- Skip to main content
- Skip to primary sidebar
- Skip to footer
- QuestionPro

- Solutions Industries Gaming Automotive Sports and events Education Government Travel & Hospitality Financial Services Healthcare Cannabis Technology Use Case AskWhy Communities Audience Contactless surveys Mobile LivePolls Member Experience GDPR Positive People Science 360 Feedback Surveys
- Resources Blog eBooks Survey Templates Case Studies Training Help center
Home Market Research

Data Analysis in Research: Types & Methods
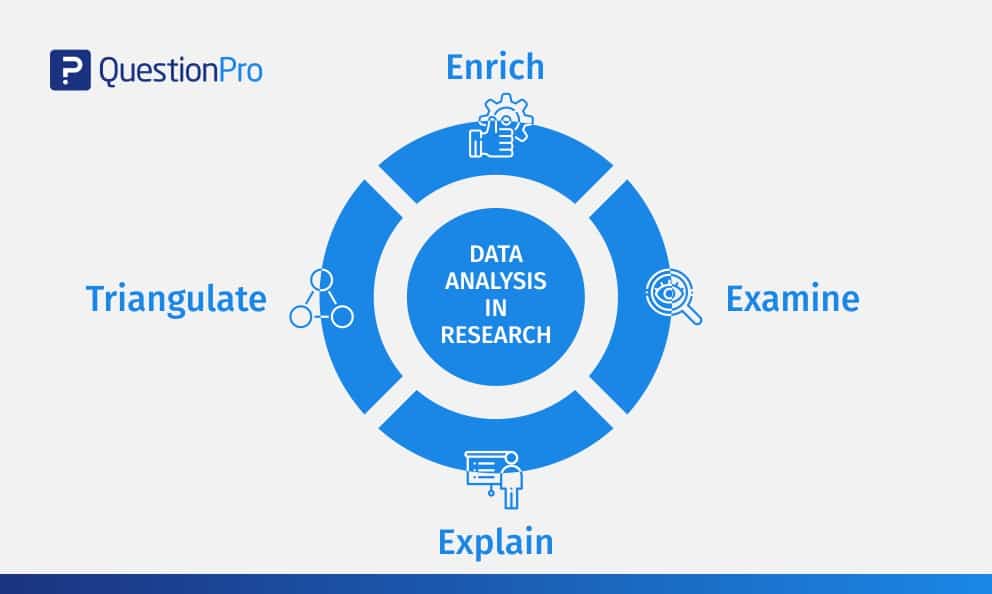
Content Index
Why analyze data in research?
Types of data in research, finding patterns in the qualitative data, methods used for data analysis in qualitative research, preparing data for analysis, methods used for data analysis in quantitative research, considerations in research data analysis, what is data analysis in research.
Definition of research in data analysis: According to LeCompte and Schensul, research data analysis is a process used by researchers to reduce data to a story and interpret it to derive insights. The data analysis process helps reduce a large chunk of data into smaller fragments, which makes sense.
Three essential things occur during the data analysis process — the first is data organization . Summarization and categorization together contribute to becoming the second known method used for data reduction. It helps find patterns and themes in the data for easy identification and linking. The third and last way is data analysis – researchers do it in both top-down and bottom-up fashion.
LEARN ABOUT: Research Process Steps
On the other hand, Marshall and Rossman describe data analysis as a messy, ambiguous, and time-consuming but creative and fascinating process through which a mass of collected data is brought to order, structure and meaning.
We can say that “the data analysis and data interpretation is a process representing the application of deductive and inductive logic to the research and data analysis.”
Researchers rely heavily on data as they have a story to tell or research problems to solve. It starts with a question, and data is nothing but an answer to that question. But, what if there is no question to ask? Well! It is possible to explore data even without a problem – we call it ‘Data Mining’, which often reveals some interesting patterns within the data that are worth exploring.
Irrelevant to the type of data researchers explore, their mission and audiences’ vision guide them to find the patterns to shape the story they want to tell. One of the essential things expected from researchers while analyzing data is to stay open and remain unbiased toward unexpected patterns, expressions, and results. Remember, sometimes, data analysis tells the most unforeseen yet exciting stories that were not expected when initiating data analysis. Therefore, rely on the data you have at hand and enjoy the journey of exploratory research.
Create a Free Account
Every kind of data has a rare quality of describing things after assigning a specific value to it. For analysis, you need to organize these values, processed and presented in a given context, to make it useful. Data can be in different forms; here are the primary data types.
- Qualitative data: When the data presented has words and descriptions, then we call it qualitative data . Although you can observe this data, it is subjective and harder to analyze data in research, especially for comparison. Example: Quality data represents everything describing taste, experience, texture, or an opinion that is considered quality data. This type of data is usually collected through focus groups, personal qualitative interviews , qualitative observation or using open-ended questions in surveys.
- Quantitative data: Any data expressed in numbers of numerical figures are called quantitative data . This type of data can be distinguished into categories, grouped, measured, calculated, or ranked. Example: questions such as age, rank, cost, length, weight, scores, etc. everything comes under this type of data. You can present such data in graphical format, charts, or apply statistical analysis methods to this data. The (Outcomes Measurement Systems) OMS questionnaires in surveys are a significant source of collecting numeric data.
- Categorical data: It is data presented in groups. However, an item included in the categorical data cannot belong to more than one group. Example: A person responding to a survey by telling his living style, marital status, smoking habit, or drinking habit comes under the categorical data. A chi-square test is a standard method used to analyze this data.
Learn More : Examples of Qualitative Data in Education
Data analysis in qualitative research
Data analysis and qualitative data research work a little differently from the numerical data as the quality data is made up of words, descriptions, images, objects, and sometimes symbols. Getting insight from such complicated information is a complicated process. Hence it is typically used for exploratory research and data analysis .
Although there are several ways to find patterns in the textual information, a word-based method is the most relied and widely used global technique for research and data analysis. Notably, the data analysis process in qualitative research is manual. Here the researchers usually read the available data and find repetitive or commonly used words.
For example, while studying data collected from African countries to understand the most pressing issues people face, researchers might find “food” and “hunger” are the most commonly used words and will highlight them for further analysis.
LEARN ABOUT: Level of Analysis
The keyword context is another widely used word-based technique. In this method, the researcher tries to understand the concept by analyzing the context in which the participants use a particular keyword.
For example , researchers conducting research and data analysis for studying the concept of ‘diabetes’ amongst respondents might analyze the context of when and how the respondent has used or referred to the word ‘diabetes.’
The scrutiny-based technique is also one of the highly recommended text analysis methods used to identify a quality data pattern. Compare and contrast is the widely used method under this technique to differentiate how a specific text is similar or different from each other.
For example: To find out the “importance of resident doctor in a company,” the collected data is divided into people who think it is necessary to hire a resident doctor and those who think it is unnecessary. Compare and contrast is the best method that can be used to analyze the polls having single-answer questions types .
Metaphors can be used to reduce the data pile and find patterns in it so that it becomes easier to connect data with theory.
Variable Partitioning is another technique used to split variables so that researchers can find more coherent descriptions and explanations from the enormous data.
LEARN ABOUT: Qualitative Research Questions and Questionnaires
There are several techniques to analyze the data in qualitative research, but here are some commonly used methods,
- Content Analysis: It is widely accepted and the most frequently employed technique for data analysis in research methodology. It can be used to analyze the documented information from text, images, and sometimes from the physical items. It depends on the research questions to predict when and where to use this method.
- Narrative Analysis: This method is used to analyze content gathered from various sources such as personal interviews, field observation, and surveys . The majority of times, stories, or opinions shared by people are focused on finding answers to the research questions.
- Discourse Analysis: Similar to narrative analysis, discourse analysis is used to analyze the interactions with people. Nevertheless, this particular method considers the social context under which or within which the communication between the researcher and respondent takes place. In addition to that, discourse analysis also focuses on the lifestyle and day-to-day environment while deriving any conclusion.
- Grounded Theory: When you want to explain why a particular phenomenon happened, then using grounded theory for analyzing quality data is the best resort. Grounded theory is applied to study data about the host of similar cases occurring in different settings. When researchers are using this method, they might alter explanations or produce new ones until they arrive at some conclusion.
LEARN ABOUT: 12 Best Tools for Researchers
Data analysis in quantitative research
The first stage in research and data analysis is to make it for the analysis so that the nominal data can be converted into something meaningful. Data preparation consists of the below phases.
Phase I: Data Validation
Data validation is done to understand if the collected data sample is per the pre-set standards, or it is a biased data sample again divided into four different stages
- Fraud: To ensure an actual human being records each response to the survey or the questionnaire
- Screening: To make sure each participant or respondent is selected or chosen in compliance with the research criteria
- Procedure: To ensure ethical standards were maintained while collecting the data sample
- Completeness: To ensure that the respondent has answered all the questions in an online survey. Else, the interviewer had asked all the questions devised in the questionnaire.
Phase II: Data Editing
More often, an extensive research data sample comes loaded with errors. Respondents sometimes fill in some fields incorrectly or sometimes skip them accidentally. Data editing is a process wherein the researchers have to confirm that the provided data is free of such errors. They need to conduct necessary checks and outlier checks to edit the raw edit and make it ready for analysis.
Phase III: Data Coding
Out of all three, this is the most critical phase of data preparation associated with grouping and assigning values to the survey responses . If a survey is completed with a 1000 sample size, the researcher will create an age bracket to distinguish the respondents based on their age. Thus, it becomes easier to analyze small data buckets rather than deal with the massive data pile.
LEARN ABOUT: Steps in Qualitative Research
After the data is prepared for analysis, researchers are open to using different research and data analysis methods to derive meaningful insights. For sure, statistical analysis plans are the most favored to analyze numerical data. In statistical analysis, distinguishing between categorical data and numerical data is essential, as categorical data involves distinct categories or labels, while numerical data consists of measurable quantities. The method is again classified into two groups. First, ‘Descriptive Statistics’ used to describe data. Second, ‘Inferential statistics’ that helps in comparing the data .
Descriptive statistics
This method is used to describe the basic features of versatile types of data in research. It presents the data in such a meaningful way that pattern in the data starts making sense. Nevertheless, the descriptive analysis does not go beyond making conclusions. The conclusions are again based on the hypothesis researchers have formulated so far. Here are a few major types of descriptive analysis methods.
Measures of Frequency
- Count, Percent, Frequency
- It is used to denote home often a particular event occurs.
- Researchers use it when they want to showcase how often a response is given.
Measures of Central Tendency
- Mean, Median, Mode
- The method is widely used to demonstrate distribution by various points.
- Researchers use this method when they want to showcase the most commonly or averagely indicated response.
Measures of Dispersion or Variation
- Range, Variance, Standard deviation
- Here the field equals high/low points.
- Variance standard deviation = difference between the observed score and mean
- It is used to identify the spread of scores by stating intervals.
- Researchers use this method to showcase data spread out. It helps them identify the depth until which the data is spread out that it directly affects the mean.
Measures of Position
- Percentile ranks, Quartile ranks
- It relies on standardized scores helping researchers to identify the relationship between different scores.
- It is often used when researchers want to compare scores with the average count.
For quantitative research use of descriptive analysis often give absolute numbers, but the in-depth analysis is never sufficient to demonstrate the rationale behind those numbers. Nevertheless, it is necessary to think of the best method for research and data analysis suiting your survey questionnaire and what story researchers want to tell. For example, the mean is the best way to demonstrate the students’ average scores in schools. It is better to rely on the descriptive statistics when the researchers intend to keep the research or outcome limited to the provided sample without generalizing it. For example, when you want to compare average voting done in two different cities, differential statistics are enough.
Descriptive analysis is also called a ‘univariate analysis’ since it is commonly used to analyze a single variable.
Inferential statistics
Inferential statistics are used to make predictions about a larger population after research and data analysis of the representing population’s collected sample. For example, you can ask some odd 100 audiences at a movie theater if they like the movie they are watching. Researchers then use inferential statistics on the collected sample to reason that about 80-90% of people like the movie.
Here are two significant areas of inferential statistics.
- Estimating parameters: It takes statistics from the sample research data and demonstrates something about the population parameter.
- Hypothesis test: I t’s about sampling research data to answer the survey research questions. For example, researchers might be interested to understand if the new shade of lipstick recently launched is good or not, or if the multivitamin capsules help children to perform better at games.
These are sophisticated analysis methods used to showcase the relationship between different variables instead of describing a single variable. It is often used when researchers want something beyond absolute numbers to understand the relationship between variables.
Here are some of the commonly used methods for data analysis in research.
- Correlation: When researchers are not conducting experimental research or quasi-experimental research wherein the researchers are interested to understand the relationship between two or more variables, they opt for correlational research methods.
- Cross-tabulation: Also called contingency tables, cross-tabulation is used to analyze the relationship between multiple variables. Suppose provided data has age and gender categories presented in rows and columns. A two-dimensional cross-tabulation helps for seamless data analysis and research by showing the number of males and females in each age category.
- Regression analysis: For understanding the strong relationship between two variables, researchers do not look beyond the primary and commonly used regression analysis method, which is also a type of predictive analysis used. In this method, you have an essential factor called the dependent variable. You also have multiple independent variables in regression analysis. You undertake efforts to find out the impact of independent variables on the dependent variable. The values of both independent and dependent variables are assumed as being ascertained in an error-free random manner.
- Frequency tables: The statistical procedure is used for testing the degree to which two or more vary or differ in an experiment. A considerable degree of variation means research findings were significant. In many contexts, ANOVA testing and variance analysis are similar.
- Analysis of variance: The statistical procedure is used for testing the degree to which two or more vary or differ in an experiment. A considerable degree of variation means research findings were significant. In many contexts, ANOVA testing and variance analysis are similar.
- Researchers must have the necessary research skills to analyze and manipulation the data , Getting trained to demonstrate a high standard of research practice. Ideally, researchers must possess more than a basic understanding of the rationale of selecting one statistical method over the other to obtain better data insights.
- Usually, research and data analytics projects differ by scientific discipline; therefore, getting statistical advice at the beginning of analysis helps design a survey questionnaire, select data collection methods , and choose samples.
LEARN ABOUT: Best Data Collection Tools
- The primary aim of data research and analysis is to derive ultimate insights that are unbiased. Any mistake in or keeping a biased mind to collect data, selecting an analysis method, or choosing audience sample il to draw a biased inference.
- Irrelevant to the sophistication used in research data and analysis is enough to rectify the poorly defined objective outcome measurements. It does not matter if the design is at fault or intentions are not clear, but lack of clarity might mislead readers, so avoid the practice.
- The motive behind data analysis in research is to present accurate and reliable data. As far as possible, avoid statistical errors, and find a way to deal with everyday challenges like outliers, missing data, data altering, data mining , or developing graphical representation.
LEARN MORE: Descriptive Research vs Correlational Research The sheer amount of data generated daily is frightening. Especially when data analysis has taken center stage. in 2018. In last year, the total data supply amounted to 2.8 trillion gigabytes. Hence, it is clear that the enterprises willing to survive in the hypercompetitive world must possess an excellent capability to analyze complex research data, derive actionable insights, and adapt to the new market needs.
LEARN ABOUT: Average Order Value
QuestionPro is an online survey platform that empowers organizations in data analysis and research and provides them a medium to collect data by creating appealing surveys.
MORE LIKE THIS

Customer Experience Lessons from 13,000 Feet — Tuesday CX Thoughts
Aug 20, 2024

Insight: Definition & meaning, types and examples
Aug 19, 2024

Employee Loyalty: Strategies for Long-Term Business Success
Jotform vs SurveyMonkey: Which Is Best in 2024
Aug 15, 2024
Other categories
- Academic Research
- Artificial Intelligence
- Assessments
- Brand Awareness
- Case Studies
- Communities
- Consumer Insights
- Customer effort score
- Customer Engagement
- Customer Experience
- Customer Loyalty
- Customer Research
- Customer Satisfaction
- Employee Benefits
- Employee Engagement
- Employee Retention
- Friday Five
- General Data Protection Regulation
- Insights Hub
- Life@QuestionPro
- Market Research
- Mobile diaries
- Mobile Surveys
- New Features
- Online Communities
- Question Types
- Questionnaire
- QuestionPro Products
- Release Notes
- Research Tools and Apps
- Revenue at Risk
- Survey Templates
- Training Tips
- Tuesday CX Thoughts (TCXT)
- Uncategorized
- What’s Coming Up
- Workforce Intelligence
Different Types of Data Analysis; Data Analysis Methods and Techniques in Research Projects
International Journal of Academic Research in Management, 9(1):1-9, 2022 http://elvedit.com/journals/IJARM/wp-content/uploads/Different-Types-of-Data-Analysis-Data-Analysis-Methods-and-Tec
9 Pages Posted: 18 Aug 2022
Hamed Taherdoost
Hamta Group
Date Written: August 1, 2022
This article is concentrated to define data analysis and the concept of data preparation. Then, the data analysis methods will be discussed. For doing so, the first six main categories are described briefly. Then, the statistical tools of the most commonly used methods including descriptive, explanatory, and inferential analyses are investigated in detail. Finally, we focus more on qualitative data analysis to get familiar with the data preparation and strategies in this concept.
Keywords: Data Analysis, Data Preparation, Data Analysis Methods, Data Analysis Types, Descriptive Analysis, Explanatory Analysis, Inferential Analysis, Predictive Analysis, Explanatory Analysis, Causal Analysis and Mechanistic Analysis, Statistical Analysis.
Suggested Citation: Suggested Citation
Hamed Taherdoost (Contact Author)
Hamta group ( email ).
Vancouver Canada
Do you have a job opening that you would like to promote on SSRN?
Paper statistics, related ejournals, data science, data analytics & informatics ejournal.
Subscribe to this fee journal for more curated articles on this topic
Exploring Data Analysis and Visualization Techniques for Project Tracking: Insights from the ITC
- Conference paper
- First Online: 13 September 2023
- Cite this conference paper

- André Barrocas 9 ,
- Alberto Rodrigues da Silva 10 &
- João Saraiva 11
Part of the book series: Communications in Computer and Information Science ((CCIS,volume 1871))
Included in the following conference series:
- International Conference on the Quality of Information and Communications Technology
285 Accesses
Data analysis has emerged as a cornerstone in facilitating informed decision-making across myriad fields, in particular in software development and project management. This integrative practice proves instrumental in enhancing operational efficiency, cutting expenditures, mitigating potential risks, and delivering superior results, all while sustaining structured organization and robust control. This paper presents ITC, a synergistic platform architected to streamline multi-organizational and multi-workspace collaboration for project management and technical documentation. ITC serves as a powerful tool, equipping users with the capability to swiftly establish and manage workspaces and documentation, thereby fostering the derivation of invaluable insights pivotal to both technical and business-oriented decisions. ITC boasts a plethora of features, from support for a diverse range of technologies and languages, synchronization of data, and customizable templates to reusable libraries and task automation, including data extraction, validation, and document automation. This paper also delves into the predictive analytics aspect of the ITC platform. It demonstrates how ITC harnesses predictive data models, such as Random Forest Regression, to anticipate project outcomes and risks, enhancing decision-making in project management. This feature plays a critical role in the strategic allocation of resources, optimizing project timelines, and promoting overall project success. In an effort to substantiate the efficacy and usability of ITC, we have also incorporated the results and feedback garnered from a comprehensive user assessment conducted in 2022. The feedback suggests promising potential for the platform’s application, setting the stage for further development and refinement. The insights provided in this paper not only underline the successful implementation of the ITC platform but also shed light on the transformative impact of predictive analytics in information systems.
This is a preview of subscription content, log in via an institution to check access.
Access this chapter
Subscribe and save.
- Get 10 units per month
- Download Article/Chapter or eBook
- 1 Unit = 1 Article or 1 Chapter
- Cancel anytime
- Available as PDF
- Read on any device
- Instant download
- Own it forever
- Available as EPUB and PDF
- Compact, lightweight edition
- Dispatched in 3 to 5 business days
- Free shipping worldwide - see info
Tax calculation will be finalised at checkout
Purchases are for personal use only
Institutional subscriptions
Similar content being viewed by others

