023) FedEmo: A Privacy-Preserving Framework for Emotion Recognition using EEG Physiological Data. In: 2023 15th International conference on communication systems & NETworkS (COMSNETS), pp 119–124. https://doi.org/10.1109/COMSNETS56262.2023.10041308 . IEEE
Hamer et al (2020) Fedboost: A Communication-Efficient Algorithm for Federated Learning. In: International conference on machine learning, pp 3973–3983. PMLR
Wang et al (2020) Federated learning with matched averaging. arXiv preprint arXiv:2002.06440
Arivazhagan et al (2019) Federated learning with personalization layers. arXiv preprint arXiv:1912.00818
Bonawitz et al (2017) Practical Secure Aggregation for Privacy-Preserving Machine Learning. In: Proceedings of the 2017 ACM SIGSAC conference on computer and communications security, pp 1175–1191. https://doi.org/10.1145/3133956.3133982
Google (2022) Tensorflow federated. https://www.tensorflow.org/federated . Accessed 7 Aug 2022
Ryffel et al (2018) A Generic Framework for Privacy Preserving Deep Learning. arXiv preprint arXiv:1811.04017 . https://doi.org/10.48550/arXiv.1811.04017
Federated AI Technology Enabler (2022). https://www.fedai.org/cn/ . Accessed 7 Aug 2022
Flower A Friendly Federated Learning Framework (2023). https://flower.dev/ . Accessed 11 Aug 2023
Xu et al (2021) Federated Learning for Healthcare Informatics. J Healthcare Inf Res 5(1):1–19. https://doi.org/10.1007/s41666-020-00082-4
Lee et al (2018) Privacy-Preserving Patient Similarity Learning in a Federated Environment: Development and Analysis. JMIR Med Inf 6(2):7744. https://doi.org/10.2196/medinform.7744
Kim et al (2017) Federated Tensor Factorization For Computational Phenotyping. In: Proceedings of the 23rd ACM SIGKDD international conference on knowledge discovery and data mining, pp 887–895. https://doi.org/10.1145/3097983.3098118
Brisimi et al (2018) Federated Learning of Predictive Models From Federated Electronic Health Records. Int J Med Inf 112:59–67. https://doi.org/10.1016/j.ijmedinf.2018.01.007
Gu T, Wang Z, Xu X, Li D, Yang H, Du W (2022) Frame-level teacher-student learning with data privacy for eeg emotion recognition. IEEE Trans Neural Netw Learn Syst. https://doi.org/10.1109/TNNLS.2022.3168935
Agrawal M, Anwar MA, Jindal R (2023) FedCER-Emotion Recognition Using 2D-CNN in Decentralized Federated Learning Environment. In: 2023 6th International conference on information systems and computer networks (ISCON), pp 1–5. https://doi.org/10.1109/ISCON57294.2023.10112028 . IEEE
Vepakomma et al (2018) Split Learning for Health: Distributed Deep Learning Without Sharing Raw Patient Data. arXiv preprint arXiv:1812.00564 . https://doi.org/10.48550/arXiv.1812.00564
Gupta et al (2018) Distributed learning of deep neural network over multiple agents. J Netw Comput App 116:1–8. https://doi.org/10.1016/j.jnca.2018.05.003
Huang et al (2019) Patient Clustering Improves Efficiency of Federated Machine Learning to Predict Mortality and Hospital Stay Time using Distributed Electronic Medical Records. J Biomed Informat 99:103291. https://doi.org/10.1016/j.jbi.2019.103291
Peyvandi et al (2022) Privacy-Preserving Federated Learning for Scalable and High Data Quality Computational-Intelligence-as-a-Service in Society 5.0. Multimed Tools App 1–22. https://doi.org/10.1007/s11042-022-12900-5
Morris JD (1995) Observations: SAM: the Self-Assessment Manikin; an efficient cross-cultural measurement of emotional response. J Advert Res 35(6):63–68
MathSciNet Google Scholar
Chen et al (2020) Fedhealth: A Federated Transfer Learning Framework for Wearable Healthcare. IEEE Intell Syst 35(4):83–93. https://doi.org/10.1109/MIS.2020.2988604
Sun et al (2022) A Scalable and Transferable Federated Learning System for Classifying Healthcare Sensor Data. IEEE J Biomed Health Informat. https://doi.org/10.1109/JBHI.2022.3171402
Wu et al (2020) Fedhome: Cloud-edge Based Personalized Federated Learning for In-home Health Monitoring. IEEE Trans Mobile Comput. https://doi.org/10.1109/TMC.2020.3045266
Chuah et al (2021) The Future of Service: The Power of Emotion in Human-Robot Interaction. J Retail Consumer Services 61:102551. https://doi.org/10.1016/j.jretconser.2021.102551