Importance of Project Management in Business Analytics: Academia and Real World

Project Management: Basics

Data Mining-Based Metrics for the Systematic Evaluation of Software Project Management Methodologies
Patanakul, P., Iewwongcharoen, B., Milosevic, D.: An empirical study on the use of project management tools and techniques across project life-cycle and their impact on project success. J. Gen. Manag. 35 (3), 41–66 (2010)
Google Scholar
el Emam, K., Koru, A.G.: A replicated survey of IT software project failures. IEEE Softw. 25 (5), 84–90 (2008)
Article Google Scholar
da Silva, R.: ITLingo Research Initiative in 2022,” arXiv preprint arXiv:2206.14553 (2022)
Nayebi, M., Ruhe, G., Mota, R.C., Mufti, M.: Analytics for software project management - Where are we and where do we go? In: Proceedings - 2015 30th IEEE/ACM International Conference on Automated Software Engineering Workshops, ASEW 2015, March 2016, pp. 18–21 (2016). https://doi.org/10.1109/ASEW.2015.28
Kanakaris, N., Karacapilidis, N., Kournetas, G., Lazanas, A.: Combining machine learning and operations research methods to advance the project management practice. In: Operations Research and Enterprise Systems, pp. 135–155 (2020). https://doi.org/10.1007/978-3-030-37584-3_7
Novitzká, V., et al.: Informatics 2017 : 2017 IEEE 14th International Scientific Conference on Informatics : Proceedings : 14–16 November 2017, Poprad, Slovakia (2017)
Gamito, I., da Silva, A.R.: From rigorous requirements and user interfaces specifications into software business applications. In: International Conference on the Quality of Information and Communications Technology, pp. 459–473 (2020). https://doi.org/10.1007/978-3-030-58793-2_37
da Silva, R.: Rigorous specification of use cases with the RSL language. In: International Conference on Information Systems Development’2019, AIS (2019)
Smith, A., Gupta, J.N.D.: Neural networks in business: techniques and applications for the operations researcher. Comput. Oper. Res. 27 (11), 1023–1044 (2000). https://doi.org/10.1016/S0305-0548(99)00141-0
Article MATH Google Scholar
Wang, Y.-R., Yu, C.-Y., Chan, H.-H.: Predicting construction cost and schedule success using artificial neural networks ensemble and support vector machines classification models. Int. J. Project Manage. 30 (4), 470–478 (2012). https://doi.org/10.1016/j.ijproman.2011.09.002
Costantino, F., di Gravio, G., Nonino, F.: Project selection in project portfolio management: an artificial neural network model based on critical success factors. Int. J. Project Manage. 33 (8), 1744–1754 (2015). https://doi.org/10.1016/j.ijproman.2015.07.003
Sadiku, M., Shadare, A.E., Musa, S.M., Akujuobi, C.M., Perry, R.: Data visualization. Int. J. Eng. Res. Adv. Technol. (IJERAT) 2 (12), 11–16 (2016)
Zheng, G.: Data visualization in business intelligence. In: Global Business Intelligence, Routledge , pp. 67–81 (2017)
Hardin, M., Hom, D., Perez, R., Williams, L.: Which chart or graph is right for you? Tell Impactful Stories with Data. Tableau Software (2012)
Vetter, T.R.: Fundamentals of research data and variables: the devil is in the details. Anesth. Analg. 125 (4), 1375–1380 (2017)
Cabri, A., Griffiths, M.: Earned value and agile reporting. In: AGILE 2006 (AGILE 2006), pp. 6-p (2006)
Project Management Institute., A guide to the project management body of knowledge (PMBOK Guide). Project Management Institute (2008)
Stellingwerf, R., Zandhuis, A.: ISO 21500 Guidance on project management–A Pocket Guide. Van Haren (2013)
Vukomanović, M., Young, M., Huynink, S.: IPMA ICB 4.0—A global standard for project, programme and portfolio management competences. Int. J. Project Manage. 34 (8), 1703–1705 (2016)
E. Commission and D.-G. for Informatics, PM2 project management methodology : guide 3.0.1. Publications Office (2021). https://doi.org/10.2799/022317
Charvat, J.: Project management methodologies: selecting, implementing, and supporting methodologies and processes for projects (2003)
Thesing, T., Feldmann, C., Burchardt, M.: Agile versus waterfall project management: decision model for selecting the appropriate approach to a project. Procedia Comput. Sci. 181 , 746–756 (2021). https://doi.org/10.1016/j.procs.2021.01.227
Špundak, M.: Mixed agile/traditional project management methodology–reality or illusion? Procedia-Soc. Behav. Sci. 119 , 939–948 (2014)
Beck, K., et al.: Manifesto for agile software development (2001)
Alqudah, M., Razali, R.: A comparison of scrum and Kanban for identifying their selection factors. In: 2017 6th International Conference on Electrical Engineering and Informatics (ICEEI), pp. 1–6 (2017). https://doi.org/10.1109/ICEEI.2017.8312434
de Carvalho Bragança, D.: Document Automation in ITLingo PSL Excel Template: MSc dissertation. Instituto Superior Técnico, Universidade de Lisboa (2021)
Ravindran, A.: Django Design Patterns and Best Practices. Packt Publishing Ltd (2015)
Nielsen, J., Landauer, T.K.: A mathematical model of the finding of usability problems. In: Proceedings of the INTERACT 1993 and CHI 1993 Conference on Human Factors in Computing Systems, pp. 206–213 (1993)
Breiman, L.: Random Forests. Mach. Learn. 45 (1), 5–32 (2001)
Liaw, A., Wiener, M.: Classification and regression by Random Forest. R. News 2 (3), 18–22 (2002)
Genuer, R., Poggi, J.M., Tuleau-Malot, C.: Variable selection using Random Forests. Pattern Recogn. Lett. 31 (14), 2225–2236 (2010)
Download references
Author information
Authors and affiliations.
Instituto Superior Técnico, Universidade de Lisboa, Lisbon, Portugal
André Barrocas
INESC-ID, Instituto Superior Técnico, Universidade de Lisboa, Lisbon, Portugal
Alberto Rodrigues da Silva
DefineScope, Setúba, Portugal
João Saraiva
You can also search for this author in PubMed Google Scholar
Corresponding author
Correspondence to Alberto Rodrigues da Silva .
Editor information
Editors and affiliations.
University of Aveiro, Aveiro, Portugal
José Maria Fernandes
Federal University of Rio de Janeiro, Rio de Janeiro, Brazil
Guilherme H. Travassos
University of Oulu, Oulu, Finland
Valentina Lenarduzzi
Xiaozhou Li
Rights and permissions
Reprints and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper.
Barrocas, A., da Silva, A.R., Saraiva, J. (2023). Exploring Data Analysis and Visualization Techniques for Project Tracking: Insights from the ITC. In: Fernandes, J.M., Travassos, G.H., Lenarduzzi, V., Li, X. (eds) Quality of Information and Communications Technology. QUATIC 2023. Communications in Computer and Information Science, vol 1871. Springer, Cham. https://doi.org/10.1007/978-3-031-43703-8_11
Download citation
DOI : https://doi.org/10.1007/978-3-031-43703-8_11
Published : 13 September 2023
Publisher Name : Springer, Cham
Print ISBN : 978-3-031-43702-1
Online ISBN : 978-3-031-43703-8
eBook Packages : Computer Science Computer Science (R0)
Share this paper
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Publish with us
Policies and ethics
- Find a journal
- Track your research

Qualitative Data Analysis Methods 101:
The “big 6” methods + examples.
By: Kerryn Warren (PhD) | Reviewed By: Eunice Rautenbach (D.Tech) | May 2020 (Updated April 2023)
Qualitative data analysis methods. Wow, that’s a mouthful.
If you’re new to the world of research, qualitative data analysis can look rather intimidating. So much bulky terminology and so many abstract, fluffy concepts. It certainly can be a minefield!
Don’t worry – in this post, we’ll unpack the most popular analysis methods , one at a time, so that you can approach your analysis with confidence and competence – whether that’s for a dissertation, thesis or really any kind of research project.
What (exactly) is qualitative data analysis?
To understand qualitative data analysis, we need to first understand qualitative data – so let’s step back and ask the question, “what exactly is qualitative data?”.
Qualitative data refers to pretty much any data that’s “not numbers” . In other words, it’s not the stuff you measure using a fixed scale or complex equipment, nor do you analyse it using complex statistics or mathematics.
So, if it’s not numbers, what is it?
Words, you guessed? Well… sometimes , yes. Qualitative data can, and often does, take the form of interview transcripts, documents and open-ended survey responses – but it can also involve the interpretation of images and videos. In other words, qualitative isn’t just limited to text-based data.
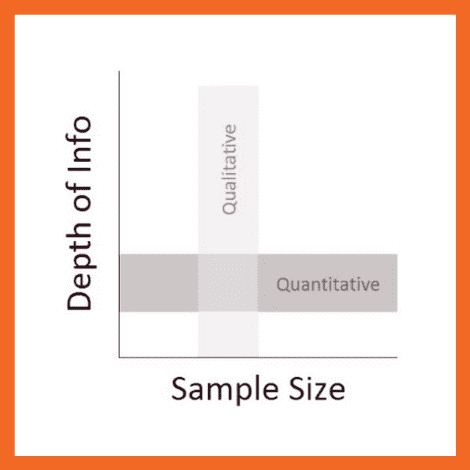
So, how’s that different from quantitative data, you ask?
Simply put, qualitative research focuses on words, descriptions, concepts or ideas – while quantitative research focuses on numbers and statistics . Qualitative research investigates the “softer side” of things to explore and describe , while quantitative research focuses on the “hard numbers”, to measure differences between variables and the relationships between them. If you’re keen to learn more about the differences between qual and quant, we’ve got a detailed post over here .
So, qualitative analysis is easier than quantitative, right?
Not quite. In many ways, qualitative data can be challenging and time-consuming to analyse and interpret. At the end of your data collection phase (which itself takes a lot of time), you’ll likely have many pages of text-based data or hours upon hours of audio to work through. You might also have subtle nuances of interactions or discussions that have danced around in your mind, or that you scribbled down in messy field notes. All of this needs to work its way into your analysis.
Making sense of all of this is no small task and you shouldn’t underestimate it. Long story short – qualitative analysis can be a lot of work! Of course, quantitative analysis is no piece of cake either, but it’s important to recognise that qualitative analysis still requires a significant investment in terms of time and effort.
Need a helping hand?
In this post, we’ll explore qualitative data analysis by looking at some of the most common analysis methods we encounter. We’re not going to cover every possible qualitative method and we’re not going to go into heavy detail – we’re just going to give you the big picture. That said, we will of course includes links to loads of extra resources so that you can learn more about whichever analysis method interests you.
Without further delay, let’s get into it.
The “Big 6” Qualitative Analysis Methods
There are many different types of qualitative data analysis, all of which serve different purposes and have unique strengths and weaknesses . We’ll start by outlining the analysis methods and then we’ll dive into the details for each.
The 6 most popular methods (or at least the ones we see at Grad Coach) are:
- Content analysis
- Narrative analysis
- Discourse analysis
- Thematic analysis
- Grounded theory (GT)
- Interpretive phenomenological analysis (IPA)
Let’s take a look at each of them…
QDA Method #1: Qualitative Content Analysis
Content analysis is possibly the most common and straightforward QDA method. At the simplest level, content analysis is used to evaluate patterns within a piece of content (for example, words, phrases or images) or across multiple pieces of content or sources of communication. For example, a collection of newspaper articles or political speeches.
With content analysis, you could, for instance, identify the frequency with which an idea is shared or spoken about – like the number of times a Kardashian is mentioned on Twitter. Or you could identify patterns of deeper underlying interpretations – for instance, by identifying phrases or words in tourist pamphlets that highlight India as an ancient country.
Because content analysis can be used in such a wide variety of ways, it’s important to go into your analysis with a very specific question and goal, or you’ll get lost in the fog. With content analysis, you’ll group large amounts of text into codes , summarise these into categories, and possibly even tabulate the data to calculate the frequency of certain concepts or variables. Because of this, content analysis provides a small splash of quantitative thinking within a qualitative method.
Naturally, while content analysis is widely useful, it’s not without its drawbacks . One of the main issues with content analysis is that it can be very time-consuming , as it requires lots of reading and re-reading of the texts. Also, because of its multidimensional focus on both qualitative and quantitative aspects, it is sometimes accused of losing important nuances in communication.
Content analysis also tends to concentrate on a very specific timeline and doesn’t take into account what happened before or after that timeline. This isn’t necessarily a bad thing though – just something to be aware of. So, keep these factors in mind if you’re considering content analysis. Every analysis method has its limitations , so don’t be put off by these – just be aware of them ! If you’re interested in learning more about content analysis, the video below provides a good starting point.
QDA Method #2: Narrative Analysis
As the name suggests, narrative analysis is all about listening to people telling stories and analysing what that means . Since stories serve a functional purpose of helping us make sense of the world, we can gain insights into the ways that people deal with and make sense of reality by analysing their stories and the ways they’re told.
You could, for example, use narrative analysis to explore whether how something is being said is important. For instance, the narrative of a prisoner trying to justify their crime could provide insight into their view of the world and the justice system. Similarly, analysing the ways entrepreneurs talk about the struggles in their careers or cancer patients telling stories of hope could provide powerful insights into their mindsets and perspectives . Simply put, narrative analysis is about paying attention to the stories that people tell – and more importantly, the way they tell them.
Of course, the narrative approach has its weaknesses , too. Sample sizes are generally quite small due to the time-consuming process of capturing narratives. Because of this, along with the multitude of social and lifestyle factors which can influence a subject, narrative analysis can be quite difficult to reproduce in subsequent research. This means that it’s difficult to test the findings of some of this research.
Similarly, researcher bias can have a strong influence on the results here, so you need to be particularly careful about the potential biases you can bring into your analysis when using this method. Nevertheless, narrative analysis is still a very useful qualitative analysis method – just keep these limitations in mind and be careful not to draw broad conclusions . If you’re keen to learn more about narrative analysis, the video below provides a great introduction to this qualitative analysis method.
QDA Method #3: Discourse Analysis
Discourse is simply a fancy word for written or spoken language or debate . So, discourse analysis is all about analysing language within its social context. In other words, analysing language – such as a conversation, a speech, etc – within the culture and society it takes place. For example, you could analyse how a janitor speaks to a CEO, or how politicians speak about terrorism.
To truly understand these conversations or speeches, the culture and history of those involved in the communication are important factors to consider. For example, a janitor might speak more casually with a CEO in a company that emphasises equality among workers. Similarly, a politician might speak more about terrorism if there was a recent terrorist incident in the country.
So, as you can see, by using discourse analysis, you can identify how culture , history or power dynamics (to name a few) have an effect on the way concepts are spoken about. So, if your research aims and objectives involve understanding culture or power dynamics, discourse analysis can be a powerful method.
Because there are many social influences in terms of how we speak to each other, the potential use of discourse analysis is vast . Of course, this also means it’s important to have a very specific research question (or questions) in mind when analysing your data and looking for patterns and themes, or you might land up going down a winding rabbit hole.
Discourse analysis can also be very time-consuming as you need to sample the data to the point of saturation – in other words, until no new information and insights emerge. But this is, of course, part of what makes discourse analysis such a powerful technique. So, keep these factors in mind when considering this QDA method. Again, if you’re keen to learn more, the video below presents a good starting point.
QDA Method #4: Thematic Analysis
Thematic analysis looks at patterns of meaning in a data set – for example, a set of interviews or focus group transcripts. But what exactly does that… mean? Well, a thematic analysis takes bodies of data (which are often quite large) and groups them according to similarities – in other words, themes . These themes help us make sense of the content and derive meaning from it.
Let’s take a look at an example.
With thematic analysis, you could analyse 100 online reviews of a popular sushi restaurant to find out what patrons think about the place. By reviewing the data, you would then identify the themes that crop up repeatedly within the data – for example, “fresh ingredients” or “friendly wait staff”.
So, as you can see, thematic analysis can be pretty useful for finding out about people’s experiences , views, and opinions . Therefore, if your research aims and objectives involve understanding people’s experience or view of something, thematic analysis can be a great choice.
Since thematic analysis is a bit of an exploratory process, it’s not unusual for your research questions to develop , or even change as you progress through the analysis. While this is somewhat natural in exploratory research, it can also be seen as a disadvantage as it means that data needs to be re-reviewed each time a research question is adjusted. In other words, thematic analysis can be quite time-consuming – but for a good reason. So, keep this in mind if you choose to use thematic analysis for your project and budget extra time for unexpected adjustments.

QDA Method #5: Grounded theory (GT)
Grounded theory is a powerful qualitative analysis method where the intention is to create a new theory (or theories) using the data at hand, through a series of “ tests ” and “ revisions ”. Strictly speaking, GT is more a research design type than an analysis method, but we’ve included it here as it’s often referred to as a method.
What’s most important with grounded theory is that you go into the analysis with an open mind and let the data speak for itself – rather than dragging existing hypotheses or theories into your analysis. In other words, your analysis must develop from the ground up (hence the name).
Let’s look at an example of GT in action.
Assume you’re interested in developing a theory about what factors influence students to watch a YouTube video about qualitative analysis. Using Grounded theory , you’d start with this general overarching question about the given population (i.e., graduate students). First, you’d approach a small sample – for example, five graduate students in a department at a university. Ideally, this sample would be reasonably representative of the broader population. You’d interview these students to identify what factors lead them to watch the video.
After analysing the interview data, a general pattern could emerge. For example, you might notice that graduate students are more likely to read a post about qualitative methods if they are just starting on their dissertation journey, or if they have an upcoming test about research methods.
From here, you’ll look for another small sample – for example, five more graduate students in a different department – and see whether this pattern holds true for them. If not, you’ll look for commonalities and adapt your theory accordingly. As this process continues, the theory would develop . As we mentioned earlier, what’s important with grounded theory is that the theory develops from the data – not from some preconceived idea.
So, what are the drawbacks of grounded theory? Well, some argue that there’s a tricky circularity to grounded theory. For it to work, in principle, you should know as little as possible regarding the research question and population, so that you reduce the bias in your interpretation. However, in many circumstances, it’s also thought to be unwise to approach a research question without knowledge of the current literature . In other words, it’s a bit of a “chicken or the egg” situation.
Regardless, grounded theory remains a popular (and powerful) option. Naturally, it’s a very useful method when you’re researching a topic that is completely new or has very little existing research about it, as it allows you to start from scratch and work your way from the ground up .

QDA Method #6: Interpretive Phenomenological Analysis (IPA)
Interpretive. Phenomenological. Analysis. IPA . Try saying that three times fast…
Let’s just stick with IPA, okay?
IPA is designed to help you understand the personal experiences of a subject (for example, a person or group of people) concerning a major life event, an experience or a situation . This event or experience is the “phenomenon” that makes up the “P” in IPA. Such phenomena may range from relatively common events – such as motherhood, or being involved in a car accident – to those which are extremely rare – for example, someone’s personal experience in a refugee camp. So, IPA is a great choice if your research involves analysing people’s personal experiences of something that happened to them.
It’s important to remember that IPA is subject – centred . In other words, it’s focused on the experiencer . This means that, while you’ll likely use a coding system to identify commonalities, it’s important not to lose the depth of experience or meaning by trying to reduce everything to codes. Also, keep in mind that since your sample size will generally be very small with IPA, you often won’t be able to draw broad conclusions about the generalisability of your findings. But that’s okay as long as it aligns with your research aims and objectives.
Another thing to be aware of with IPA is personal bias . While researcher bias can creep into all forms of research, self-awareness is critically important with IPA, as it can have a major impact on the results. For example, a researcher who was a victim of a crime himself could insert his own feelings of frustration and anger into the way he interprets the experience of someone who was kidnapped. So, if you’re going to undertake IPA, you need to be very self-aware or you could muddy the analysis.

How to choose the right analysis method
In light of all of the qualitative analysis methods we’ve covered so far, you’re probably asking yourself the question, “ How do I choose the right one? ”
Much like all the other methodological decisions you’ll need to make, selecting the right qualitative analysis method largely depends on your research aims, objectives and questions . In other words, the best tool for the job depends on what you’re trying to build. For example:
- Perhaps your research aims to analyse the use of words and what they reveal about the intention of the storyteller and the cultural context of the time.
- Perhaps your research aims to develop an understanding of the unique personal experiences of people that have experienced a certain event, or
- Perhaps your research aims to develop insight regarding the influence of a certain culture on its members.
As you can probably see, each of these research aims are distinctly different , and therefore different analysis methods would be suitable for each one. For example, narrative analysis would likely be a good option for the first aim, while grounded theory wouldn’t be as relevant.
It’s also important to remember that each method has its own set of strengths, weaknesses and general limitations. No single analysis method is perfect . So, depending on the nature of your research, it may make sense to adopt more than one method (this is called triangulation ). Keep in mind though that this will of course be quite time-consuming.
As we’ve seen, all of the qualitative analysis methods we’ve discussed make use of coding and theme-generating techniques, but the intent and approach of each analysis method differ quite substantially. So, it’s very important to come into your research with a clear intention before you decide which analysis method (or methods) to use.
Start by reviewing your research aims , objectives and research questions to assess what exactly you’re trying to find out – then select a qualitative analysis method that fits. Never pick a method just because you like it or have experience using it – your analysis method (or methods) must align with your broader research aims and objectives.

Let’s recap on QDA methods…
In this post, we looked at six popular qualitative data analysis methods:
- First, we looked at content analysis , a straightforward method that blends a little bit of quant into a primarily qualitative analysis.
- Then we looked at narrative analysis , which is about analysing how stories are told.
- Next up was discourse analysis – which is about analysing conversations and interactions.
- Then we moved on to thematic analysis – which is about identifying themes and patterns.
- From there, we went south with grounded theory – which is about starting from scratch with a specific question and using the data alone to build a theory in response to that question.
- And finally, we looked at IPA – which is about understanding people’s unique experiences of a phenomenon.
Of course, these aren’t the only options when it comes to qualitative data analysis, but they’re a great starting point if you’re dipping your toes into qualitative research for the first time.
If you’re still feeling a bit confused, consider our private coaching service , where we hold your hand through the research process to help you develop your best work.

Psst... there’s more!
This post was based on one of our popular Research Bootcamps . If you're working on a research project, you'll definitely want to check this out ...
87 Comments
This has been very helpful. Thank you.
Thank you madam,
Thank you so much for this information
I wonder it so clear for understand and good for me. can I ask additional query?
Very insightful and useful
Good work done with clear explanations. Thank you.
Thanks so much for the write-up, it’s really good.
Thanks madam . It is very important .
thank you very good
Great presentation
This has been very well explained in simple language . It is useful even for a new researcher.
Great to hear that. Good luck with your qualitative data analysis, Pramod!
This is very useful information. And it was very a clear language structured presentation. Thanks a lot.
Thank you so much.
very informative sequential presentation
Precise explanation of method.
Hi, may we use 2 data analysis methods in our qualitative research?
Thanks for your comment. Most commonly, one would use one type of analysis method, but it depends on your research aims and objectives.
You explained it in very simple language, everyone can understand it. Thanks so much.
Thank you very much, this is very helpful. It has been explained in a very simple manner that even a layman understands
Thank nicely explained can I ask is Qualitative content analysis the same as thematic analysis?
Thanks for your comment. No, QCA and thematic are two different types of analysis. This article might help clarify – https://onlinelibrary.wiley.com/doi/10.1111/nhs.12048
This is my first time to come across a well explained data analysis. so helpful.
I have thoroughly enjoyed your explanation of the six qualitative analysis methods. This is very helpful. Thank you!
Thank you very much, this is well explained and useful
i need a citation of your book.
Thanks a lot , remarkable indeed, enlighting to the best
Hi Derek, What other theories/methods would you recommend when the data is a whole speech?
Keep writing useful artikel.
It is important concept about QDA and also the way to express is easily understandable, so thanks for all.
Thank you, this is well explained and very useful.
Very helpful .Thanks.
Hi there! Very well explained. Simple but very useful style of writing. Please provide the citation of the text. warm regards
The session was very helpful and insightful. Thank you
This was very helpful and insightful. Easy to read and understand
As a professional academic writer, this has been so informative and educative. Keep up the good work Grad Coach you are unmatched with quality content for sure.
Keep up the good work Grad Coach you are unmatched with quality content for sure.
Its Great and help me the most. A Million Thanks you Dr.
It is a very nice work
Very insightful. Please, which of this approach could be used for a research that one is trying to elicit students’ misconceptions in a particular concept ?
This is Amazing and well explained, thanks
great overview
What do we call a research data analysis method that one use to advise or determining the best accounting tool or techniques that should be adopted in a company.
Informative video, explained in a clear and simple way. Kudos
Waoo! I have chosen method wrong for my data analysis. But I can revise my work according to this guide. Thank you so much for this helpful lecture.
This has been very helpful. It gave me a good view of my research objectives and how to choose the best method. Thematic analysis it is.
Very helpful indeed. Thanku so much for the insight.
This was incredibly helpful.
Very helpful.
very educative
Nicely written especially for novice academic researchers like me! Thank you.
choosing a right method for a paper is always a hard job for a student, this is a useful information, but it would be more useful personally for me, if the author provide me with a little bit more information about the data analysis techniques in type of explanatory research. Can we use qualitative content analysis technique for explanatory research ? or what is the suitable data analysis method for explanatory research in social studies?
that was very helpful for me. because these details are so important to my research. thank you very much
I learnt a lot. Thank you
Relevant and Informative, thanks !
Well-planned and organized, thanks much! 🙂
I have reviewed qualitative data analysis in a simplest way possible. The content will highly be useful for developing my book on qualitative data analysis methods. Cheers!
Clear explanation on qualitative and how about Case study
This was helpful. Thank you
This was really of great assistance, it was just the right information needed. Explanation very clear and follow.
Wow, Thanks for making my life easy
This was helpful thanks .
Very helpful…. clear and written in an easily understandable manner. Thank you.
This was so helpful as it was easy to understand. I’m a new to research thank you so much.
so educative…. but Ijust want to know which method is coding of the qualitative or tallying done?
Thank you for the great content, I have learnt a lot. So helpful
precise and clear presentation with simple language and thank you for that.
very informative content, thank you.
You guys are amazing on YouTube on this platform. Your teachings are great, educative, and informative. kudos!
Brilliant Delivery. You made a complex subject seem so easy. Well done.
Beautifully explained.
Thanks a lot
Is there a video the captures the practical process of coding using automated applications?
Thanks for the comment. We don’t recommend using automated applications for coding, as they are not sufficiently accurate in our experience.
content analysis can be qualitative research?
THANK YOU VERY MUCH.
Thank you very much for such a wonderful content
do you have any material on Data collection
What a powerful explanation of the QDA methods. Thank you.
Great explanation both written and Video. i have been using of it on a day to day working of my thesis project in accounting and finance. Thank you very much for your support.
very helpful, thank you so much
The tutorial is useful. I benefited a lot.
This is an eye opener for me and very informative, I have used some of your guidance notes on my Thesis, I wonder if you can assist with your 1. name of your book, year of publication, topic etc., this is for citing in my Bibliography,
I certainly hope to hear from you
Submit a Comment Cancel reply
Your email address will not be published. Required fields are marked *
Save my name, email, and website in this browser for the next time I comment.
- Print Friendly

Data Analysis Techniques in Research – Methods, Tools & Examples
Varun Saharawat is a seasoned professional in the fields of SEO and content writing. With a profound knowledge of the intricate aspects of these disciplines, Varun has established himself as a valuable asset in the world of digital marketing and online content creation.

Data analysis techniques in research are essential because they allow researchers to derive meaningful insights from data sets to support their hypotheses or research objectives.
Data Analysis Techniques in Research : While various groups, institutions, and professionals may have diverse approaches to data analysis, a universal definition captures its essence. Data analysis involves refining, transforming, and interpreting raw data to derive actionable insights that guide informed decision-making for businesses.

A straightforward illustration of data analysis emerges when we make everyday decisions, basing our choices on past experiences or predictions of potential outcomes.
If you want to learn more about this topic and acquire valuable skills that will set you apart in today’s data-driven world, we highly recommend enrolling in the Data Analytics Course by Physics Wallah . And as a special offer for our readers, use the coupon code “READER” to get a discount on this course.
Table of Contents
What is Data Analysis?
Data analysis is the systematic process of inspecting, cleaning, transforming, and interpreting data with the objective of discovering valuable insights and drawing meaningful conclusions. This process involves several steps:
- Inspecting : Initial examination of data to understand its structure, quality, and completeness.
- Cleaning : Removing errors, inconsistencies, or irrelevant information to ensure accurate analysis.
- Transforming : Converting data into a format suitable for analysis, such as normalization or aggregation.
- Interpreting : Analyzing the transformed data to identify patterns, trends, and relationships.
Types of Data Analysis Techniques in Research
Data analysis techniques in research are categorized into qualitative and quantitative methods, each with its specific approaches and tools. These techniques are instrumental in extracting meaningful insights, patterns, and relationships from data to support informed decision-making, validate hypotheses, and derive actionable recommendations. Below is an in-depth exploration of the various types of data analysis techniques commonly employed in research:
1) Qualitative Analysis:
Definition: Qualitative analysis focuses on understanding non-numerical data, such as opinions, concepts, or experiences, to derive insights into human behavior, attitudes, and perceptions.
- Content Analysis: Examines textual data, such as interview transcripts, articles, or open-ended survey responses, to identify themes, patterns, or trends.
- Narrative Analysis: Analyzes personal stories or narratives to understand individuals’ experiences, emotions, or perspectives.
- Ethnographic Studies: Involves observing and analyzing cultural practices, behaviors, and norms within specific communities or settings.
2) Quantitative Analysis:
Quantitative analysis emphasizes numerical data and employs statistical methods to explore relationships, patterns, and trends. It encompasses several approaches:
Descriptive Analysis:
- Frequency Distribution: Represents the number of occurrences of distinct values within a dataset.
- Central Tendency: Measures such as mean, median, and mode provide insights into the central values of a dataset.
- Dispersion: Techniques like variance and standard deviation indicate the spread or variability of data.
Diagnostic Analysis:
- Regression Analysis: Assesses the relationship between dependent and independent variables, enabling prediction or understanding causality.
- ANOVA (Analysis of Variance): Examines differences between groups to identify significant variations or effects.
Predictive Analysis:
- Time Series Forecasting: Uses historical data points to predict future trends or outcomes.
- Machine Learning Algorithms: Techniques like decision trees, random forests, and neural networks predict outcomes based on patterns in data.
Prescriptive Analysis:
- Optimization Models: Utilizes linear programming, integer programming, or other optimization techniques to identify the best solutions or strategies.
- Simulation: Mimics real-world scenarios to evaluate various strategies or decisions and determine optimal outcomes.
Specific Techniques:
- Monte Carlo Simulation: Models probabilistic outcomes to assess risk and uncertainty.
- Factor Analysis: Reduces the dimensionality of data by identifying underlying factors or components.
- Cohort Analysis: Studies specific groups or cohorts over time to understand trends, behaviors, or patterns within these groups.
- Cluster Analysis: Classifies objects or individuals into homogeneous groups or clusters based on similarities or attributes.
- Sentiment Analysis: Uses natural language processing and machine learning techniques to determine sentiment, emotions, or opinions from textual data.
Also Read: AI and Predictive Analytics: Examples, Tools, Uses, Ai Vs Predictive Analytics
Data Analysis Techniques in Research Examples
To provide a clearer understanding of how data analysis techniques are applied in research, let’s consider a hypothetical research study focused on evaluating the impact of online learning platforms on students’ academic performance.
Research Objective:
Determine if students using online learning platforms achieve higher academic performance compared to those relying solely on traditional classroom instruction.
Data Collection:
- Quantitative Data: Academic scores (grades) of students using online platforms and those using traditional classroom methods.
- Qualitative Data: Feedback from students regarding their learning experiences, challenges faced, and preferences.
Data Analysis Techniques Applied:
1) Descriptive Analysis:
- Calculate the mean, median, and mode of academic scores for both groups.
- Create frequency distributions to represent the distribution of grades in each group.
2) Diagnostic Analysis:
- Conduct an Analysis of Variance (ANOVA) to determine if there’s a statistically significant difference in academic scores between the two groups.
- Perform Regression Analysis to assess the relationship between the time spent on online platforms and academic performance.
3) Predictive Analysis:
- Utilize Time Series Forecasting to predict future academic performance trends based on historical data.
- Implement Machine Learning algorithms to develop a predictive model that identifies factors contributing to academic success on online platforms.
4) Prescriptive Analysis:
- Apply Optimization Models to identify the optimal combination of online learning resources (e.g., video lectures, interactive quizzes) that maximize academic performance.
- Use Simulation Techniques to evaluate different scenarios, such as varying student engagement levels with online resources, to determine the most effective strategies for improving learning outcomes.
5) Specific Techniques:
- Conduct Factor Analysis on qualitative feedback to identify common themes or factors influencing students’ perceptions and experiences with online learning.
- Perform Cluster Analysis to segment students based on their engagement levels, preferences, or academic outcomes, enabling targeted interventions or personalized learning strategies.
- Apply Sentiment Analysis on textual feedback to categorize students’ sentiments as positive, negative, or neutral regarding online learning experiences.
By applying a combination of qualitative and quantitative data analysis techniques, this research example aims to provide comprehensive insights into the effectiveness of online learning platforms.
Also Read: Learning Path to Become a Data Analyst in 2024
Data Analysis Techniques in Quantitative Research
Quantitative research involves collecting numerical data to examine relationships, test hypotheses, and make predictions. Various data analysis techniques are employed to interpret and draw conclusions from quantitative data. Here are some key data analysis techniques commonly used in quantitative research:
1) Descriptive Statistics:
- Description: Descriptive statistics are used to summarize and describe the main aspects of a dataset, such as central tendency (mean, median, mode), variability (range, variance, standard deviation), and distribution (skewness, kurtosis).
- Applications: Summarizing data, identifying patterns, and providing initial insights into the dataset.
2) Inferential Statistics:
- Description: Inferential statistics involve making predictions or inferences about a population based on a sample of data. This technique includes hypothesis testing, confidence intervals, t-tests, chi-square tests, analysis of variance (ANOVA), regression analysis, and correlation analysis.
- Applications: Testing hypotheses, making predictions, and generalizing findings from a sample to a larger population.
3) Regression Analysis:
- Description: Regression analysis is a statistical technique used to model and examine the relationship between a dependent variable and one or more independent variables. Linear regression, multiple regression, logistic regression, and nonlinear regression are common types of regression analysis .
- Applications: Predicting outcomes, identifying relationships between variables, and understanding the impact of independent variables on the dependent variable.
4) Correlation Analysis:
- Description: Correlation analysis is used to measure and assess the strength and direction of the relationship between two or more variables. The Pearson correlation coefficient, Spearman rank correlation coefficient, and Kendall’s tau are commonly used measures of correlation.
- Applications: Identifying associations between variables and assessing the degree and nature of the relationship.
5) Factor Analysis:
- Description: Factor analysis is a multivariate statistical technique used to identify and analyze underlying relationships or factors among a set of observed variables. It helps in reducing the dimensionality of data and identifying latent variables or constructs.
- Applications: Identifying underlying factors or constructs, simplifying data structures, and understanding the underlying relationships among variables.
6) Time Series Analysis:
- Description: Time series analysis involves analyzing data collected or recorded over a specific period at regular intervals to identify patterns, trends, and seasonality. Techniques such as moving averages, exponential smoothing, autoregressive integrated moving average (ARIMA), and Fourier analysis are used.
- Applications: Forecasting future trends, analyzing seasonal patterns, and understanding time-dependent relationships in data.
7) ANOVA (Analysis of Variance):
- Description: Analysis of variance (ANOVA) is a statistical technique used to analyze and compare the means of two or more groups or treatments to determine if they are statistically different from each other. One-way ANOVA, two-way ANOVA, and MANOVA (Multivariate Analysis of Variance) are common types of ANOVA.
- Applications: Comparing group means, testing hypotheses, and determining the effects of categorical independent variables on a continuous dependent variable.
8) Chi-Square Tests:
- Description: Chi-square tests are non-parametric statistical tests used to assess the association between categorical variables in a contingency table. The Chi-square test of independence, goodness-of-fit test, and test of homogeneity are common chi-square tests.
- Applications: Testing relationships between categorical variables, assessing goodness-of-fit, and evaluating independence.
These quantitative data analysis techniques provide researchers with valuable tools and methods to analyze, interpret, and derive meaningful insights from numerical data. The selection of a specific technique often depends on the research objectives, the nature of the data, and the underlying assumptions of the statistical methods being used.
Also Read: Analysis vs. Analytics: How Are They Different?
Data Analysis Methods
Data analysis methods refer to the techniques and procedures used to analyze, interpret, and draw conclusions from data. These methods are essential for transforming raw data into meaningful insights, facilitating decision-making processes, and driving strategies across various fields. Here are some common data analysis methods:
- Description: Descriptive statistics summarize and organize data to provide a clear and concise overview of the dataset. Measures such as mean, median, mode, range, variance, and standard deviation are commonly used.
- Description: Inferential statistics involve making predictions or inferences about a population based on a sample of data. Techniques such as hypothesis testing, confidence intervals, and regression analysis are used.
3) Exploratory Data Analysis (EDA):
- Description: EDA techniques involve visually exploring and analyzing data to discover patterns, relationships, anomalies, and insights. Methods such as scatter plots, histograms, box plots, and correlation matrices are utilized.
- Applications: Identifying trends, patterns, outliers, and relationships within the dataset.
4) Predictive Analytics:
- Description: Predictive analytics use statistical algorithms and machine learning techniques to analyze historical data and make predictions about future events or outcomes. Techniques such as regression analysis, time series forecasting, and machine learning algorithms (e.g., decision trees, random forests, neural networks) are employed.
- Applications: Forecasting future trends, predicting outcomes, and identifying potential risks or opportunities.
5) Prescriptive Analytics:
- Description: Prescriptive analytics involve analyzing data to recommend actions or strategies that optimize specific objectives or outcomes. Optimization techniques, simulation models, and decision-making algorithms are utilized.
- Applications: Recommending optimal strategies, decision-making support, and resource allocation.
6) Qualitative Data Analysis:
- Description: Qualitative data analysis involves analyzing non-numerical data, such as text, images, videos, or audio, to identify themes, patterns, and insights. Methods such as content analysis, thematic analysis, and narrative analysis are used.
- Applications: Understanding human behavior, attitudes, perceptions, and experiences.
7) Big Data Analytics:
- Description: Big data analytics methods are designed to analyze large volumes of structured and unstructured data to extract valuable insights. Technologies such as Hadoop, Spark, and NoSQL databases are used to process and analyze big data.
- Applications: Analyzing large datasets, identifying trends, patterns, and insights from big data sources.
8) Text Analytics:
- Description: Text analytics methods involve analyzing textual data, such as customer reviews, social media posts, emails, and documents, to extract meaningful information and insights. Techniques such as sentiment analysis, text mining, and natural language processing (NLP) are used.
- Applications: Analyzing customer feedback, monitoring brand reputation, and extracting insights from textual data sources.
These data analysis methods are instrumental in transforming data into actionable insights, informing decision-making processes, and driving organizational success across various sectors, including business, healthcare, finance, marketing, and research. The selection of a specific method often depends on the nature of the data, the research objectives, and the analytical requirements of the project or organization.
Also Read: Quantitative Data Analysis: Types, Analysis & Examples
Data Analysis Tools
Data analysis tools are essential instruments that facilitate the process of examining, cleaning, transforming, and modeling data to uncover useful information, make informed decisions, and drive strategies. Here are some prominent data analysis tools widely used across various industries:
1) Microsoft Excel:
- Description: A spreadsheet software that offers basic to advanced data analysis features, including pivot tables, data visualization tools, and statistical functions.
- Applications: Data cleaning, basic statistical analysis, visualization, and reporting.
2) R Programming Language:
- Description: An open-source programming language specifically designed for statistical computing and data visualization.
- Applications: Advanced statistical analysis, data manipulation, visualization, and machine learning.
3) Python (with Libraries like Pandas, NumPy, Matplotlib, and Seaborn):
- Description: A versatile programming language with libraries that support data manipulation, analysis, and visualization.
- Applications: Data cleaning, statistical analysis, machine learning, and data visualization.
4) SPSS (Statistical Package for the Social Sciences):
- Description: A comprehensive statistical software suite used for data analysis, data mining, and predictive analytics.
- Applications: Descriptive statistics, hypothesis testing, regression analysis, and advanced analytics.
5) SAS (Statistical Analysis System):
- Description: A software suite used for advanced analytics, multivariate analysis, and predictive modeling.
- Applications: Data management, statistical analysis, predictive modeling, and business intelligence.
6) Tableau:
- Description: A data visualization tool that allows users to create interactive and shareable dashboards and reports.
- Applications: Data visualization , business intelligence , and interactive dashboard creation.
7) Power BI:
- Description: A business analytics tool developed by Microsoft that provides interactive visualizations and business intelligence capabilities.
- Applications: Data visualization, business intelligence, reporting, and dashboard creation.
8) SQL (Structured Query Language) Databases (e.g., MySQL, PostgreSQL, Microsoft SQL Server):
- Description: Database management systems that support data storage, retrieval, and manipulation using SQL queries.
- Applications: Data retrieval, data cleaning, data transformation, and database management.
9) Apache Spark:
- Description: A fast and general-purpose distributed computing system designed for big data processing and analytics.
- Applications: Big data processing, machine learning, data streaming, and real-time analytics.
10) IBM SPSS Modeler:
- Description: A data mining software application used for building predictive models and conducting advanced analytics.
- Applications: Predictive modeling, data mining, statistical analysis, and decision optimization.
These tools serve various purposes and cater to different data analysis needs, from basic statistical analysis and data visualization to advanced analytics, machine learning, and big data processing. The choice of a specific tool often depends on the nature of the data, the complexity of the analysis, and the specific requirements of the project or organization.
Also Read: How to Analyze Survey Data: Methods & Examples
Importance of Data Analysis in Research
The importance of data analysis in research cannot be overstated; it serves as the backbone of any scientific investigation or study. Here are several key reasons why data analysis is crucial in the research process:
- Data analysis helps ensure that the results obtained are valid and reliable. By systematically examining the data, researchers can identify any inconsistencies or anomalies that may affect the credibility of the findings.
- Effective data analysis provides researchers with the necessary information to make informed decisions. By interpreting the collected data, researchers can draw conclusions, make predictions, or formulate recommendations based on evidence rather than intuition or guesswork.
- Data analysis allows researchers to identify patterns, trends, and relationships within the data. This can lead to a deeper understanding of the research topic, enabling researchers to uncover insights that may not be immediately apparent.
- In empirical research, data analysis plays a critical role in testing hypotheses. Researchers collect data to either support or refute their hypotheses, and data analysis provides the tools and techniques to evaluate these hypotheses rigorously.
- Transparent and well-executed data analysis enhances the credibility of research findings. By clearly documenting the data analysis methods and procedures, researchers allow others to replicate the study, thereby contributing to the reproducibility of research findings.
- In fields such as business or healthcare, data analysis helps organizations allocate resources more efficiently. By analyzing data on consumer behavior, market trends, or patient outcomes, organizations can make strategic decisions about resource allocation, budgeting, and planning.
- In public policy and social sciences, data analysis is instrumental in developing and evaluating policies and interventions. By analyzing data on social, economic, or environmental factors, policymakers can assess the effectiveness of existing policies and inform the development of new ones.
- Data analysis allows for continuous improvement in research methods and practices. By analyzing past research projects, identifying areas for improvement, and implementing changes based on data-driven insights, researchers can refine their approaches and enhance the quality of future research endeavors.
However, it is important to remember that mastering these techniques requires practice and continuous learning. That’s why we highly recommend the Data Analytics Course by Physics Wallah . Not only does it cover all the fundamentals of data analysis, but it also provides hands-on experience with various tools such as Excel, Python, and Tableau. Plus, if you use the “ READER ” coupon code at checkout, you can get a special discount on the course.
For Latest Tech Related Information, Join Our Official Free Telegram Group : PW Skills Telegram Group
Data Analysis Techniques in Research FAQs
What are the 5 techniques for data analysis.
The five techniques for data analysis include: Descriptive Analysis Diagnostic Analysis Predictive Analysis Prescriptive Analysis Qualitative Analysis
What are techniques of data analysis in research?
Techniques of data analysis in research encompass both qualitative and quantitative methods. These techniques involve processes like summarizing raw data, investigating causes of events, forecasting future outcomes, offering recommendations based on predictions, and examining non-numerical data to understand concepts or experiences.
What are the 3 methods of data analysis?
The three primary methods of data analysis are: Qualitative Analysis Quantitative Analysis Mixed-Methods Analysis
What are the four types of data analysis techniques?
The four types of data analysis techniques are: Descriptive Analysis Diagnostic Analysis Predictive Analysis Prescriptive Analysis

- Book About Data Analytics: Top 15 Books You Should Read

In a world with abundant data in different formats, extracting important insights from data is the work of data analytics.…
- Data Analytics Internships – What Does A Data Analyst Intern Do?