Christou et al (2020) Tourists’ Perceptions Regarding the Use of Anthropomorphic Robots in Tourism and Hospitality. International Journal of Contemporary Hospitality Management. https://doi.org/10.1108/IJCHM-05-2020-0423
EMO Robot (2022). https://living.ai/emo/ . Accessed 7 Aug 2022
Mood tracking app (2024) https://dribbble.com/shots/9524341-Employee-Engagement-App-Mood-Tracker-UI
Marmpena et al (2022) Data-Driven Emotional Body Language Generation for Social Robotics. arXiv preprint arXiv:2205.00763
Data Breach (2023). https://tinyurl.com/2p8b57ax . Accessed 3 Feb 2023
Tarnowski et al (2018) Combined Analysis of GSR and EEG Signals for Emotion Recognition. In: 2018 International interdisciplinary phd workshop (IIPhDW), pp 137–141. https://doi.org/10.1109/IIPHDW.2018.8388342 . IEEE
Zhang et al (2016) Emotion Recognition of GSR Based on An Improved Quantum Neural Network. In: 2016 8th International conference on intelligent human-machine systems and cybernetics (IHMSC), vol 1, pp 488–492. https://doi.org/10.1109/IHMSC.2016.66 . IEEE
Das et al (2016) Emotion Recognition Employing ECG and GSR Signals as Markers of ANS. In: 2016 Conference on advances in signal processing (CASP), pp 37–42. IEEE
Lang PJ, Bradley MM, Cuthbert BN (1990) Emotion, attention, and the startle reflex. Psycho Rev 97(3):377
Busso C, Deng Z, Yildirim S, Bulut M, Lee CM, Kazemzadeh A, Lee S, Neumann U, Narayanan S (2004) Analysis of Emotion Recognition using Facial Expressions, Speech and Multimodal Information. In: Proceedings of the 6th international conference on multimodal interfaces, pp 205–211. https://doi.org/10.1145/1027933.1027968
McMahan et al (2017) Communication-efficient learning of deep networks from decentralized data. In: Artificial intelligence and statistics, pp 1273–1282. PMLR
Li et al (2020) Federated optimization in heterogeneous networks. Proc Mach Learn Syst 2:429–450
Download references
Author information
Authors and affiliations.
Department of Software Engineering, Delhi Technological University, Rohini, New Delhi, 10042, Delhi, India
Neha Gahlan & Divyashikha Sethia
You can also search for this author in PubMed Google Scholar
Corresponding author
Correspondence to Divyashikha Sethia .
Ethics declarations
Funding and or conflicts of interests/competing interests.
The authors have no competing interests to declare relevant to this article’s content. The authors have no relevant financial or non-financial interests to disclose.
Additional information
Publisher's note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Reprints and permissions
About this article
Gahlan, N., Sethia, D. Federated learning in Emotion Recognition Systems based on physiological signals for privacy preservation: a review. Multimed Tools Appl (2024). https://doi.org/10.1007/s11042-024-19467-3
Download citation
Received : 16 September 2022
Revised : 16 February 2024
Accepted : 21 May 2024
Published : 03 June 2024
DOI : https://doi.org/10.1007/s11042-024-19467-3
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Emotion recognition
- Federated learning
- Physiological signals
- Wearable sensors
Advertisement
- Find a journal
- Publish with us
- Track your research
Main Navigation
- Contact NeurIPS
- Code of Ethics
- Code of Conduct
- Create Profile
- Journal To Conference Track
- Diversity & Inclusion
- Proceedings
- Future Meetings
- Exhibitor Information
- Privacy Policy
NeurIPS 2024, the Thirty-eighth Annual Conference on Neural Information Processing Systems, will be held at the Vancouver Convention Center
Monday Dec 9 through Sunday Dec 15. Monday is an industry expo.

Registration
Pricing » Registration 2024 Registration Cancellation Policy » . Certificate of Attendance
Our Hotel Reservation page is currently under construction and will be released shortly. NeurIPS has contracted Hotel guest rooms for the Conference at group pricing, requiring reservations only through this page. Please do not make room reservations through any other channel, as it only impedes us from putting on the best Conference for you. We thank you for your assistance in helping us protect the NeurIPS conference.