Data analytics internships involve working with data to identify trends, create visualizations, and generate reports. Data Analytics Internships learn to…
- 10 Most Popular Analytic Tools in Big Data

Many online analytics tools can be used to perform data analytics. Let us know some of the most frequently used…
Related Articles
- BI & Analytics: What’s The Difference?
- What is Business Analytics?
- Mastering Business Analytics: Strategies and Insights
- Big Data: What Do You Mean By Big Data?
- Why is Data Analytics Skills Important?
- Best 5 Unique Strategies to Use Artificial Intelligence Data Analytics
- BI And Analytics: Understanding The Differences
- Privacy Policy

Home » Data Analysis – Process, Methods and Types
Data Analysis – Process, Methods and Types
Table of Contents

Data Analysis
Definition:
Data analysis refers to the process of inspecting, cleaning, transforming, and modeling data with the goal of discovering useful information, drawing conclusions, and supporting decision-making. It involves applying various statistical and computational techniques to interpret and derive insights from large datasets. The ultimate aim of data analysis is to convert raw data into actionable insights that can inform business decisions, scientific research, and other endeavors.
Data Analysis Process
The following are step-by-step guides to the data analysis process:
Define the Problem
The first step in data analysis is to clearly define the problem or question that needs to be answered. This involves identifying the purpose of the analysis, the data required, and the intended outcome.
Collect the Data
The next step is to collect the relevant data from various sources. This may involve collecting data from surveys, databases, or other sources. It is important to ensure that the data collected is accurate, complete, and relevant to the problem being analyzed.
Clean and Organize the Data
Once the data has been collected, it needs to be cleaned and organized. This involves removing any errors or inconsistencies in the data, filling in missing values, and ensuring that the data is in a format that can be easily analyzed.
Analyze the Data
The next step is to analyze the data using various statistical and analytical techniques. This may involve identifying patterns in the data, conducting statistical tests, or using machine learning algorithms to identify trends and insights.
Interpret the Results
After analyzing the data, the next step is to interpret the results. This involves drawing conclusions based on the analysis and identifying any significant findings or trends.
Communicate the Findings
Once the results have been interpreted, they need to be communicated to stakeholders. This may involve creating reports, visualizations, or presentations to effectively communicate the findings and recommendations.
Take Action
The final step in the data analysis process is to take action based on the findings. This may involve implementing new policies or procedures, making strategic decisions, or taking other actions based on the insights gained from the analysis.
Types of Data Analysis
Types of Data Analysis are as follows:
Descriptive Analysis
This type of analysis involves summarizing and describing the main characteristics of a dataset, such as the mean, median, mode, standard deviation, and range.
Inferential Analysis
This type of analysis involves making inferences about a population based on a sample. Inferential analysis can help determine whether a certain relationship or pattern observed in a sample is likely to be present in the entire population.
Diagnostic Analysis
This type of analysis involves identifying and diagnosing problems or issues within a dataset. Diagnostic analysis can help identify outliers, errors, missing data, or other anomalies in the dataset.
Predictive Analysis
This type of analysis involves using statistical models and algorithms to predict future outcomes or trends based on historical data. Predictive analysis can help businesses and organizations make informed decisions about the future.
Prescriptive Analysis
This type of analysis involves recommending a course of action based on the results of previous analyses. Prescriptive analysis can help organizations make data-driven decisions about how to optimize their operations, products, or services.
Exploratory Analysis
This type of analysis involves exploring the relationships and patterns within a dataset to identify new insights and trends. Exploratory analysis is often used in the early stages of research or data analysis to generate hypotheses and identify areas for further investigation.
Data Analysis Methods
Data Analysis Methods are as follows:
Statistical Analysis
This method involves the use of mathematical models and statistical tools to analyze and interpret data. It includes measures of central tendency, correlation analysis, regression analysis, hypothesis testing, and more.
Machine Learning
This method involves the use of algorithms to identify patterns and relationships in data. It includes supervised and unsupervised learning, classification, clustering, and predictive modeling.
Data Mining
This method involves using statistical and machine learning techniques to extract information and insights from large and complex datasets.
Text Analysis
This method involves using natural language processing (NLP) techniques to analyze and interpret text data. It includes sentiment analysis, topic modeling, and entity recognition.
Network Analysis
This method involves analyzing the relationships and connections between entities in a network, such as social networks or computer networks. It includes social network analysis and graph theory.
Time Series Analysis
This method involves analyzing data collected over time to identify patterns and trends. It includes forecasting, decomposition, and smoothing techniques.
Spatial Analysis
This method involves analyzing geographic data to identify spatial patterns and relationships. It includes spatial statistics, spatial regression, and geospatial data visualization.
Data Visualization
This method involves using graphs, charts, and other visual representations to help communicate the findings of the analysis. It includes scatter plots, bar charts, heat maps, and interactive dashboards.
Qualitative Analysis
This method involves analyzing non-numeric data such as interviews, observations, and open-ended survey responses. It includes thematic analysis, content analysis, and grounded theory.

Multi-criteria Decision Analysis
This method involves analyzing multiple criteria and objectives to support decision-making. It includes techniques such as the analytical hierarchy process, TOPSIS, and ELECTRE.
Data Analysis Tools
There are various data analysis tools available that can help with different aspects of data analysis. Below is a list of some commonly used data analysis tools:
- Microsoft Excel: A widely used spreadsheet program that allows for data organization, analysis, and visualization.
- SQL : A programming language used to manage and manipulate relational databases.
- R : An open-source programming language and software environment for statistical computing and graphics.
- Python : A general-purpose programming language that is widely used in data analysis and machine learning.
- Tableau : A data visualization software that allows for interactive and dynamic visualizations of data.
- SAS : A statistical analysis software used for data management, analysis, and reporting.
- SPSS : A statistical analysis software used for data analysis, reporting, and modeling.
- Matlab : A numerical computing software that is widely used in scientific research and engineering.
- RapidMiner : A data science platform that offers a wide range of data analysis and machine learning tools.
Applications of Data Analysis
Data analysis has numerous applications across various fields. Below are some examples of how data analysis is used in different fields:
- Business : Data analysis is used to gain insights into customer behavior, market trends, and financial performance. This includes customer segmentation, sales forecasting, and market research.
- Healthcare : Data analysis is used to identify patterns and trends in patient data, improve patient outcomes, and optimize healthcare operations. This includes clinical decision support, disease surveillance, and healthcare cost analysis.
- Education : Data analysis is used to measure student performance, evaluate teaching effectiveness, and improve educational programs. This includes assessment analytics, learning analytics, and program evaluation.
- Finance : Data analysis is used to monitor and evaluate financial performance, identify risks, and make investment decisions. This includes risk management, portfolio optimization, and fraud detection.
- Government : Data analysis is used to inform policy-making, improve public services, and enhance public safety. This includes crime analysis, disaster response planning, and social welfare program evaluation.
- Sports : Data analysis is used to gain insights into athlete performance, improve team strategy, and enhance fan engagement. This includes player evaluation, scouting analysis, and game strategy optimization.
- Marketing : Data analysis is used to measure the effectiveness of marketing campaigns, understand customer behavior, and develop targeted marketing strategies. This includes customer segmentation, marketing attribution analysis, and social media analytics.
- Environmental science : Data analysis is used to monitor and evaluate environmental conditions, assess the impact of human activities on the environment, and develop environmental policies. This includes climate modeling, ecological forecasting, and pollution monitoring.
When to Use Data Analysis
Data analysis is useful when you need to extract meaningful insights and information from large and complex datasets. It is a crucial step in the decision-making process, as it helps you understand the underlying patterns and relationships within the data, and identify potential areas for improvement or opportunities for growth.
Here are some specific scenarios where data analysis can be particularly helpful:
- Problem-solving : When you encounter a problem or challenge, data analysis can help you identify the root cause and develop effective solutions.
- Optimization : Data analysis can help you optimize processes, products, or services to increase efficiency, reduce costs, and improve overall performance.
- Prediction: Data analysis can help you make predictions about future trends or outcomes, which can inform strategic planning and decision-making.
- Performance evaluation : Data analysis can help you evaluate the performance of a process, product, or service to identify areas for improvement and potential opportunities for growth.
- Risk assessment : Data analysis can help you assess and mitigate risks, whether it is financial, operational, or related to safety.
- Market research : Data analysis can help you understand customer behavior and preferences, identify market trends, and develop effective marketing strategies.
- Quality control: Data analysis can help you ensure product quality and customer satisfaction by identifying and addressing quality issues.
Purpose of Data Analysis
The primary purposes of data analysis can be summarized as follows:
- To gain insights: Data analysis allows you to identify patterns and trends in data, which can provide valuable insights into the underlying factors that influence a particular phenomenon or process.
- To inform decision-making: Data analysis can help you make informed decisions based on the information that is available. By analyzing data, you can identify potential risks, opportunities, and solutions to problems.
- To improve performance: Data analysis can help you optimize processes, products, or services by identifying areas for improvement and potential opportunities for growth.
- To measure progress: Data analysis can help you measure progress towards a specific goal or objective, allowing you to track performance over time and adjust your strategies accordingly.
- To identify new opportunities: Data analysis can help you identify new opportunities for growth and innovation by identifying patterns and trends that may not have been visible before.
Examples of Data Analysis
Some Examples of Data Analysis are as follows:
- Social Media Monitoring: Companies use data analysis to monitor social media activity in real-time to understand their brand reputation, identify potential customer issues, and track competitors. By analyzing social media data, businesses can make informed decisions on product development, marketing strategies, and customer service.
- Financial Trading: Financial traders use data analysis to make real-time decisions about buying and selling stocks, bonds, and other financial instruments. By analyzing real-time market data, traders can identify trends and patterns that help them make informed investment decisions.
- Traffic Monitoring : Cities use data analysis to monitor traffic patterns and make real-time decisions about traffic management. By analyzing data from traffic cameras, sensors, and other sources, cities can identify congestion hotspots and make changes to improve traffic flow.
- Healthcare Monitoring: Healthcare providers use data analysis to monitor patient health in real-time. By analyzing data from wearable devices, electronic health records, and other sources, healthcare providers can identify potential health issues and provide timely interventions.
- Online Advertising: Online advertisers use data analysis to make real-time decisions about advertising campaigns. By analyzing data on user behavior and ad performance, advertisers can make adjustments to their campaigns to improve their effectiveness.
- Sports Analysis : Sports teams use data analysis to make real-time decisions about strategy and player performance. By analyzing data on player movement, ball position, and other variables, coaches can make informed decisions about substitutions, game strategy, and training regimens.
- Energy Management : Energy companies use data analysis to monitor energy consumption in real-time. By analyzing data on energy usage patterns, companies can identify opportunities to reduce energy consumption and improve efficiency.
Characteristics of Data Analysis
Characteristics of Data Analysis are as follows:
- Objective : Data analysis should be objective and based on empirical evidence, rather than subjective assumptions or opinions.
- Systematic : Data analysis should follow a systematic approach, using established methods and procedures for collecting, cleaning, and analyzing data.
- Accurate : Data analysis should produce accurate results, free from errors and bias. Data should be validated and verified to ensure its quality.
- Relevant : Data analysis should be relevant to the research question or problem being addressed. It should focus on the data that is most useful for answering the research question or solving the problem.
- Comprehensive : Data analysis should be comprehensive and consider all relevant factors that may affect the research question or problem.
- Timely : Data analysis should be conducted in a timely manner, so that the results are available when they are needed.
- Reproducible : Data analysis should be reproducible, meaning that other researchers should be able to replicate the analysis using the same data and methods.
- Communicable : Data analysis should be communicated clearly and effectively to stakeholders and other interested parties. The results should be presented in a way that is understandable and useful for decision-making.
Advantages of Data Analysis
Advantages of Data Analysis are as follows:
- Better decision-making: Data analysis helps in making informed decisions based on facts and evidence, rather than intuition or guesswork.
- Improved efficiency: Data analysis can identify inefficiencies and bottlenecks in business processes, allowing organizations to optimize their operations and reduce costs.
- Increased accuracy: Data analysis helps to reduce errors and bias, providing more accurate and reliable information.
- Better customer service: Data analysis can help organizations understand their customers better, allowing them to provide better customer service and improve customer satisfaction.
- Competitive advantage: Data analysis can provide organizations with insights into their competitors, allowing them to identify areas where they can gain a competitive advantage.
- Identification of trends and patterns : Data analysis can identify trends and patterns in data that may not be immediately apparent, helping organizations to make predictions and plan for the future.
- Improved risk management : Data analysis can help organizations identify potential risks and take proactive steps to mitigate them.
- Innovation: Data analysis can inspire innovation and new ideas by revealing new opportunities or previously unknown correlations in data.
Limitations of Data Analysis
- Data quality: The quality of data can impact the accuracy and reliability of analysis results. If data is incomplete, inconsistent, or outdated, the analysis may not provide meaningful insights.
- Limited scope: Data analysis is limited by the scope of the data available. If data is incomplete or does not capture all relevant factors, the analysis may not provide a complete picture.
- Human error : Data analysis is often conducted by humans, and errors can occur in data collection, cleaning, and analysis.
- Cost : Data analysis can be expensive, requiring specialized tools, software, and expertise.
- Time-consuming : Data analysis can be time-consuming, especially when working with large datasets or conducting complex analyses.
- Overreliance on data: Data analysis should be complemented with human intuition and expertise. Overreliance on data can lead to a lack of creativity and innovation.
- Privacy concerns: Data analysis can raise privacy concerns if personal or sensitive information is used without proper consent or security measures.
About the author
Muhammad Hassan
Researcher, Academic Writer, Web developer
You may also like

Data Verification – Process, Types and Examples

Research Report – Example, Writing Guide and...

Research Summary – Structure, Examples and...

Significance of the Study – Examples and Writing...

Narrative Analysis – Types, Methods and Examples

Descriptive Statistics – Types, Methods and...
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- Can J Hosp Pharm
- v.68(3); May-Jun 2015

Qualitative Research: Data Collection, Analysis, and Management
Introduction.
In an earlier paper, 1 we presented an introduction to using qualitative research methods in pharmacy practice. In this article, we review some principles of the collection, analysis, and management of qualitative data to help pharmacists interested in doing research in their practice to continue their learning in this area. Qualitative research can help researchers to access the thoughts and feelings of research participants, which can enable development of an understanding of the meaning that people ascribe to their experiences. Whereas quantitative research methods can be used to determine how many people undertake particular behaviours, qualitative methods can help researchers to understand how and why such behaviours take place. Within the context of pharmacy practice research, qualitative approaches have been used to examine a diverse array of topics, including the perceptions of key stakeholders regarding prescribing by pharmacists and the postgraduation employment experiences of young pharmacists (see “Further Reading” section at the end of this article).
In the previous paper, 1 we outlined 3 commonly used methodologies: ethnography 2 , grounded theory 3 , and phenomenology. 4 Briefly, ethnography involves researchers using direct observation to study participants in their “real life” environment, sometimes over extended periods. Grounded theory and its later modified versions (e.g., Strauss and Corbin 5 ) use face-to-face interviews and interactions such as focus groups to explore a particular research phenomenon and may help in clarifying a less-well-understood problem, situation, or context. Phenomenology shares some features with grounded theory (such as an exploration of participants’ behaviour) and uses similar techniques to collect data, but it focuses on understanding how human beings experience their world. It gives researchers the opportunity to put themselves in another person’s shoes and to understand the subjective experiences of participants. 6 Some researchers use qualitative methodologies but adopt a different standpoint, and an example of this appears in the work of Thurston and others, 7 discussed later in this paper.
Qualitative work requires reflection on the part of researchers, both before and during the research process, as a way of providing context and understanding for readers. When being reflexive, researchers should not try to simply ignore or avoid their own biases (as this would likely be impossible); instead, reflexivity requires researchers to reflect upon and clearly articulate their position and subjectivities (world view, perspectives, biases), so that readers can better understand the filters through which questions were asked, data were gathered and analyzed, and findings were reported. From this perspective, bias and subjectivity are not inherently negative but they are unavoidable; as a result, it is best that they be articulated up-front in a manner that is clear and coherent for readers.
THE PARTICIPANT’S VIEWPOINT
What qualitative study seeks to convey is why people have thoughts and feelings that might affect the way they behave. Such study may occur in any number of contexts, but here, we focus on pharmacy practice and the way people behave with regard to medicines use (e.g., to understand patients’ reasons for nonadherence with medication therapy or to explore physicians’ resistance to pharmacists’ clinical suggestions). As we suggested in our earlier article, 1 an important point about qualitative research is that there is no attempt to generalize the findings to a wider population. Qualitative research is used to gain insights into people’s feelings and thoughts, which may provide the basis for a future stand-alone qualitative study or may help researchers to map out survey instruments for use in a quantitative study. It is also possible to use different types of research in the same study, an approach known as “mixed methods” research, and further reading on this topic may be found at the end of this paper.
The role of the researcher in qualitative research is to attempt to access the thoughts and feelings of study participants. This is not an easy task, as it involves asking people to talk about things that may be very personal to them. Sometimes the experiences being explored are fresh in the participant’s mind, whereas on other occasions reliving past experiences may be difficult. However the data are being collected, a primary responsibility of the researcher is to safeguard participants and their data. Mechanisms for such safeguarding must be clearly articulated to participants and must be approved by a relevant research ethics review board before the research begins. Researchers and practitioners new to qualitative research should seek advice from an experienced qualitative researcher before embarking on their project.
DATA COLLECTION
Whatever philosophical standpoint the researcher is taking and whatever the data collection method (e.g., focus group, one-to-one interviews), the process will involve the generation of large amounts of data. In addition to the variety of study methodologies available, there are also different ways of making a record of what is said and done during an interview or focus group, such as taking handwritten notes or video-recording. If the researcher is audio- or video-recording data collection, then the recordings must be transcribed verbatim before data analysis can begin. As a rough guide, it can take an experienced researcher/transcriber 8 hours to transcribe one 45-minute audio-recorded interview, a process than will generate 20–30 pages of written dialogue.
Many researchers will also maintain a folder of “field notes” to complement audio-taped interviews. Field notes allow the researcher to maintain and comment upon impressions, environmental contexts, behaviours, and nonverbal cues that may not be adequately captured through the audio-recording; they are typically handwritten in a small notebook at the same time the interview takes place. Field notes can provide important context to the interpretation of audio-taped data and can help remind the researcher of situational factors that may be important during data analysis. Such notes need not be formal, but they should be maintained and secured in a similar manner to audio tapes and transcripts, as they contain sensitive information and are relevant to the research. For more information about collecting qualitative data, please see the “Further Reading” section at the end of this paper.
DATA ANALYSIS AND MANAGEMENT
If, as suggested earlier, doing qualitative research is about putting oneself in another person’s shoes and seeing the world from that person’s perspective, the most important part of data analysis and management is to be true to the participants. It is their voices that the researcher is trying to hear, so that they can be interpreted and reported on for others to read and learn from. To illustrate this point, consider the anonymized transcript excerpt presented in Appendix 1 , which is taken from a research interview conducted by one of the authors (J.S.). We refer to this excerpt throughout the remainder of this paper to illustrate how data can be managed, analyzed, and presented.
Interpretation of Data
Interpretation of the data will depend on the theoretical standpoint taken by researchers. For example, the title of the research report by Thurston and others, 7 “Discordant indigenous and provider frames explain challenges in improving access to arthritis care: a qualitative study using constructivist grounded theory,” indicates at least 2 theoretical standpoints. The first is the culture of the indigenous population of Canada and the place of this population in society, and the second is the social constructivist theory used in the constructivist grounded theory method. With regard to the first standpoint, it can be surmised that, to have decided to conduct the research, the researchers must have felt that there was anecdotal evidence of differences in access to arthritis care for patients from indigenous and non-indigenous backgrounds. With regard to the second standpoint, it can be surmised that the researchers used social constructivist theory because it assumes that behaviour is socially constructed; in other words, people do things because of the expectations of those in their personal world or in the wider society in which they live. (Please see the “Further Reading” section for resources providing more information about social constructivist theory and reflexivity.) Thus, these 2 standpoints (and there may have been others relevant to the research of Thurston and others 7 ) will have affected the way in which these researchers interpreted the experiences of the indigenous population participants and those providing their care. Another standpoint is feminist standpoint theory which, among other things, focuses on marginalized groups in society. Such theories are helpful to researchers, as they enable us to think about things from a different perspective. Being aware of the standpoints you are taking in your own research is one of the foundations of qualitative work. Without such awareness, it is easy to slip into interpreting other people’s narratives from your own viewpoint, rather than that of the participants.
To analyze the example in Appendix 1 , we will adopt a phenomenological approach because we want to understand how the participant experienced the illness and we want to try to see the experience from that person’s perspective. It is important for the researcher to reflect upon and articulate his or her starting point for such analysis; for example, in the example, the coder could reflect upon her own experience as a female of a majority ethnocultural group who has lived within middle class and upper middle class settings. This personal history therefore forms the filter through which the data will be examined. This filter does not diminish the quality or significance of the analysis, since every researcher has his or her own filters; however, by explicitly stating and acknowledging what these filters are, the researcher makes it easer for readers to contextualize the work.
Transcribing and Checking
For the purposes of this paper it is assumed that interviews or focus groups have been audio-recorded. As mentioned above, transcribing is an arduous process, even for the most experienced transcribers, but it must be done to convert the spoken word to the written word to facilitate analysis. For anyone new to conducting qualitative research, it is beneficial to transcribe at least one interview and one focus group. It is only by doing this that researchers realize how difficult the task is, and this realization affects their expectations when asking others to transcribe. If the research project has sufficient funding, then a professional transcriber can be hired to do the work. If this is the case, then it is a good idea to sit down with the transcriber, if possible, and talk through the research and what the participants were talking about. This background knowledge for the transcriber is especially important in research in which people are using jargon or medical terms (as in pharmacy practice). Involving your transcriber in this way makes the work both easier and more rewarding, as he or she will feel part of the team. Transcription editing software is also available, but it is expensive. For example, ELAN (more formally known as EUDICO Linguistic Annotator, developed at the Technical University of Berlin) 8 is a tool that can help keep data organized by linking media and data files (particularly valuable if, for example, video-taping of interviews is complemented by transcriptions). It can also be helpful in searching complex data sets. Products such as ELAN do not actually automatically transcribe interviews or complete analyses, and they do require some time and effort to learn; nonetheless, for some research applications, it may be a valuable to consider such software tools.
All audio recordings should be transcribed verbatim, regardless of how intelligible the transcript may be when it is read back. Lines of text should be numbered. Once the transcription is complete, the researcher should read it while listening to the recording and do the following: correct any spelling or other errors; anonymize the transcript so that the participant cannot be identified from anything that is said (e.g., names, places, significant events); insert notations for pauses, laughter, looks of discomfort; insert any punctuation, such as commas and full stops (periods) (see Appendix 1 for examples of inserted punctuation), and include any other contextual information that might have affected the participant (e.g., temperature or comfort of the room).
Dealing with the transcription of a focus group is slightly more difficult, as multiple voices are involved. One way of transcribing such data is to “tag” each voice (e.g., Voice A, Voice B). In addition, the focus group will usually have 2 facilitators, whose respective roles will help in making sense of the data. While one facilitator guides participants through the topic, the other can make notes about context and group dynamics. More information about group dynamics and focus groups can be found in resources listed in the “Further Reading” section.
Reading between the Lines
During the process outlined above, the researcher can begin to get a feel for the participant’s experience of the phenomenon in question and can start to think about things that could be pursued in subsequent interviews or focus groups (if appropriate). In this way, one participant’s narrative informs the next, and the researcher can continue to interview until nothing new is being heard or, as it says in the text books, “saturation is reached”. While continuing with the processes of coding and theming (described in the next 2 sections), it is important to consider not just what the person is saying but also what they are not saying. For example, is a lengthy pause an indication that the participant is finding the subject difficult, or is the person simply deciding what to say? The aim of the whole process from data collection to presentation is to tell the participants’ stories using exemplars from their own narratives, thus grounding the research findings in the participants’ lived experiences.
Smith 9 suggested a qualitative research method known as interpretative phenomenological analysis, which has 2 basic tenets: first, that it is rooted in phenomenology, attempting to understand the meaning that individuals ascribe to their lived experiences, and second, that the researcher must attempt to interpret this meaning in the context of the research. That the researcher has some knowledge and expertise in the subject of the research means that he or she can have considerable scope in interpreting the participant’s experiences. Larkin and others 10 discussed the importance of not just providing a description of what participants say. Rather, interpretative phenomenological analysis is about getting underneath what a person is saying to try to truly understand the world from his or her perspective.
Once all of the research interviews have been transcribed and checked, it is time to begin coding. Field notes compiled during an interview can be a useful complementary source of information to facilitate this process, as the gap in time between an interview, transcribing, and coding can result in memory bias regarding nonverbal or environmental context issues that may affect interpretation of data.
Coding refers to the identification of topics, issues, similarities, and differences that are revealed through the participants’ narratives and interpreted by the researcher. This process enables the researcher to begin to understand the world from each participant’s perspective. Coding can be done by hand on a hard copy of the transcript, by making notes in the margin or by highlighting and naming sections of text. More commonly, researchers use qualitative research software (e.g., NVivo, QSR International Pty Ltd; www.qsrinternational.com/products_nvivo.aspx ) to help manage their transcriptions. It is advised that researchers undertake a formal course in the use of such software or seek supervision from a researcher experienced in these tools.
Returning to Appendix 1 and reading from lines 8–11, a code for this section might be “diagnosis of mental health condition”, but this would just be a description of what the participant is talking about at that point. If we read a little more deeply, we can ask ourselves how the participant might have come to feel that the doctor assumed he or she was aware of the diagnosis or indeed that they had only just been told the diagnosis. There are a number of pauses in the narrative that might suggest the participant is finding it difficult to recall that experience. Later in the text, the participant says “nobody asked me any questions about my life” (line 19). This could be coded simply as “health care professionals’ consultation skills”, but that would not reflect how the participant must have felt never to be asked anything about his or her personal life, about the participant as a human being. At the end of this excerpt, the participant just trails off, recalling that no-one showed any interest, which makes for very moving reading. For practitioners in pharmacy, it might also be pertinent to explore the participant’s experience of akathisia and why this was left untreated for 20 years.
One of the questions that arises about qualitative research relates to the reliability of the interpretation and representation of the participants’ narratives. There are no statistical tests that can be used to check reliability and validity as there are in quantitative research. However, work by Lincoln and Guba 11 suggests that there are other ways to “establish confidence in the ‘truth’ of the findings” (p. 218). They call this confidence “trustworthiness” and suggest that there are 4 criteria of trustworthiness: credibility (confidence in the “truth” of the findings), transferability (showing that the findings have applicability in other contexts), dependability (showing that the findings are consistent and could be repeated), and confirmability (the extent to which the findings of a study are shaped by the respondents and not researcher bias, motivation, or interest).
One way of establishing the “credibility” of the coding is to ask another researcher to code the same transcript and then to discuss any similarities and differences in the 2 resulting sets of codes. This simple act can result in revisions to the codes and can help to clarify and confirm the research findings.
Theming refers to the drawing together of codes from one or more transcripts to present the findings of qualitative research in a coherent and meaningful way. For example, there may be examples across participants’ narratives of the way in which they were treated in hospital, such as “not being listened to” or “lack of interest in personal experiences” (see Appendix 1 ). These may be drawn together as a theme running through the narratives that could be named “the patient’s experience of hospital care”. The importance of going through this process is that at its conclusion, it will be possible to present the data from the interviews using quotations from the individual transcripts to illustrate the source of the researchers’ interpretations. Thus, when the findings are organized for presentation, each theme can become the heading of a section in the report or presentation. Underneath each theme will be the codes, examples from the transcripts, and the researcher’s own interpretation of what the themes mean. Implications for real life (e.g., the treatment of people with chronic mental health problems) should also be given.
DATA SYNTHESIS
In this final section of this paper, we describe some ways of drawing together or “synthesizing” research findings to represent, as faithfully as possible, the meaning that participants ascribe to their life experiences. This synthesis is the aim of the final stage of qualitative research. For most readers, the synthesis of data presented by the researcher is of crucial significance—this is usually where “the story” of the participants can be distilled, summarized, and told in a manner that is both respectful to those participants and meaningful to readers. There are a number of ways in which researchers can synthesize and present their findings, but any conclusions drawn by the researchers must be supported by direct quotations from the participants. In this way, it is made clear to the reader that the themes under discussion have emerged from the participants’ interviews and not the mind of the researcher. The work of Latif and others 12 gives an example of how qualitative research findings might be presented.
Planning and Writing the Report
As has been suggested above, if researchers code and theme their material appropriately, they will naturally find the headings for sections of their report. Qualitative researchers tend to report “findings” rather than “results”, as the latter term typically implies that the data have come from a quantitative source. The final presentation of the research will usually be in the form of a report or a paper and so should follow accepted academic guidelines. In particular, the article should begin with an introduction, including a literature review and rationale for the research. There should be a section on the chosen methodology and a brief discussion about why qualitative methodology was most appropriate for the study question and why one particular methodology (e.g., interpretative phenomenological analysis rather than grounded theory) was selected to guide the research. The method itself should then be described, including ethics approval, choice of participants, mode of recruitment, and method of data collection (e.g., semistructured interviews or focus groups), followed by the research findings, which will be the main body of the report or paper. The findings should be written as if a story is being told; as such, it is not necessary to have a lengthy discussion section at the end. This is because much of the discussion will take place around the participants’ quotes, such that all that is needed to close the report or paper is a summary, limitations of the research, and the implications that the research has for practice. As stated earlier, it is not the intention of qualitative research to allow the findings to be generalized, and therefore this is not, in itself, a limitation.
Planning out the way that findings are to be presented is helpful. It is useful to insert the headings of the sections (the themes) and then make a note of the codes that exemplify the thoughts and feelings of your participants. It is generally advisable to put in the quotations that you want to use for each theme, using each quotation only once. After all this is done, the telling of the story can begin as you give your voice to the experiences of the participants, writing around their quotations. Do not be afraid to draw assumptions from the participants’ narratives, as this is necessary to give an in-depth account of the phenomena in question. Discuss these assumptions, drawing on your participants’ words to support you as you move from one code to another and from one theme to the next. Finally, as appropriate, it is possible to include examples from literature or policy documents that add support for your findings. As an exercise, you may wish to code and theme the sample excerpt in Appendix 1 and tell the participant’s story in your own way. Further reading about “doing” qualitative research can be found at the end of this paper.
CONCLUSIONS
Qualitative research can help researchers to access the thoughts and feelings of research participants, which can enable development of an understanding of the meaning that people ascribe to their experiences. It can be used in pharmacy practice research to explore how patients feel about their health and their treatment. Qualitative research has been used by pharmacists to explore a variety of questions and problems (see the “Further Reading” section for examples). An understanding of these issues can help pharmacists and other health care professionals to tailor health care to match the individual needs of patients and to develop a concordant relationship. Doing qualitative research is not easy and may require a complete rethink of how research is conducted, particularly for researchers who are more familiar with quantitative approaches. There are many ways of conducting qualitative research, and this paper has covered some of the practical issues regarding data collection, analysis, and management. Further reading around the subject will be essential to truly understand this method of accessing peoples’ thoughts and feelings to enable researchers to tell participants’ stories.
Appendix 1. Excerpt from a sample transcript
The participant (age late 50s) had suffered from a chronic mental health illness for 30 years. The participant had become a “revolving door patient,” someone who is frequently in and out of hospital. As the participant talked about past experiences, the researcher asked:
- What was treatment like 30 years ago?
- Umm—well it was pretty much they could do what they wanted with you because I was put into the er, the er kind of system er, I was just on
- endless section threes.
- Really…
- But what I didn’t realize until later was that if you haven’t actually posed a threat to someone or yourself they can’t really do that but I didn’t know
- that. So wh-when I first went into hospital they put me on the forensic ward ’cause they said, “We don’t think you’ll stay here we think you’ll just
- run-run away.” So they put me then onto the acute admissions ward and – er – I can remember one of the first things I recall when I got onto that
- ward was sitting down with a er a Dr XXX. He had a book this thick [gestures] and on each page it was like three questions and he went through
- all these questions and I answered all these questions. So we’re there for I don’t maybe two hours doing all that and he asked me he said “well
- when did somebody tell you then that you have schizophrenia” I said “well nobody’s told me that” so he seemed very surprised but nobody had
- actually [pause] whe-when I first went up there under police escort erm the senior kind of consultants people I’d been to where I was staying and
- ermm so er [pause] I . . . the, I can remember the very first night that I was there and given this injection in this muscle here [gestures] and just
- having dreadful side effects the next day I woke up [pause]
- . . . and I suffered that akathesia I swear to you, every minute of every day for about 20 years.
- Oh how awful.
- And that side of it just makes life impossible so the care on the wards [pause] umm I don’t know it’s kind of, it’s kind of hard to put into words
- [pause]. Because I’m not saying they were sort of like not friendly or interested but then nobody ever seemed to want to talk about your life [pause]
- nobody asked me any questions about my life. The only questions that came into was they asked me if I’d be a volunteer for these student exams
- and things and I said “yeah” so all the questions were like “oh what jobs have you done,” er about your relationships and things and er but
- nobody actually sat down and had a talk and showed some interest in you as a person you were just there basically [pause] um labelled and you
- know there was there was [pause] but umm [pause] yeah . . .
This article is the 10th in the CJHP Research Primer Series, an initiative of the CJHP Editorial Board and the CSHP Research Committee. The planned 2-year series is intended to appeal to relatively inexperienced researchers, with the goal of building research capacity among practising pharmacists. The articles, presenting simple but rigorous guidance to encourage and support novice researchers, are being solicited from authors with appropriate expertise.
Previous articles in this series:
Bond CM. The research jigsaw: how to get started. Can J Hosp Pharm . 2014;67(1):28–30.
Tully MP. Research: articulating questions, generating hypotheses, and choosing study designs. Can J Hosp Pharm . 2014;67(1):31–4.
Loewen P. Ethical issues in pharmacy practice research: an introductory guide. Can J Hosp Pharm. 2014;67(2):133–7.
Tsuyuki RT. Designing pharmacy practice research trials. Can J Hosp Pharm . 2014;67(3):226–9.
Bresee LC. An introduction to developing surveys for pharmacy practice research. Can J Hosp Pharm . 2014;67(4):286–91.
Gamble JM. An introduction to the fundamentals of cohort and case–control studies. Can J Hosp Pharm . 2014;67(5):366–72.
Austin Z, Sutton J. Qualitative research: getting started. C an J Hosp Pharm . 2014;67(6):436–40.
Houle S. An introduction to the fundamentals of randomized controlled trials in pharmacy research. Can J Hosp Pharm . 2014; 68(1):28–32.
Charrois TL. Systematic reviews: What do you need to know to get started? Can J Hosp Pharm . 2014;68(2):144–8.
Competing interests: None declared.
Further Reading
Examples of qualitative research in pharmacy practice.
- Farrell B, Pottie K, Woodend K, Yao V, Dolovich L, Kennie N, et al. Shifts in expectations: evaluating physicians’ perceptions as pharmacists integrated into family practice. J Interprof Care. 2010; 24 (1):80–9. [ PubMed ] [ Google Scholar ]
- Gregory P, Austin Z. Postgraduation employment experiences of new pharmacists in Ontario in 2012–2013. Can Pharm J. 2014; 147 (5):290–9. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Marks PZ, Jennnings B, Farrell B, Kennie-Kaulbach N, Jorgenson D, Pearson-Sharpe J, et al. “I gained a skill and a change in attitude”: a case study describing how an online continuing professional education course for pharmacists supported achievement of its transfer to practice outcomes. Can J Univ Contin Educ. 2014; 40 (2):1–18. [ Google Scholar ]
- Nair KM, Dolovich L, Brazil K, Raina P. It’s all about relationships: a qualitative study of health researchers’ perspectives on interdisciplinary research. BMC Health Serv Res. 2008; 8 :110. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Pojskic N, MacKeigan L, Boon H, Austin Z. Initial perceptions of key stakeholders in Ontario regarding independent prescriptive authority for pharmacists. Res Soc Adm Pharm. 2014; 10 (2):341–54. [ PubMed ] [ Google Scholar ]
Qualitative Research in General
- Breakwell GM, Hammond S, Fife-Schaw C. Research methods in psychology. Thousand Oaks (CA): Sage Publications; 1995. [ Google Scholar ]
- Given LM. 100 questions (and answers) about qualitative research. Thousand Oaks (CA): Sage Publications; 2015. [ Google Scholar ]
- Miles B, Huberman AM. Qualitative data analysis. Thousand Oaks (CA): Sage Publications; 2009. [ Google Scholar ]
- Patton M. Qualitative research and evaluation methods. Thousand Oaks (CA): Sage Publications; 2002. [ Google Scholar ]
- Willig C. Introducing qualitative research in psychology. Buckingham (UK): Open University Press; 2001. [ Google Scholar ]
Group Dynamics in Focus Groups
- Farnsworth J, Boon B. Analysing group dynamics within the focus group. Qual Res. 2010; 10 (5):605–24. [ Google Scholar ]
Social Constructivism
- Social constructivism. Berkeley (CA): University of California, Berkeley, Berkeley Graduate Division, Graduate Student Instruction Teaching & Resource Center; [cited 2015 June 4]. Available from: http://gsi.berkeley.edu/gsi-guide-contents/learning-theory-research/social-constructivism/ [ Google Scholar ]
Mixed Methods
- Creswell J. Research design: qualitative, quantitative, and mixed methods approaches. Thousand Oaks (CA): Sage Publications; 2009. [ Google Scholar ]
Collecting Qualitative Data
- Arksey H, Knight P. Interviewing for social scientists: an introductory resource with examples. Thousand Oaks (CA): Sage Publications; 1999. [ Google Scholar ]
- Guest G, Namey EE, Mitchel ML. Collecting qualitative data: a field manual for applied research. Thousand Oaks (CA): Sage Publications; 2013. [ Google Scholar ]
Constructivist Grounded Theory
- Charmaz K. Grounded theory: objectivist and constructivist methods. In: Denzin N, Lincoln Y, editors. Handbook of qualitative research. 2nd ed. Thousand Oaks (CA): Sage Publications; 2000. pp. 509–35. [ Google Scholar ]
Information
- Author Services
Initiatives
You are accessing a machine-readable page. In order to be human-readable, please install an RSS reader.
All articles published by MDPI are made immediately available worldwide under an open access license. No special permission is required to reuse all or part of the article published by MDPI, including figures and tables. For articles published under an open access Creative Common CC BY license, any part of the article may be reused without permission provided that the original article is clearly cited. For more information, please refer to https://www.mdpi.com/openaccess .
Feature papers represent the most advanced research with significant potential for high impact in the field. A Feature Paper should be a substantial original Article that involves several techniques or approaches, provides an outlook for future research directions and describes possible research applications.
Feature papers are submitted upon individual invitation or recommendation by the scientific editors and must receive positive feedback from the reviewers.
Editor’s Choice articles are based on recommendations by the scientific editors of MDPI journals from around the world. Editors select a small number of articles recently published in the journal that they believe will be particularly interesting to readers, or important in the respective research area. The aim is to provide a snapshot of some of the most exciting work published in the various research areas of the journal.
Original Submission Date Received: .
- Active Journals
- Find a Journal
- Proceedings Series
- For Authors
- For Reviewers
- For Editors
- For Librarians
- For Publishers
- For Societies
- For Conference Organizers
- Open Access Policy
- Institutional Open Access Program
- Special Issues Guidelines
- Editorial Process
- Research and Publication Ethics
- Article Processing Charges
- Testimonials
- Preprints.org
- SciProfiles
- Encyclopedia

Article Menu

- Subscribe SciFeed
- Recommended Articles
- Author Biographies
- Google Scholar
- on Google Scholar
- Table of Contents
Find support for a specific problem in the support section of our website.
Please let us know what you think of our products and services.
Visit our dedicated information section to learn more about MDPI.
JSmol Viewer
Advancements in deep learning techniques for time series forecasting in maritime applications: a comprehensive review.