Announcements
- The call for High School Projects has been released
- The Call For Papers has been released
- See the Visa Information page for changes to the visa process for 2024.
Latest NeurIPS Blog Entries [ All Entries ]
Important dates.
If you have questions about supporting the conference, please contact us .
View NeurIPS 2024 exhibitors » Become an 2024 Exhibitor Exhibitor Info »
Organizing Committee
General chair, program chair, workshop chair, workshop chair assistant, tutorial chair, competition chair, data and benchmark chair, diversity, inclusion and accessibility chair, affinity chair, ethics review chair, communication chair, social chair, journal chair, creative ai chair, workflow manager, logistics and it, mission statement.
The Neural Information Processing Systems Foundation is a non-profit corporation whose purpose is to foster the exchange of research advances in Artificial Intelligence and Machine Learning, principally by hosting an annual interdisciplinary academic conference with the highest ethical standards for a diverse and inclusive community.
About the Conference
The conference was founded in 1987 and is now a multi-track interdisciplinary annual meeting that includes invited talks, demonstrations, symposia, and oral and poster presentations of refereed papers. Along with the conference is a professional exposition focusing on machine learning in practice, a series of tutorials, and topical workshops that provide a less formal setting for the exchange of ideas.
More about the Neural Information Processing Systems foundation »

IMAGES
VIDEO
COMMENTS
NaiboWang/EasySpider • ACM The Web Conference 2023. As such, web-crawling is an essential tool for both computational and non-computational scientists to conduct research. Data Integration Marketing. 26,735. 0.94 stars / hour. Paper. Code. Papers With Code highlights trending Machine Learning research and the code to implement it.
Since ML appeared in the 1990s, all published documents (i.e., journal papers, reviews, conference papers, preprints, code repositories and more) related to this field from 1990 to 2020 have been selected, and specifically, within the search fields, the following keywords were used: "machine learning" OR "machine learning-based approach" OR ...
To discuss the applicability of machine learning-based solutions in various real-world application domains. To highlight and summarize the potential research directions within the scope of our study for intelligent data analysis and services. The rest of the paper is organized as follows.
The Journal of Machine Learning Research (JMLR), , provides an international forum for the electronic and paper publication of high-quality scholarly articles in all areas of machine learning. All published papers are freely available online. JMLR has a commitment to rigorous yet rapid reviewing. Final versions are (ISSN 1533-7928) immediately ...
Machine learning is the ability of a machine to improve its performance based on previous results. Machine learning methods enable computers to learn without being explicitly programmed and have ...
Title: HC-GAE: The Hierarchical Cluster-based Graph Auto-Encoder for Graph Representation Learning Zhuo Xu, Lu Bai, Lixin Cui, Ming Li, Yue Wang ... Subjects: Machine Learning (cs.LG); Disordered Systems and Neural Networks (cond-mat.dis-nn); Statistical Mechanics (cond-mat.stat-mech)
This article examines the background to the problem and outlines a project that TNA undertook to research the feasibility of using commercially available artificial intelligence tools to aid selection. ... this research aims to predict user's personalities based on Indonesian text from social media using machine learning techniques. This ...
Explore the latest full-text research PDFs, articles, conference papers, preprints and more on MACHINE LEARNING. Find methods information, sources, references or conduct a literature review on ...
Machine Learning is an international forum focusing on computational approaches to learning. ... Improves how machine learning research is conducted. Prioritizes verifiable and replicable supporting evidence in all published papers. Editor-in-Chief. Hendrik Blockeel; Impact factor 7.5 (2022) 5 year impact factor
The corpus of scientific literature grows at an ever-increasing speed. Specifically, in the field of artificial intelligence (AI) and machine learning (ML), the number of papers every month is ...
Today, intelligent systems that offer artificial intelligence capabilities often rely on machine learning. Machine learning describes the capacity of systems to learn from problem-specific training data to automate the process of analytical model building and solve associated tasks. Deep learning is a machine learning concept based on artificial neural networks. For many applications, deep ...
JMLR Papers. Select a volume number to see its table of contents with links to the papers. Volume 23 (January 2022 - Present) . Volume 22 (January 2021 - December 2021) . Volume 21 (January 2020 - December 2020) . Volume 20 (January 2019 - December 2019) . Volume 19 (August 2018 - December 2018) . Volume 18 (February 2017 - August 2018) . Volume 17 (January 2016 - January 2017)
It is a challenging task for any research field to screen the literature and determine what needs to be included in a systematic review in a transparent way. A new open source machine learning ...