1. Introduction
2. literature collection procedure.
- Search scope: Titles, Keywords, and Abstracts
- Keywords 1: ‘deep’ AND ‘learning’, AND
- Keywords 2: ‘time AND series’, AND
- Keywords 3: ‘maritime’, OR
- Keywords 4: ‘vessel’, OR
- Keywords 5: ‘shipping’, OR
- Keywords 6: ‘marine’, OR
- Keywords 7: ‘ship’, OR
- Keywords 8: ‘port’, OR
- Keywords 9: ‘terminal’
- Retain only articles related to maritime operations. For example, studies on ship-surrounding weather and risk prediction based on ship data will be kept, while research solely focused on marine weather or wave prediction that is unrelated to any aspect of maritime operations will be excluded.
- Exclude neural network studies that do not employ deep learning techniques, such as ANN or MLP with only one hidden layer.
- The language of the publications must be English.
- The original data used in the papers must include time series sequences.
3. Deep Learning Algorithms
3.1. artificial neural network (ann), 3.1.1. multilayer perceptron (mlp)/deep neural networks (dnn), 3.1.2. wavenet, 3.1.3. randomized neural network, 3.2. convolutional neural network (cnn), 3.3. recurrent neural network (rnn), 3.3.1. long short-term memory (lstm), 3.3.2. gated recurrent unit (gru), 3.4. attention mechanism (am)/transformer, 3.5. overview of algorithms usage, 4. time series forecasting in maritime applications, 4.1. ship operation-related applications, 4.1.1. ship trajectory prediction, 4.1.2. meteorological factor prediction, 4.1.3. ship fuel consumption prediction, 4.1.4. others, 4.2. port operation-related applications, 4.3. shipping market-related applications, 4.4. overview of time series forecasting in maritime applications, 5. overall analysis, 5.1. literature description, 5.1.1. literature distribution, 5.1.2. literature classification, 5.2. data utilized in maritime research, 5.2.1. automatic identification system data (ais data), 5.2.2. high-frequency radar data and sensor data, 5.2.3. container throughput data, 5.2.4. other datasets, 5.3. evaluation parameters, 5.4. real-world application examples, 5.5. future research directions, 5.5.1. data processing and feature extraction, 5.5.2. model optimization and application of new technologies, 5.5.3. specific application scenarios, 5.5.4. practical applications and long-term predictions, 5.5.5. environmental impact, fault prediction, and cross-domain applications, 6. conclusions, author contributions, institutional review board statement, informed consent statement, data availability statement, conflicts of interest.
- UNCTAD. Review of Maritime Transport 2023 ; United Nations Conference on Trade and Development: Geneva, Switzerland, 2023; Available online: https://www.un-ilibrary.org/content/books/9789213584569 (accessed on 1 April 2024).
- Liang, M.; Liu, R.W.; Zhan, Y.; Li, H.; Zhu, F.; Wang, F.Y. Fine-Grained Vessel Traffic Flow Prediction With a Spatio-Temporal Multigraph Convolutional Network. IEEE Trans. Intell. Transp. Syst. 2022 , 23 , 23694–23707. [ Google Scholar ] [ CrossRef ]
- Liu, R.W.; Liang, M.; Nie, J.; Lim, W.Y.B.; Zhang, Y.; Guizani, M. Deep Learning-Powered Vessel Trajectory Prediction for Improving Smart Traffic Services in Maritime Internet of Things. IEEE Trans. Netw. Sci. Eng. 2022 , 9 , 3080–3094. [ Google Scholar ] [ CrossRef ]
- Dui, H.; Zheng, X.; Wu, S. Resilience analysis of maritime transportation systems based on importance measures. Reliab. Eng. Syst. Saf. 2021 , 209 , 107461. [ Google Scholar ] [ CrossRef ]
- Liang, M.; Li, H.; Liu, R.W.; Lam, J.S.L.; Yang, Z. PiracyAnalyzer: Spatial temporal patterns analysis of global piracy incidents. Reliab. Eng. Syst. Saf. 2024 , 243 , 109877. [ Google Scholar ] [ CrossRef ]
- Chen, Z.S.; Lam, J.S.L.; Xiao, Z. Prediction of harbour vessel emissions based on machine learning approach. Transp. Res. Part D Transp. Environ. 2024 , 131 , 104214. [ Google Scholar ] [ CrossRef ]
- Chen, Z.S.; Lam, J.S.L.; Xiao, Z. Prediction of harbour vessel fuel consumption based on machine learning approach. Ocean Eng. 2023 , 278 , 114483. [ Google Scholar ] [ CrossRef ]
- Liang, M.; Weng, L.; Gao, R.; Li, Y.; Du, L. Unsupervised maritime anomaly detection for intelligent situational awareness using AIS data. Knowl.-Based Syst. 2024 , 284 , 111313. [ Google Scholar ] [ CrossRef ]
- Dave, V.S.; Dutta, K. Neural network based models for software effort estimation: A review. Artif. Intell. Rev. 2014 , 42 , 295–307. [ Google Scholar ] [ CrossRef ]
- Uslu, S.; Celik, M.B. Prediction of engine emissions and performance with artificial neural networks in a single cylinder diesel engine using diethyl ether. Eng. Sci. Technol. Int. J. 2018 , 21 , 1194–1201. [ Google Scholar ] [ CrossRef ]
- Chaudhary, L.; Sharma, S.; Sajwan, M. Systematic Literature Review of Various Neural Network Techniques for Sea Surface Temperature Prediction Using Remote Sensing Data. Arch. Comput. Methods Eng. 2023 , 30 , 5071–5103. [ Google Scholar ] [ CrossRef ]
- Dharia, A.; Adeli, H. Neural network model for rapid forecasting of freeway link travel time. Eng. Appl. Artif. Intell. 2003 , 16 , 607–613. [ Google Scholar ] [ CrossRef ]
- Hecht-Nielsen, R. Applications of counterpropagation networks. Neural Netw. 1988 , 1 , 131–139. [ Google Scholar ] [ CrossRef ]
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learning ; MIT Press: Cambridge, MA, USA, 2016. [ Google Scholar ]
- Veerappa, M.; Anneken, M.; Burkart, N. Evaluation of Interpretable Association Rule Mining Methods on Time-Series in the Maritime Domain. Springer International Publishing: Cham, Switzerland, 2021; pp. 204–218. [ Google Scholar ]
- Frizzell, J.; Furth, M. Prediction of Vessel RAOs: Applications of Deep Learning to Assist in Design. In Proceedings of the SNAME 27th Offshore Symposium, Houston, TX, USA, 22 February 2022. [ Google Scholar ] [ CrossRef ]
- Van Den Oord, A.; Dieleman, S.; Zen, H.; Simonyan, K.; Vinyals, O.; Graves, A.; Kalchbrenner, N.; Senior, A.; Kavukcuoglu, K. Wavenet: A generative model for raw audio. arXiv 2016 , arXiv:1609.03499. [ Google Scholar ]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [ Google Scholar ]
- Ning, C.X.; Xie, Y.Z.; Sun, L.J. LSTM, WaveNet, and 2D CNN for nonlinear time history prediction of seismic responses. Eng. Struct. 2023 , 286 , 116083. [ Google Scholar ] [ CrossRef ]
- Schmidt, W.F.; Kraaijveld, M.A.; Duin, R.P. Feed forward neural networks with random weights. In International Conference on Pattern Recognition ; IEEE Computer Society Press: Washington, DC, USA, 1992; pp. 1–4. [ Google Scholar ]
- Huang, G.B.; Zhu, Q.Y.; Siew, C.K. Extreme learning machine: Theory and applications. Neurocomputing 2006 , 70 , 489–501. [ Google Scholar ] [ CrossRef ]
- Pao, Y.H.; Park, G.H.; Sobajic, D.J. Learning and generalization characteristics of the random vector functional-link net. Neurocomputing 1994 , 6 , 163–180. [ Google Scholar ] [ CrossRef ]
- Zhang, L.; Suganthan, P.N. A comprehensive evaluation of random vector functional link networks. Inf. Sci. 2016 , 367 , 1094–1105. [ Google Scholar ] [ CrossRef ]
- Huang, G.; Huang, G.B.; Song, S.J.; You, K.Y. Trends in extreme learning machines: A review. Neural Netw. 2015 , 61 , 32–48. [ Google Scholar ] [ CrossRef ]
- Shi, Q.S.; Katuwal, R.; Suganthan, P.N.; Tanveer, M. Random vector functional link neural network based ensemble deep learning. Pattern Recognit. 2021 , 117 , 107978. [ Google Scholar ] [ CrossRef ]
- Du, L.; Gao, R.B.; Suganthan, P.N.; Wang, D.Z.W. Graph ensemble deep random vector functional link network for traffic forecasting. Appl. Soft Comput. 2022 , 131 , 109809. [ Google Scholar ] [ CrossRef ]
- Rehman, A.; Xing, H.L.; Hussain, M.; Gulzar, N.; Khan, M.A.; Hussain, A.; Mahmood, S. HCDP-DELM: Heterogeneous chronic disease prediction with temporal perspective enabled deep extreme learning machine. Knowl.-Based Syst. 2024 , 284 , 111316. [ Google Scholar ] [ CrossRef ]
- Gao, R.B.; Li, R.L.; Hu, M.H.; Suganthan, P.N.; Yuen, K.F. Online dynamic ensemble deep random vector functional link neural network for forecasting. Neural Netw. 2023 , 166 , 51–69. [ Google Scholar ] [ CrossRef ]
- Lecun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-based learning applied to document recognition. Proc. IEEE 1998 , 86 , 2278–2324. [ Google Scholar ] [ CrossRef ]
- Palaz, D.; Magimai-Doss, M.; Collobert, R. End-to-end acoustic modeling using convolutional neural networks for HMM-based automatic speech recognition. Speech Commun. 2019 , 108 , 15–32. [ Google Scholar ] [ CrossRef ]
- Fang, W.; Love, P.E.D.; Luo, H.; Ding, L. Computer vision for behaviour-based safety in construction: A review and future directions. Adv. Eng. Inform. 2020 , 43 , 100980. [ Google Scholar ] [ CrossRef ]
- Qin, L.; Yu, N.; Zhao, D. Applying the convolutional neural network deep learning technology to behavioural recognition in intelligent video. Teh. Vjesn. 2018 , 25 , 528–535. [ Google Scholar ]
- Hoseinzade, E.; Haratizadeh, S. CNNpred: CNN-based stock market prediction using a diverse set of variables. Expert Syst. Appl. 2019 , 129 , 273–285. [ Google Scholar ] [ CrossRef ]
- Rasp, S.; Dueben, P.D.; Scher, S.; Weyn, J.A.; Mouatadid, S.; Thuerey, N. WeatherBench: A Benchmark Data Set for Data-Driven Weather Forecasting. J. Adv. Model. Earth Syst. 2020 , 12 , e2020MS002203. [ Google Scholar ] [ CrossRef ]
- Crivellari, A.; Beinat, E.; Caetano, S.; Seydoux, A.; Cardoso, T. Multi-target CNN-LSTM regressor for predicting urban distribution of short-term food delivery demand. J. Bus. Res. 2022 , 144 , 844–853. [ Google Scholar ] [ CrossRef ]
- Bai, S.; Kolter, J.Z.; Koltun, V. An empirical evaluation of generic convolutional and recurrent networks for sequence modeling. arXiv 2018 , arXiv:1803.01271. [ Google Scholar ]
- Lin, Z.; Yue, W.; Huang, J.; Wan, J. Ship Trajectory Prediction Based on the TTCN-Attention-GRU Model. Electronics 2023 , 12 , 2556. [ Google Scholar ] [ CrossRef ]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Identity Mappings in Deep Residual Networks. In Computer Vision–ECCV 2016 ; Leibe, B., Matas, J., Sebe, N., Welling, M., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 630–645. [ Google Scholar ]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 2014 , 15 , 1929–1958. [ Google Scholar ]
- Bin Syed, M.A.; Ahmed, I. A CNN-LSTM Architecture for Marine Vessel Track Association Using Automatic Identification System (AIS) Data. Sensors 2023 , 23 , 6400. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Li, M.-W.; Xu, D.-Y.; Geng, J.; Hong, W.-C. A hybrid approach for forecasting ship motion using CNN–GRU–AM and GCWOA. Appl. Soft Comput. 2022 , 114 , 108084. [ Google Scholar ] [ CrossRef ]
- Zhang, B.; Wang, S.; Deng, L.; Jia, M.; Xu, J. Ship motion attitude prediction model based on IWOA-TCN-Attention. Ocean Eng. 2023 , 272 , 113911. [ Google Scholar ] [ CrossRef ]
- Elman, J.L. Finding Structure in Time. Cogn. Sci. 1990 , 14 , 179–211. [ Google Scholar ] [ CrossRef ]
- Shan, F.; He, X.; Armaghani, D.J.; Sheng, D. Effects of data smoothing and recurrent neural network (RNN) algorithms for real-time forecasting of tunnel boring machine (TBM) performance. J. Rock Mech. Geotech. Eng. 2024 , 16 , 1538–1551. [ Google Scholar ] [ CrossRef ]
- Apaydin, H.; Feizi, H.; Sattari, M.T.; Colak, M.S.; Shamshirband, S.; Chau, K.-W. Comparative Analysis of Recurrent Neural Network Architectures for Reservoir Inflow Forecasting. Water 2020 , 12 , 1500. [ Google Scholar ] [ CrossRef ]
- Ma, Z.; Zhang, H.; Liu, J. MM-RNN: A Multimodal RNN for Precipitation Nowcasting. IEEE Trans. Geosci. Remote Sens. 2023 , 61 , 1–14. [ Google Scholar ] [ CrossRef ]
- Lu, M.; Xu, X. TRNN: An efficient time-series recurrent neural network for stock price prediction. Inf. Sci. 2024 , 657 , 119951. [ Google Scholar ] [ CrossRef ]
- Bengio, Y.; Simard, P.; Frasconi, P. Learning long-term dependencies with gradient descent is difficult. IEEE Trans. Neural Netw. 1994 , 5 , 157–166. [ Google Scholar ] [ CrossRef ]
- Kratzert, F.; Klotz, D.; Brenner, C.; Schulz, K.; Herrnegger, M. Rainfall–runoff modelling using Long Short-Term Memory (LSTM) networks. Hydrol. Earth Syst. Sci. 2018 , 22 , 6005–6022. [ Google Scholar ] [ CrossRef ]
- Hochreiter, S. Untersuchungen zu dynamischen neuronalen Netzen. Diploma Tech. Univ. München 1991 , 91 , 31. [ Google Scholar ]
- Hochreiter, S.; Schmidhuber, J. Long Short-Term Memory. Neural Comput. 1997 , 9 , 1735–1780. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Gers, F.A.; Schmidhuber, J.; Cummins, F. Learning to Forget: Continual Prediction with LSTM. Neural Comput. 2000 , 12 , 2451–2471. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Witten, I.H.; Frank, E. Data mining: Practical machine learning tools and techniques with Java implementations. SIGMOD Rec. 2002 , 31 , 76–77. [ Google Scholar ] [ CrossRef ]
- Mo, J.X.; Gao, R.B.; Liu, J.H.; Du, L.; Yuen, K.F. Annual dilated convolutional LSTM network for time charter rate forecasting. Appl. Soft Comput. 2022 , 126 , 109259. [ Google Scholar ] [ CrossRef ]
- Cho, K.; Van Merriënboer, B.; Bahdanau, D.; Bengio, Y. On the properties of neural machine translation: Encoder-decoder approaches. arXiv 2014 , arXiv:1409.1259. [ Google Scholar ]
- Yang, S.; Yu, X.; Zhou, Y. LSTM and GRU Neural Network Performance Comparison Study: Taking Yelp Review Dataset as an Example. In Proceedings of the 2020 International Workshop on Electronic Communication and Artificial Intelligence (IWECAI), Shanghai, China, 12–14 June 2020; pp. 98–101. [ Google Scholar ] [ CrossRef ]
- Zhao, Z.N.; Yun, S.N.; Jia, L.Y.; Guo, J.X.; Meng, Y.; He, N.; Li, X.J.; Shi, J.R.; Yang, L. Hybrid VMD-CNN-GRU-based model for short-term forecasting of wind power considering spatio-temporal features. Eng. Appl. Artif. Intell. 2023 , 121 , 105982. [ Google Scholar ] [ CrossRef ]
- Pan, N.; Ding, Y.; Fu, J.; Wang, J.; Zheng, H. Research on Ship Arrival Law Based on Route Matching and Deep Learning. J. Phys. Conf. Ser. 2021 , 1952 , 022023. [ Google Scholar ] [ CrossRef ]
- Ma, J.; Li, W.K.; Jia, C.F.; Zhang, C.W.; Zhang, Y. Risk Prediction for Ship Encounter Situation Awareness Using Long Short-Term Memory Based Deep Learning on Intership Behaviors. J. Adv. Transp. 2020 , 2020 , 8897700. [ Google Scholar ] [ CrossRef ]
- Suo, Y.F.; Chen, W.K.; Claramunt, C.; Yang, S.H. A Ship Trajectory Prediction Framework Based on a Recurrent Neural Network. Sensors 2020 , 20 , 5133. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Bahdanau, D.; Cho, K.; Bengio, Y. Neural machine translation by jointly learning to align and translate. arXiv 2014 , arXiv:1409.0473. [ Google Scholar ] [ CrossRef ]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. Adv. Neural Inf. Process. Syst. 2017 , 30 , 03762. [ Google Scholar ]
- Nascimento, E.G.S.; de Melo, T.A.C.; Moreira, D.M. A transformer-based deep neural network with wavelet transform for forecasting wind speed and wind energy. Energy 2023 , 278 , 127678. [ Google Scholar ] [ CrossRef ]
- Zhang, L.; Zhang, J.; Niu, J.; Wu, Q.M.J.; Li, G. Track Prediction for HF Radar Vessels Submerged in Strong Clutter Based on MSCNN Fusion with GRU-AM and AR Model. Remote Sens. 2021 , 13 , 2164. [ Google Scholar ] [ CrossRef ]
- Zhang, X.; Fu, X.; Xiao, Z.; Xu, H.; Zhang, W.; Koh, J.; Qin, Z. A Dynamic Context-Aware Approach for Vessel Trajectory Prediction Based on Multi-Stage Deep Learning. IEEE Trans. Intell. Veh. 2024 , 1–16. [ Google Scholar ] [ CrossRef ]
- Jiang, D.; Shi, G.; Li, N.; Ma, L.; Li, W.; Shi, J. TRFM-LS: Transformer-Based Deep Learning Method for Vessel Trajectory Prediction. J. Mar. Sci. Eng. 2023 , 11 , 880. [ Google Scholar ] [ CrossRef ]
- Violos, J.; Tsanakas, S.; Androutsopoulou, M.; Palaiokrassas, G.; Varvarigou, T. Next Position Prediction Using LSTM Neural Networks. In Proceedings of the 11th Hellenic Conference on Artificial Intelligence, Athens, Greece, 2–4 September 2020; pp. 232–240. [ Google Scholar ] [ CrossRef ]
- Hoque, X.; Sharma, S.K. Ensembled Deep Learning Approach for Maritime Anomaly Detection System. In Proceedings of the 1st International Conference on Emerging Trends in Information Technology (ICETIT), Inst Informat Technol & Management, New Delhi, India, 21–22 June 2020; In Lecture Notes in Electrical Engineering. Volume 605, pp. 862–869. [ Google Scholar ]
- Wang, Y.; Zhang, M.; Fu, H.; Wang, Q. Research on Prediction Method of Ship Rolling Motion Based on Deep Learning. In Proceedings of the 2020 39th Chinese Control Conference (CCC), Shenyang, China, 27–29 July 2020; pp. 7182–7187. [ Google Scholar ] [ CrossRef ]
- Choi, J. Predicting the Frequency of Marine Accidents by Navigators’ Watch Duty Time in South Korea Using LSTM. Appl. Sci. 2022 , 12 , 11724. [ Google Scholar ] [ CrossRef ]
- Li, T.; Li, Y.B. Prediction of ship trajectory based on deep learning. J. Phys. Conf. Ser. 2023 , 2613 , 012023. [ Google Scholar ] [ CrossRef ]
- Chondrodima, E.; Pelekis, N.; Pikrakis, A.; Theodoridis, Y. An Efficient LSTM Neural Network-Based Framework for Vessel Location Forecasting. IEEE Trans. Intell. Transp. Syst. 2023 , 24 , 4872–4888. [ Google Scholar ] [ CrossRef ]
- Long, Z.; Suyuan, W.; Zhongma, C.; Jiaqi, F.; Xiaoting, Y.; Wei, D. Lira-YOLO: A lightweight model for ship detection in radar images. J. Syst. Eng. Electron. 2020 , 31 , 950–956. [ Google Scholar ] [ CrossRef ]
- Cheng, X.; Li, G.; Skulstad, R.; Zhang, H.; Chen, S. SpectralSeaNet: Spectrogram and Convolutional Network-based Sea State Estimation. In Proceedings of the IECON 2020 the 46th Annual Conference of the IEEE Industrial Electronics Society, Singapore, 18–21 October 2020; pp. 5069–5074. [ Google Scholar ] [ CrossRef ]
- Wang, K.; Cheng, X.; Shi, F. Learning Dynamic Graph Structures for Sea State Estimation with Deep Neural Networks. In Proceedings of the 2023 6th International Conference on Intelligent Autonomous Systems (ICoIAS), Qinhuangdao, China, 22–24 September 2023; pp. 161–166. [ Google Scholar ]
- Yu, J.; Huang, D.; Shi, X.; Li, W.; Wang, X. Real-Time Moving Ship Detection from Low-Resolution Large-Scale Remote Sensing Image Sequence. Appl. Sci. 2023 , 13 , 2584. [ Google Scholar ] [ CrossRef ]
- Ilias, L.; Kapsalis, P.; Mouzakitis, S.; Askounis, D. A Multitask Learning Framework for Predicting Ship Fuel Oil Consumption. IEEE Access 2023 , 11 , 132576–132589. [ Google Scholar ] [ CrossRef ]
- Selimovic, D.; Hrzic, F.; Prpic-Orsic, J.; Lerga, J. Estimation of sea state parameters from ship motion responses using attention-based neural networks. Ocean Eng. 2023 , 281 , 114915. [ Google Scholar ] [ CrossRef ]
- Ma, J.; Jia, C.; Yang, X.; Cheng, X.; Li, W.; Zhang, C. A Data-Driven Approach for Collision Risk Early Warning in Vessel Encounter Situations Using Attention-BiLSTM. IEEE Access 2020 , 8 , 188771–188783. [ Google Scholar ] [ CrossRef ]
- Ji, Z.; Gan, H.; Liu, B. A Deep Learning-Based Fault Warning Model for Exhaust Temperature Prediction and Fault Warning of Marine Diesel Engine. J. Mar. Sci. Eng. 2023 , 11 , 1509. [ Google Scholar ] [ CrossRef ]
- Liu, Y.; Gan, H.; Cong, Y.; Hu, G. Research on fault prediction of marine diesel engine based on attention-LSTM. Proc. Inst. Mech. Eng. Part M J. Eng. Marit. Environ. 2023 , 237 , 508–519. [ Google Scholar ] [ CrossRef ]
- Li, M.W.; Xu, D.Y.; Geng, J.; Hong, W.C. A ship motion forecasting approach based on empirical mode decomposition method hybrid deep learning network and quantum butterfly optimization algorithm. Nonlinear Dyn. 2022 , 107 , 2447–2467. [ Google Scholar ] [ CrossRef ]
- Yang, C.H.; Chang, P.Y. Forecasting the Demand for Container Throughput Using a Mixed-Precision Neural Architecture Based on CNN–LSTM. Mathematics 2020 , 8 , 1784. [ Google Scholar ] [ CrossRef ]
- Zhang, W.; Wu, P.; Peng, Y.; Liu, D. Roll Motion Prediction of Unmanned Surface Vehicle Based on Coupled CNN and LSTM. Future Internet 2019 , 11 , 243. [ Google Scholar ] [ CrossRef ]
- Kamal, I.M.; Bae, H.; Sunghyun, S.; Yun, H. DERN: Deep Ensemble Learning Model for Short- and Long-Term Prediction of Baltic Dry Index. Appl. Sci. 2020 , 10 , 1504. [ Google Scholar ] [ CrossRef ]
- Li, M.Z.; Li, B.; Qi, Z.G.; Li, J.S.; Wu, J.W. Enhancing Maritime Navigational Safety: Ship Trajectory Prediction Using ACoAtt–LSTM and AIS Data. ISPRS Int. J. Geo-Inform. 2024 , 13 , 85. [ Google Scholar ] [ CrossRef ]
- Yu, T.; Zhang, Y.; Zhao, S.; Yang, J.; Li, W.; Guo, W. Vessel trajectory prediction based on modified LSTM with attention mechanism. In Proceedings of the 2024 4th International Conference on Neural Networks, Information and Communication Engineering, NNICE, Guangzhou, China, 19–21 January 2024; pp. 912–918. [ Google Scholar ] [ CrossRef ]
- Xia, C.; Peng, Y.; Qu, D. A pre-trained model specialized for ship trajectory prediction. In Proceedings of the IEEE Advanced Information Technology, Electronic and Automation Control Conference (IAEAC), Chongqing, China, 15–17 March 2024; pp. 1857–1860. [ Google Scholar ] [ CrossRef ]
- Cheng, X.; Li, G.; Skulstad, R.; Chen, S.; Hildre, H.P.; Zhang, H. Modeling and Analysis of Motion Data from Dynamically Positioned Vessels for Sea State Estimation. In Proceedings of the 2019 International Conference on Robotics and Automation (ICRA), Montreal, QC, Canada, 20–24 May 2019; pp. 6644–6650. [ Google Scholar ] [ CrossRef ]
- Xia, C.; Qu, D.; Zheng, Y. TATBformer: A Divide-and-Conquer Approach to Ship Trajectory Prediction Modeling. In Proceedings of the 2023 IEEE 11th Joint International Information Technology and Artificial Intelligence Conference (ITAIC), Chongqing, China, 8–10 December 2023; pp. 335–339. [ Google Scholar ] [ CrossRef ]
- Ran, Y.; Shi, G.; Li, W. Ship Track Prediction Model based on Automatic Identification System Data and Bidirectional Cyclic Neural Network. In Proceedings of the 2021 4th International Symposium on Traffic Transportation and Civil Architecture, ISTTCA, Suzhou, China, 12–14 November 2021; pp. 297–301. [ Google Scholar ] [ CrossRef ]
- Yang, C.H.; Wu, C.H.; Shao, J.C.; Wang, Y.C.; Hsieh, C.M. AIS-Based Intelligent Vessel Trajectory Prediction Using Bi-LSTM. IEEE Access 2022 , 10 , 24302–24315. [ Google Scholar ] [ CrossRef ]
- Sadeghi, Z.; Matwin, S. Anomaly detection for maritime navigation based on probability density function of error of reconstruction. J. Intell. Syst. 2023 , 32 , 20220270. [ Google Scholar ] [ CrossRef ]
- Perumal, V.; Murugaiyan, S.; Ravichandran, P.; Venkatesan, R.; Sundar, R. Real time identification of anomalous events in coastal regions using deep learning techniques. Concurr. Comput. Pract. Exp. 2021 , 33 , e6421. [ Google Scholar ] [ CrossRef ]
- Xie, J.L.; Shi, W.F.; Shi, Y.Q. Research on Fault Diagnosis of Six-Phase Propulsion Motor Drive Inverter for Marine Electric Propulsion System Based on Res-BiLSTM. Machines 2022 , 10 , 736. [ Google Scholar ] [ CrossRef ]
- Han, P.; Li, G.; Skulstad, R.; Skjong, S.; Zhang, H. A Deep Learning Approach to Detect and Isolate Thruster Failures for Dynamically Positioned Vessels Using Motion Data. IEEE Trans. Instrum. Meas. 2021 , 70 , 1–11. [ Google Scholar ] [ CrossRef ]
- Cheng, X.; Wang, K.; Liu, X.; Yu, Q.; Shi, F.; Ren, Z.; Chen, S. A Novel Class-Imbalanced Ship Motion Data-Based Cross-Scale Model for Sea State Estimation. IEEE Trans. Intell. Transp. Syst. 2023 , 24 , 15907–15919. [ Google Scholar ] [ CrossRef ]
- Lei, L.; Wen, Z.; Peng, Z. Prediction of Main Engine Speed and Fuel Consumption of Inland Ships Based on Deep Learning. J. Phys. Conf. Ser. 2021 , 2025 , 012012. [ Google Scholar ]
- Ljunggren, H. Using Deep Learning for Classifying Ship Trajectories. In Proceedings of the 21st International Conference on Information Fusion (FUSION), Cambridge, UK, 10–13 July 2018; IEEE: Piscataway, NJ, USA, 2018; pp. 2158–2164. [ Google Scholar ]
- Kulshrestha, A.; Yadav, A.; Sharma, H.; Suman, S. A deep learning-based multivariate decomposition and ensemble framework for container throughput forecasting. J. Forecast. 2024 , in press . [ Google Scholar ] [ CrossRef ]
- Shankar, S.; Ilavarasan, P.V.; Punia, S.; Singh, S.P. Forecasting container throughput with long short-term memory networks. Ind. Manag. Data Syst. 2020 , 120 , 425–441. [ Google Scholar ] [ CrossRef ]
- Lee, E.; Kim, D.; Bae, H. Container Volume Prediction Using Time-Series Decomposition with a Long Short-Term Memory Models. Appl. Sci. 2021 , 11 , 8995. [ Google Scholar ] [ CrossRef ]
- Cuong, T.N.; You, S.-S.; Long, L.N.B.; Kim, H.-S. Seaport Resilience Analysis and Throughput Forecast Using a Deep Learning Approach: A Case Study of Busan Port. Sustainability 2022 , 14 , 13985. [ Google Scholar ] [ CrossRef ]
- Song, X.; Chen, Z.S. Shipping market time series forecasting via an Ensemble Deep Dual-Projection Echo State Network. Comput. Electr. Eng. 2024 , 117 , 109218. [ Google Scholar ] [ CrossRef ]
- Li, X.; Hu, Y.; Bai, Y.; Gao, X.; Chen, G. DeepDLP: Deep Reinforcement Learning based Framework for Dynamic Liner Trade Pricing. In Proceedings of the Proceedings of the 2023 17th International Conference on Ubiquitous Information Management and Communication, IMCOM, Seoul, Republic of Korea, 3–5 January 2023; pp. 1–8. [ Google Scholar ] [ CrossRef ]
- Alqatawna, A.; Abu-Salih, B.; Obeid, N.; Almiani, M. Incorporating Time-Series Forecasting Techniques to Predict Logistics Companies’ Staffing Needs and Order Volume. Computation 2023 , 11 , 141. [ Google Scholar ] [ CrossRef ]
- Lim, S.; Kim, S.J.; Park, Y.; Kwon, N. A deep learning-based time series model with missing value handling techniques to predict various types of liquid cargo traffic. Expert Syst. Appl. 2021 , 184 , 115532. [ Google Scholar ] [ CrossRef ]
- Cheng, R.; Gao, R.; Yuen, K.F. Ship order book forecasting by an ensemble deep parsimonious random vector functional link network. Eng. Appl. Artif. Intell. 2024 , 133 , 108139. [ Google Scholar ] [ CrossRef ]
- Xiao, Z.; Fu, X.J.; Zhang, L.Y.; Goh, R.S.M. Traffic Pattern Mining and Forecasting Technologies in Maritime Traffic Service Networks: A Comprehensive Survey. IEEE Trans. Intell. Transp. Syst. 2020 , 21 , 1796–1825. [ Google Scholar ] [ CrossRef ]
- Yan, R.; Wang, S.A.; Psaraftis, H.N. Data analytics for fuel consumption management in maritime transportation: Status and perspectives. Transp. Res. Part E Logist. Transp. Rev. 2021 , 155 , 102489. [ Google Scholar ] [ CrossRef ]
- Filom, S.; Amiri, A.M.; Razavi, S. Applications of machine learning methods in port operations—A systematic literature review. Transp. Res. Part E-Logist. Transp. Rev. 2022 , 161 , 102722. [ Google Scholar ] [ CrossRef ]
- Ksciuk, J.; Kuhlemann, S.; Tierney, K.; Koberstein, A. Uncertainty in maritime ship routing and scheduling: A Literature review. Eur. J. Oper. Res. 2023 , 308 , 499–524. [ Google Scholar ] [ CrossRef ]
- Jia, H.; Prakash, V.; Smith, T. Estimating vessel payloads in bulk shipping using AIS data. Int. J. Shipp. Transp. Logist. 2019 , 11 , 25–40. [ Google Scholar ] [ CrossRef ]
- Yang, D.; Wu, L.X.; Wang, S.A.; Jia, H.Y.; Li, K.X. How big data enriches maritime research—A critical review of Automatic Identification System (AIS) data applications. Transp. Rev. 2019 , 39 , 755–773. [ Google Scholar ] [ CrossRef ]
- Liu, M.; Zhao, Y.; Wang, J.; Liu, C.; Li, G. A Deep Learning Framework for Baltic Dry Index Forecasting. Procedia Comput. Sci. 2022 , 199 , 821–828. [ Google Scholar ] [ CrossRef ]
- Wang, Y.C.; Wang, H.; Zou, D.X.; Fu, H.X. Ship roll prediction algorithm based on Bi-LSTM-TPA combined model. J. Mar. Sci. Eng. 2021 , 9 , 387. [ Google Scholar ] [ CrossRef ]
- Xie, H.T.; Jiang, X.Q.; Hu, X.; Wu, Z.T.; Wang, G.Q.; Xie, K. High-efficiency and low-energy ship recognition strategy based on spiking neural network in SAR images. Front. Neurorobotics 2022 , 16 , 970832. [ Google Scholar ] [ CrossRef ]
- Muñoz, D.U.; Ruiz-Aguilar, J.J.; González-Enrique, J.; Domínguez, I.J.T. A Deep Ensemble Neural Network Approach to Improve Predictions of Container Inspection Volume. In Proceedings of the 15th International Work-Conference on Artificial Neural Networks (IWANN), Gran Canaria, Spain, 12–14 June 2019; Springer: Berlin/Heidelberg, Germany, 2019; Volume 11506, pp. 806–817. [ Google Scholar ] [ CrossRef ]
- Velasco-Gallego, C.; Lazakis, I. Mar-RUL: A remaining useful life prediction approach for fault prognostics of marine machinery. Appl. Ocean Res. 2023 , 140 , 103735. [ Google Scholar ] [ CrossRef ]
- Zhang, X.; Zheng, K.; Wang, C.; Chen, J.; Qi, H. A novel deep reinforcement learning for POMDP-based autonomous ship collision decision-making. Neural Comput. Appl. 2023 , 1–15. [ Google Scholar ] [ CrossRef ]
- Guo, X.X.; Zhang, X.T.; Lu, W.Y.; Tian, X.L.; Li, X. Real-time prediction of 6-DOF motions of a turret-moored FPSO in harsh sea state. Ocean Eng. 2022 , 265 , 112500. [ Google Scholar ] [ CrossRef ]
- Kim, D.; Kim, T.; An, M.; Cho, Y.; Baek, Y.; IEEE. Edge AI-based early anomaly detection of LNG Carrier Main Engine systems. In Proceedings of the OCEANS Conference, Limerick, Ireland, 5–8 June 2023. [ Google Scholar ] [ CrossRef ]
- Theodoropoulos, P.; Spandonidis, C.C.; Giannopoulos, F.; Fassois, S. A Deep Learning-Based Fault Detection Model for Optimization of Shipping Operations and Enhancement of Maritime Safety. Sensors 2021 , 21 , 5658. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Huang, B.; Wu, B.; Barry, M. Geographically and temporally weighted regression for modeling spatio-temporal variation in house prices. Int. J. Geogr. Inf. Sci. 2010 , 24 , 383–401. [ Google Scholar ] [ CrossRef ]
- Zhang, W.; Xu, Y.; Streets, D.G.; Wang, C. How does decarbonization of the central heating industry affect employment? A spatiotemporal analysis from the perspective of urbanization. Energy Build. 2024 , 306 , 113912. [ Google Scholar ] [ CrossRef ]
- Zhang, D.; Li, X.; Wan, C.; Man, J. A novel hybrid deep-learning framework for medium-term container throughput forecasting: An application to China’s Guangzhou, Qingdao and Shanghai hub ports. Marit. Econ. Logist. 2024 , 26 , 44–73. [ Google Scholar ] [ CrossRef ]
- Wang, Y.; Wang, H.; Zhou, B.; Fu, H. Multi-dimensional prediction method based on Bi-LSTMC for ship roll. Ocean Eng. 2021 , 242 , 110106. [ Google Scholar ] [ CrossRef ]
Click here to enlarge figure
| Ref. | Architecture | Dataset | Advantage |
|---|---|---|---|
| [ ] | MSCNN-GRU-AM | HF radar | It is applicable for high-frequency radar ship track prediction in environments with significant clutter and interference |
| [ ] | CNN-BiLSTM-Attention | 6L34DF dual fuel diesel engine | The high prediction accuracy and early warning timeliness can provide interpretable fault prediction results |
| [ ] | LSTM | Two LNG carriers | Enables early anomaly detection in new ships and new equipment |
| [ ] | LSTM | sensors | better and high-precision effects |
| [ ] | Self-Attention-BiLSTM | A real military ship | Not only can it better capture complex ship attitude changes, but it also shows greater accuracy and stability in long-term forecasting tasks |
| [ ] | CNN–GRU–AM | A C11 containership | better accuracy of forecasting |
| [ ] | GRU | A scaled model test | good prediction accuracy |
| [ ] | CNN | A bulk carrier | good prediction accuracy |
| The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
Share and Cite
Wang, M.; Guo, X.; She, Y.; Zhou, Y.; Liang, M.; Chen, Z.S. Advancements in Deep Learning Techniques for Time Series Forecasting in Maritime Applications: A Comprehensive Review. Information 2024 , 15 , 507. https://doi.org/10.3390/info15080507
Wang M, Guo X, She Y, Zhou Y, Liang M, Chen ZS. Advancements in Deep Learning Techniques for Time Series Forecasting in Maritime Applications: A Comprehensive Review. Information . 2024; 15(8):507. https://doi.org/10.3390/info15080507
Wang, Meng, Xinyan Guo, Yanling She, Yang Zhou, Maohan Liang, and Zhong Shuo Chen. 2024. "Advancements in Deep Learning Techniques for Time Series Forecasting in Maritime Applications: A Comprehensive Review" Information 15, no. 8: 507. https://doi.org/10.3390/info15080507
Article Metrics
Article access statistics, further information, mdpi initiatives, follow mdpi.

Subscribe to receive issue release notifications and newsletters from MDPI journals
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Open access
- Published: 14 August 2024
Nonlinear dynamics of multi-omics profiles during human aging
- Xiaotao Shen ORCID: orcid.org/0000-0002-9608-9964 1 , 2 , 3 na1 ,
- Chuchu Wang ORCID: orcid.org/0000-0003-2015-7331 4 , 5 na1 ,
- Xin Zhou ORCID: orcid.org/0000-0001-8089-4507 1 , 6 ,
- Wenyu Zhou 1 ,
- Daniel Hornburg ORCID: orcid.org/0000-0002-6618-7774 1 ,
- Si Wu 1 &
- Michael P. Snyder ORCID: orcid.org/0000-0003-0784-7987 1 , 6
Nature Aging ( 2024 ) Cite this article
190k Accesses
3281 Altmetric
Metrics details
- Biochemistry
- Systems biology
Aging is a complex process associated with nearly all diseases. Understanding the molecular changes underlying aging and identifying therapeutic targets for aging-related diseases are crucial for increasing healthspan. Although many studies have explored linear changes during aging, the prevalence of aging-related diseases and mortality risk accelerates after specific time points, indicating the importance of studying nonlinear molecular changes. In this study, we performed comprehensive multi-omics profiling on a longitudinal human cohort of 108 participants, aged between 25 years and 75 years. The participants resided in California, United States, and were tracked for a median period of 1.7 years, with a maximum follow-up duration of 6.8 years. The analysis revealed consistent nonlinear patterns in molecular markers of aging, with substantial dysregulation occurring at two major periods occurring at approximately 44 years and 60 years of chronological age. Distinct molecules and functional pathways associated with these periods were also identified, such as immune regulation and carbohydrate metabolism that shifted during the 60-year transition and cardiovascular disease, lipid and alcohol metabolism changes at the 40-year transition. Overall, this research demonstrates that functions and risks of aging-related diseases change nonlinearly across the human lifespan and provides insights into the molecular and biological pathways involved in these changes.
Similar content being viewed by others

Personal aging markers and ageotypes revealed by deep longitudinal profiling
Considerations for reproducible omics in aging research