Machine learning, especially its subfield of Deep Learning, had many amazing advances in the recent years, and important research papers may lead to breakthroughs in technology that get used by billio ns of people. The research in this field is developing very quickly and to help our readers monitor the progress we present the list of most important recent scientific papers published since 2014.
Machine learning (ML) methods are being widely adopted for scientific research (1-11).Compared to older statistical methods, they offer increased predictive accuracy (), the ability to process large amounts of data (), and the ability to use different types of data for scientific research, such as text, images, and video ().However, the rapid uptake of ML methods has been accompanied by ...
Deep learning (DL), a branch of machine learning (ML) and artificial intelligence (AI) is nowadays considered as a core technology of today's Fourth Industrial Revolution (4IR or Industry 4.0). Due to its learning capabilities from data, DL technology originated from artificial neural network (ANN), has become a hot topic in the context of computing, and is widely applied in various ...
Machine learning (ML), an area of artificial intelligence (AI), enables researchers, physicians, and patients to solve some of these issues. Based on relevant research, this review explains how machine learning (ML) is being used to help in the early identification of numerous diseases.
a few innovative research works and their applications in real-world, such as stock trading, medical and healthcare systems, and software automation. ... reinforcement learning-based systems; and finally, (9) evolution of few-shots, ... Machine learning (ML) is the ability of a system to automatically acquire, integrate, ...
Chen and Babar [31] provided a review of machine learning-based modern software systems (MLBSS) security. They highlight the need for joint efforts from software engineering, system security, and machine learning disciplines to address the evolving threats posed by MLBSS. ... The reviewed papers in this research were classified into four ...
In machine learning, a computer first learns to perform a task by studying a training set of examples. The computer then performs the same task with data it hasn't encountered before. This article presents a brief overview of machine-learning technologies, with a concrete case study from code analysis.
This paper demonstrated the interest attached by data scientists to this particular situation. It provided a survey of Machine Learning based research classified into two categories (Supervised Learning approaches and Deep Learning approaches) to make detection, diagnosis, or prediction of the COVID-19.
The research of machine learning-based scheduling has attracted the attention of both the academia and industry, and many related papers have been published [23, 62, 63]. Between 1957 and 2019, a total of 2207 papers on machine learning-based scheduling were published, including journal articles, conference articles, and reviews.
Machine learning (ML) provides algorithms to create computer programs based on data without explicitly programming them. In business process management (BPM), ML applications are used to analyse and improve processes efficiently. Three frequent examples of using ML are providing decision support through predictions, discovering accurate process models, and improving resource allocation.
Food production is a vital global concern and the potential for an agritech revolution through artificial intelligence (AI) remains largely unexplored. This paper presents a comprehensive review focused on the application of machine learning (ML) in agriculture, aiming to explore its transformative potential in farming practices and efficiency enhancement. To understand the extent of research ...
This paper categorize and outline several mainstream music recommendation methods: content-based and collaborative filtering recommendations, as well as weighted hybrid models of both. Machine learning is a hot topic of research in recent years and has a very wide range of applications in the fields of banking, insurance, transportation, biology, and medicine.
JMLR Papers. Select a volume number to see its table of contents with links to the papers. Volume 25 (January 2024 - Present) ... Machine Learning and Large Scale Optimization (Jul 2006 - Oct 2006) Approaches and Applications of Inductive Programming (February 2006 - Mar 2006)
The paper reviews the different emotions, benchmark datasets, machine learning, and federated learning approaches for classifying emotions. It proposes a novel multi-modal Federated Learning for Physiological signals based on Emotion Recognition Systems (Fed-PhyERS) architecture, experimenting with the AMIGOS dataset and its applications for a ...
The Neural Information Processing Systems Foundation is a non-profit corporation whose purpose is to foster the exchange of research advances in Artificial Intelligence and Machine Learning, principally by hosting an annual interdisciplinary academic conference with the highest ethical standards for a diverse and inclusive community.
With the rapid growth in urban construction in Malaysia, road breakage has challenged traditional manual inspection methods. In order to quickly and accurately detect the extent of road breakage, it is crucial to apply automated road crack detection techniques. Researchers have long studied image-based road crack detection techniques, especially the deep learning methods that have emerged in ...