Principal component-based clinical aging clocks identify signatures of healthy aging and targets for clinical intervention
Aging is a complex and multifactorial process of physiological changes strongly associated with various human diseases, including cardiovascular diseases (CVDs), diabetes, neurodegeneration and cancer 1 . The alterations of molecules (including transcripts, proteins, metabolites and cytokines) are critically important to understand the underlying mechanism of aging and discover potential therapeutic targets for aging-related diseases. Recently, the development of high-throughput omics technologies has enabled researchers to study molecular changes at the system level 2 . A growing number of studies have comprehensively explored the molecular changes that occur during aging using omics profiling 3 , 4 , and most focus on linear changes 5 . It is widely recognized that the occurrence of aging-related diseases does not follow a proportional increase with age. Instead, the risk of these diseases accelerates at specific points throughout the human lifespan 6 . For example, in the United States, the prevalence of CVDs (encompassing atherosclerosis, stroke and myocardial infarction) is approximately 40% between the ages of 40 and 59, increases to about 75% between 60 and 79 and reaches approximately 86% in individuals older than 80 years 7 . Similarly, also in the United States, the prevalence of neurodegenerative diseases, such as Parkinson’s disease and Alzheimer’s disease, exhibits an upward trend as well as human aging progresses, with distinct turning points occurring around the ages of 40 and 65, respectively 8 , 9 , 10 . Some studies also found that brain aging followed an accelerated decline in flies 11 and chimpanzees 12 that lived past middle age and advanced age.
The observation of a nonlinear increase in the prevalence of aging-related diseases implies that the process of human aging is not a simple linear trend. Consequently, investigating the nonlinear changes in molecules will likely reveal previously unreported molecular signatures and mechanistic insights. Some studies examined the nonlinear alterations of molecules during human aging 13 . For instance, nonlinear changes in RNA and protein expression related to aging have been documented 14 , 15 , 16 . Moreover, certain DNA methylation sites have exhibited nonlinear changes in methylation intensity during aging, following a power law pattern 17 . Li et al. 18 identified the 30s and 50s as transitional periods during women’s aging. Although aging patterns are thought to reflect the underlying biological mechanisms, the comprehensive landscape of nonlinear changes of different types of molecules during aging remains largely unexplored. Remarkably, the global monitoring of nonlinear changing molecular profiles throughout human aging has yet to be fully used to extract basic insights into the biology of aging.
In the present study, we conducted a comprehensive deep multi-omics profiling on a longitudinal human cohort comprising 108 individuals aged from 25 years to 75 years. The cohort was followed over a span of several years (median, 1.7 years), with the longest monitoring period for a single participant reaching 6.8 years (2,471 days). Various types of omics data were collected from the participants’ biological samples, including transcriptomics, proteomics, metabolomics, cytokines, clinical laboratory tests, lipidomics, stool microbiome, skin microbiome, oral microbiome and nasal microbiome. The investigation explored the changes occurring across different omics profiles during human aging. Remarkably, many molecular markers and biological pathways exhibited a nonlinear pattern throughout the aging process, thereby providing valuable insight into periods of dramatic alterations during human aging.
Most of the molecules change nonlinearly during aging
We collected longitudinal biological samples from 108 participants over several years, with a median tracking period of 1.7 years and a maximum period of 6.8 years, and conducted multi-omics profiling on the samples. The participants were sampled every 3–6 months while healthy and had diverse ethnic backgrounds and ages ranging from 25 years to 75 years (median, 55.7 years). The participants’ body mass index (BMI) ranged from 19.1 kg m −2 to 40.8 kg m −2 (median, 28.2 kg m −2 ). Among the participants, 51.9% were female (Fig. 1a and Extended Data Fig. 1a–d ). For each visit, we collected blood, stool, skin swab, oral swab and nasal swab samples. In total, 5,405 biological samples (including 1,440 blood samples, 926 stool samples, 1,116 skin swab samples, 1,001 oral swab samples and 922 nasal swab samples) were collected. The biological samples were used for multi-omics data acquisition (including transcriptomics from peripheral blood mononuclear cells (PBMCs), proteomics from plasma, metabolomics from plasma, cytokines from plasma, clinical laboratory tests from plasma, lipidomics from plasma, stool microbiome, skin microbiome, oral microbiome and nasal microbiome; Methods ). In total, 135,239 biological features (including 10,346 transcripts, 302 proteins, 814 metabolites, 66 cytokines, 51 clinical laboratory tests, 846 lipids, 52,460 gut microbiome taxons, 8,947 skin microbiome taxons, 8,947 oral microbiome taxons and 52,460 nasal microbiome taxons) were acquired, resulting in 246,507,456,400 data points (Fig. 1b and Extended Data Fig. 1e,f ). The average sampling period and number of samples for each participant were 626 days and 47 samples, respectively. Notably, one participant was deeply monitored for 6.8 years (2,471 days), during which 367 samples were collected (Fig. 1c ). Overall, this extensive and longitudinal multi-omics dataset enables us to examine the molecular changes that occur during the human aging process. The detailed characteristics of all participants are provided in the Supplementary Data . For each participant, the omics data were aggregated and averaged across all healthy samples to represent the individual’s mean value, as detailed in the Methods section. Compared to cross-sectional cohorts, which have only a one-time point sample from each participant, our longitudinal dataset, which includes multiple time point samples from each participant, is more robust for detecting complex aging-related changes in molecules and functions. This is because analysis of multi-time point samples can detect participants’ baseline and robustly evaluate individuals’ longitudinal molecular changes.
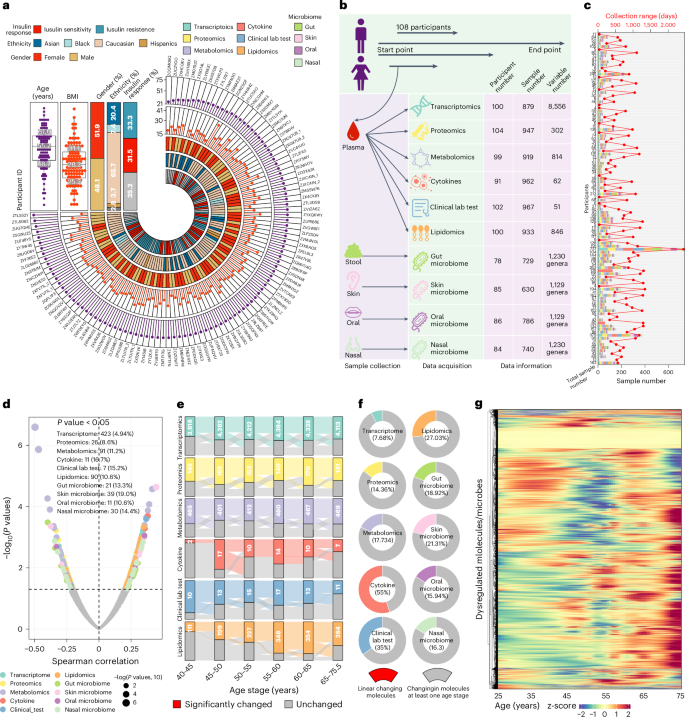
a , The demographics of the 108 participants in the study are presented. b , Sample collection and multi-omics data acquisition of the cohort. Four types of biological samples were collected, and 10 types of omics data were acquired. c , Collection time range and sample numbers for each participant. The top x axis represents the collection range for each participant (read line), and the bottom x axis represents the sample number for each participant (bar plot). Bars are color-coded by omics type. d , Significantly changed molecules and microbes during aging were detected using the Spearman correlation approach ( P < 0.05). The P values were not adjusted ( Methods ). Dots are color-coded by omics type. e , Differential expressional molecules/microbes in different age ranges compared to baseline (25–40 years old, two-sided Wilcoxon test, P < 0.05). The P values were not adjusted ( Methods ). f , The linear changing molecules comprised only a small part of dysregulated molecules in at least one age range. g , Heatmap depicting the nonlinear changing molecules and microbes during human aging.
We included samples only from healthy visits and adjusted for confounding factors (for example, BMI, sex, insulin resistance/insulin sensitivity (IRIS) and ethnicity; Extended Data Fig. 1a–d ), allowing us to discern the molecules and microbes genuinely associated with aging ( Methods ). Two common and traditional approaches, linear regression and Spearman correlation, were first used to identify the linear changing molecules during human aging 5 . The linear regression method is commonly used for linear changing molecules. As expected, both approaches have very high consistent results for each type of omics data (Supplementary Fig. 1a ). For convenience, the Spearman correlation approach was used in the analysis. Interestingly, only a small portion of all the molecules and microbes (749 out of 11,305, 6.6%; only genus level was used for microbiome data; Methods ) linearly changed during human aging (Fig. 1d and Supplementary Fig. 1b ), consistent with our previous studies 5 ( Methods ). Next, we examined nonlinear effects by categorizing all participants into distinct age stages according to their ages and investigated the dysregulated molecules within each age stage compared to the baseline (25–40 years old; Methods ). Interestingly, using this approach, 81.03% of molecules (9,106 out of 11,305) exhibited changes in at least one age stage compared to the baseline (Fig. 1e and Extended Data Fig. 2a ). Remarkably, the percentage of linear changing molecules was relatively small compared to the overall dysregulated molecules during aging (mean, 16.2%) (Fig. 1f and Extended Data Fig. 2b ). To corroborate our findings, we employed a permutation approach to calculate permutated P values, which yielded consistent results ( Methods ). The heatmap depicting all dysregulated molecules also clearly illustrates pronounced nonlinear changes (Fig. 1g ). Taken together, these findings strongly suggest that a substantial number of molecules and microbes undergo nonlinear changes throughout human aging.
Clustering reveals nonlinear multi-omics changes during aging
Next, we assessed whether the multi-omics data collected from the longitudinal cohort could serve as reliable indicators of the aging process. Our analysis revealed a substantial correlation between a significant proportion of the omics data and the ages of the participants (Fig. 2a ). Particularly noteworthy was the observation that, among all the omics data examined, metabolomics, cytokine and oral microbiome data displayed the strongest association with age (Fig. 2a and Extended Data Fig. 3a–c ). Partial least squares (PLS) regression was further used to compare the strength of the age effect across different omics data types. The results are consistent with the results presented above in Fig. 2a ( Methods ). These findings suggest the potential utility of these datasets as indicators of the aging process while acknowledging that further research is needed for validation 4 . As the omics data are not accurately matched across all the samples, we then smoothed the omics data using our previously published approach 19 ( Methods and Supplementary Fig. 2a–c ). Next, to reveal the specific patterns of molecules that change during human aging, we then grouped all the molecules with similar trajectories using an unsupervised fuzzy c-means clustering approach 19 ( Methods , Fig. 3b and Supplementary Fig. 2d,e ). We identified 11 clusters of molecular trajectories that changed during aging, which ranged in size from 638 to 1,580 molecules/microbes (Supplementary Fig. 2f and Supplementary Data ). We found that most molecular patterns exhibit nonlinear changes, indicating that aging is not a linear process (Fig. 2b ). Among the 11 identified clusters, three distinct clusters (2, 4 and 5) displayed compelling, straightforward and easily understandable patterns that spanned the entire lifespan (Fig. 2c ). Most molecules within these three clusters primarily consist of transcripts (Supplementary Fig. 2f ), which is expected because transcripts dominate the multi-omics data (8,556 out of 11,305, 75.7%). Cluster 4 exhibits a relatively stable pattern until approximately 60 years of age, after which it shows a rapid decrease (Fig. 2c ). Conversely, clusters 2 and 5 display fluctuations before 60 years of age, followed by a sharp increase and an upper inflection point at approximately 55–60 years of age (Fig. 2c ). We also attempted to observe this pattern of molecular change during aging individually. The participant with the longest follow-up period of 6.8 years (Fig. 1c ) approached the age of 60 years (range, 59.5–66.3 years; Extended Data Fig. 1g ), and it was not possible to identify obvious patterns in this short time window (Supplementary Fig. 2g ). Tracking individuals longitudinally over longer periods (decades) will be required to observe these trajectories at an individual level.
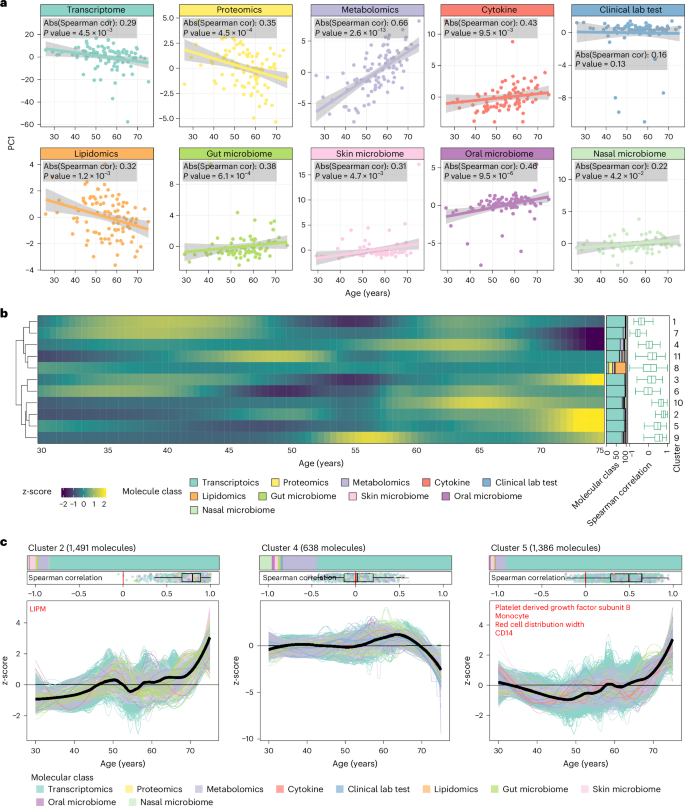
a , Spearman correlation (cor) between the first principal component and ages for each type of omics data. The shaded area around the regression line represents the 95% confidence interval. b , The heatmap shows the molecular trajectories in 11 clusters during human aging. The right stacked bar plots show the percentages of different kinds of omics data, and the right box plots show the correlation distribution between features and ages ( n = 108 participants). c , Three notable clusters of molecules that exhibit clear and straightforward nonlinear changes during human aging. The top stacked bar plots show the percentages of different kinds of omics data, and the top box plots show the correlation distribution between features and ages ( n = 108 participants). The box plot shows the median (line), interquartile range (IQR) (box) and whiskers extending to 1.5 × IQR. Bars and lines are color-coded by omics type. Abs, absolute.
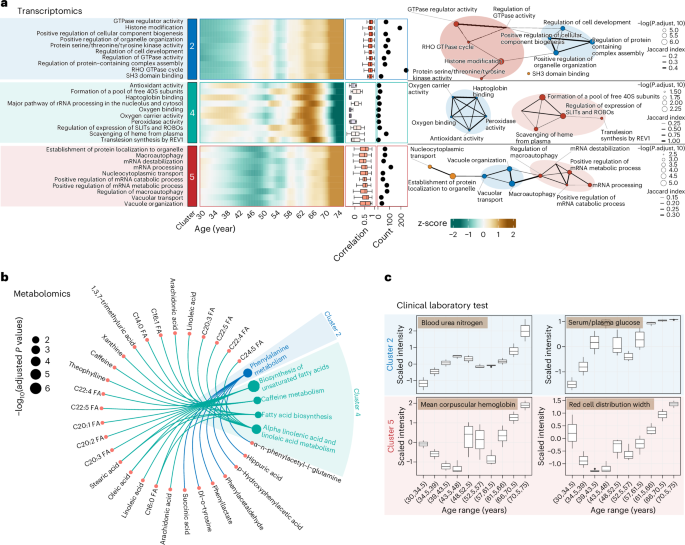
a , Pathway enrichment and module analysis for each transcriptome cluster. The left panel is the heatmap for the pathways that undergo nonlinear changes across aging. The right panel is the pathway similarity network ( Methods ) ( n = 108 participants). b , Pathway enrichment for metabolomics in each cluster. Enriched pathways and related metabolites are illustrated (Benjamini–Hochberg-adjusted P < 0.05). c , Four clinical laboratory tests that change during human aging: blood urea nitrogen, serum/plasma glucose, mean corpuscular hemoglobin and red cell distribution width ( n = 108 participants). The box plot shows the median (line), interquartile range (IQR) (box) and whiskers extending to 1.5 × IQR.
Although confounders, including sex, were corrected before analysis ( Methods ), we acknowledge that the age range for menopause in females is typically between 45 years and 55 years of age 20 , which is very close to the major transition points in all three clusters (Fig. 2c ). Therefore, we conducted further investigation into whether the menopausal status of females in the dataset contributed to the observed transition point at approximately 55 years of age (Fig. 2c ) by performing separate clustering analyses on the male and female datasets. Surprisingly, both the male and female datasets exhibited similar clusters, as illustrated in Extended Data Fig. 4a . This suggests that the transition point observed at approximately 55 years of age is not solely attributed to female menopause but, rather, represents a common phenomenon in the aging process of both sexes. This result is consistent with previous studies 14 , 15 , further supporting the notion that this transition point is a major characteristic feature of human aging. Moreover, to investigate the possibility that the transcriptomics data might skew the results toward transcriptomic changes as age-related factors, we conducted two additional clustering analyses—one focusing solely on transcriptomic data and another excluding it. Interestingly, both analyses yielded nearly identical three-cluster configurations, as observed using the complete omics dataset (Extended Data Fig. 4b ). This reinforces the robustness of the identified clusters and confirms that they are consistent across various omics platforms, not just driven by transcriptomic data.
Nonlinear changes in function and disease risk during aging
To gain further insight into the biological functions associated with the nonlinear changing molecules within the three identified clusters, we conducted separate functional analyses for transcriptomics, proteomics and metabolomics datasets for all three clusters. In brief, we constructed a similarity network using enriched pathways from various databases (Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) and Reactome) and identified modules to eliminate redundant annotations. We then used all modules from different databases to reduce redundancy further using the same approach and define the final functional modules ( Methods , Extended Data Fig. 4c and Supplementary Data ). We identified some functional modules that were reported in previous studies, but we defined their more accurate patterns of change during human aging. Additionally, we also found previously unreported potential functional modules during human aging ( Supplementary Data ). For instance, in cluster 2, we identified a transcriptomic module associated with GTPase activity (adjusted P = 1.64 × 10 −6 ) and histone modification (adjusted P = 6.36 × 10 −7 ) (Fig. 3a ). Because we lack epigenomic data in this study, our findings should be validated through additional experiments in the future. GTPase activity is closely correlated with programmed cell death (apoptosis), and some previous studies showed that this activity increases during aging 21 . Additionally, histone modifications have been demonstrated to increase during human aging 22 . In cluster 4, we identified one transcriptomics module associated with oxidative stress; this module includes antioxidant activity, oxygen carrier activity, oxygen binding and peroxidase activity (adjusted P = 0.029) (Fig. 3a ). Previous studies demonstrated that oxidative stress and many reactive oxygen species (ROS) are positively associated with increased inflammation in relation to aging 23 . In cluster 5, the first transcriptomics module is associated with mRNA stability, which includes mRNA destabilization (adjusted P = 0.0032), mRNA processing (adjusted P = 3.2 × 10 −4 ), positive regulation of the mRNA catabolic process (adjusted P = 1.46 × 10 −4 ) and positive regulation of the mRNA metabolic process (adjusted P = 0.00177) (Fig. 3a ). Previous studies showed that mRNA turnover is associated with aging 24 . The second module is associated with autophagy (Fig. 3a ), which increases during human aging, as demonstrated in previous studies 25 .
In addition, we also identified certain modules in the clusters that suggest a nonlinear increase in several disease risks during human aging. For instance, in cluster 2, where components increase gradually and then rapidly after age 60, the phenylalanine metabolism pathway (adjusted P = 4.95 × 10 −4 ) was identified (Fig. 3b ). Previous studies showed that aging is associated with a progressive increase in plasma phenylalanine levels concomitant with cardiac dysfunction, and dysregulated phenylalanine catabolism is a factor that triggers deviations from healthy cardiac aging trajectories 26 . Additionally, C-X-C motif chemokine 5 (CXCL5 or ENA78) from proteomics data, which has higher concentrations in atherosclerosis 27 , is also detected in cluster 2 ( Supplementary Data ). The clinical laboratory test blood urea nitrogen, which provides important information about kidney function, is also detected in cluster 2 (Fig. 3c ). This indicates that kidney function nonlinearly decreases during aging. Furthermore, the clinical laboratory test for serum/plasma glucose, a marker of type 2 diabetes (T2D), falls within cluster 2. This is consistent with and supported by many previous studies demonstrating that aging is a major risk factor for T2D 28 . Collectively, these findings suggest a nonlinear escalation in the risk of cardiovascular and kidney diseases and T2D with advancing age, particularly after the age of 60 years (Fig. 2c ).
The identified modules in cluster 4 also indicate a nonlinear increase in disease risks. For instance, the unsaturated fatty acids biosynthesis pathway (adjusted P = 4.71 × 10 −7 ) is decreased in cluster 4. Studies have shown that unsaturated fatty acids are helpful in reducing CVD risk and maintaining brain function 29 , 30 . The pathway of alpha-linolenic acid and linolenic acid metabolism (adjusted P = 1.32 × 10 −4 ) can reduce aging-associated diseases, such as CVD 31 . We also detected the caffeine metabolism pathway (adjusted P = 7.34 × 10 −5 ) in cluster 4, which suggests that the ability to metabolize caffeine decreases during aging. Additionally, the cytokine MCP1 (chemokine (C-C motif) ligand 2 (CCL2)), a member of the CC chemokine family, plays an important immune regulatory role and is also in cluster 4 ( Supplementary Data ). These findings further support previous observations and highlight the nonlinear increase in age-related disease risk as individuals age.
Cluster 5 comprises the clinical tests of mean corpuscular hemoglobin and red cell distribution width (Fig. 3c ). These tests assess the average hemoglobin content per red blood cell and the variability in the size and volume of red blood cells, respectively. These findings align with the aforementioned transcriptomic data, which suggest a nonlinear reduction in the oxygen-carrying capacity associated with the aging process.
Aside from these three distinct clusters (Fig. 2c ), we also conducted pathway enrichment analysis across all other eight clusters, which displayed highly nonlinear trajectories, employing the same method (Fig. 2b and Supplementary Data ). Notably, cluster 11 exhibited a consistent increase up until the age of 50, followed by a decline until the age of 56, after which no substantial changes were observed up to the age of 75. A particular transcriptomics module related to DNA repair was identified, encompassing three pathways: positive regulation of double-strand break repair (adjusted P = 0.042), peptidyl−lysine acetylation (adjusted P = 1.36 × 10 −5 ) and histone acetylation (adjusted P = 3.45 × 10 −4 ) (Extended Data Fig. 4d ). These three pathways are critical in genomic stability, gene expression and metabolic balances, with their levels diminishing across the human lifespan 32 , 33 , 34 . Our findings reveal a nonlinear alteration across the human lifespan in these pathways, indicating an enhancement in DNA repair capabilities before the age of 50, a marked reduction between the ages of 50 and 56 and stabilization after that until the age of 75. The pathway enrichment results for all clusters are detailed in the Supplementary Data .
Altogether, the comprehensive functional analysis offers valuable insights into the nonlinear changes observed in molecular profiles and their correlations with biological functions and disease risks across the human lifespan. Our findings reveal that individuals aged 60 and older exhibit increased susceptibility to CVD, kidney issues and T2D. These results carry important implications for both the diagnosis and prevention of these diseases. Notably, many clinically actionable markers were identified, which have the potential for improved healthcare management and enhanced overall well-being of the aging population.
Uncovering waves of aging-related molecules during aging
Although the trajectory clustering approaches described above effectively identify nonlinear changing molecules and microbes that exhibit clear and compelling patterns throughout human aging, it may not be as effective in capturing substantial changes that occur at specific chronological aging periods. In such cases, alternative analytical approaches may be necessary to detect and characterize these dynamics. To gain a comprehensive understanding of changes in multi-omics profiling during human aging, we used a modified version of the DE-SWAN algorithm 14 , as described in the Methods section. This algorithm identifies dysregulated molecules and microbes throughout the human lifespan by analyzing molecule levels within 20-year windows and comparing two groups in 10-year parcels while sliding the window incrementally from young to old ages 14 . Using this approach and multiomics data, we detected changes at specific stages of lifespan and uncovered the sequential effects of aging. Our analysis revealed thousands of molecules exhibiting changing patterns throughout aging, forming distinct waves, as illustrated in Fig. 3a . Notably, we observed two prominent crests occurring around the ages of 45 and 65, respectively (Fig. 4a ). Notably, too, these crests were consistent with findings from a previous study that included only proteomics data 14 . Specifically, crest 2 aligns with our previous trajectory clustering result, indicating a turning point at approximately 60 years of age (Fig. 2c ).
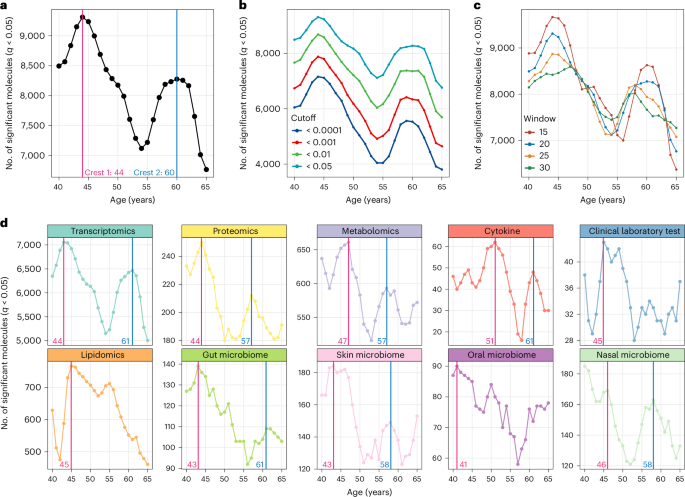
a , Number of molecules and microbes differentially expressed during aging. Two local crests at the ages of 44 years and 60 years were identified. b , c , The same waves were detected using different q value ( b ) and window ( c ) cutoffs. d , The number of molecules/microbes differentially expressed for different types of omics data during human aging.
To demonstrate the significance of the two crests, we employed different q value cutoffs and sliding window parameters, which consistently revealed the same detectable waves (Fig. 4b,c and Supplementary Fig. 4a,b ). Furthermore, when we permuted the ages of individuals, the crests disappeared (Supplementary Figs. 3a and 4c ) ( Methods ). These observations highlight the robustness of the two major waves of aging-related molecular changes across the human lifespan. Although we already accounted for confounders before our statistical analysis, we took additional steps to explore their impact. Specifically, we investigated whether confounders, such as insulin sensitivity, sex and ethnicity, differed between the two crests across various age ranges. As anticipated, these confounders did not show significant differences across other age brackets (Supplementary Fig. 4d ). This further supports our conclusion that the observed differences in the two crests are attributable to age rather than other confounding variables.
The identified crests represent notable milestones in the aging process and suggest specific age ranges where substantial molecular alterations occur. Therefore, we investigated the age-related waves for each type of omics data. Interestingly, most types of omics data exhibited two distinct crests that were highly robust (Fig. 3b and Supplementary Fig. 4 ). Notably, the proteomics data displayed two age-related crests at ages around 40 years and 60 years. Only a small overlap was observed between our dataset and the results from the previous study (1,305 proteins versus 302 proteins, with only 75 proteins overlapping). The observed pattern in our study was largely consistent with the previous findings 14 . However, our finding that many types of omics data, including transcriptomics, proteomics, metabolomics, cytokine, gut microbiome, skin microbiome and nasal microbiome, exhibit these waves, often with a similar pattern as the proteomics data (Fig. 4d ), supports the hypothesis that aging-related changes are not limited to a specific omics layer but, rather, involve a coordinated and systemic alteration across multiple molecular components. Identifying consistent crests across different omics data underscores the robustness and reliability of these molecular milestones in the aging process.
Next, we investigated the roles and functions of dysregulated molecules within two distinct crests. Notably, we found that the two crests related to aging predominantly consisted of the same molecules (Supplementary Fig. 6 ). To focus on the unique biological functions associated with each crest and eliminate commonly occurring molecules, we removed background molecules present in most stages. To explore the specific biological functions associated with each type of omics data (transcriptomics, proteomics and metabolomics) for both crests, we employed the function annotation approach described above ( Methods ). In brief, we constructed a similarity network of enriched pathways and identified modules to remove redundant annotations (Supplementary Fig. 6 and Extended Data Fig. 5a,b ). Furthermore, we applied the same approach to all modules to reduce redundancy and identify the final functional modules ( Methods and Extended Data Fig. 6a ). Our analysis revealed significant changes in multiple modules associated with the two crests (Extended Data Fig. 6b–d ). To present the results clearly, Fig. 5a displays the top 20 pathways (according to adjusted P value) for each type of omics data, and the Supplementary Data provides a comprehensive list of all identified functional modules.
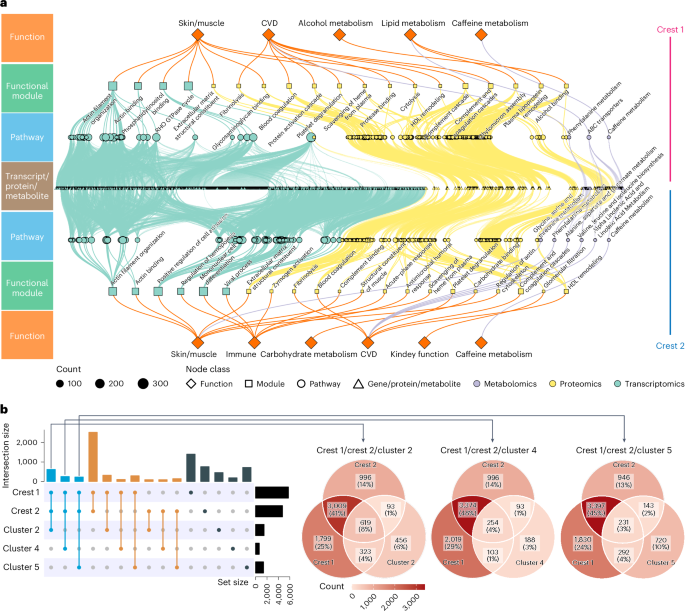
a , Pathway enrichment and biological functional module analysis for crests 1 and 2. Dots and lines are color-coded by omics type. b , The overlapping of molecules between two crests and three clusters.
Interestingly, the analysis identifies many dysregulated functional modules in crests 1 and 2, indicating a nonlinear risk for aging-related diseases. In particular, several modules associated with CVD were identified in both crest 1 and crest 2 (Fig. 5a ), which is consistent with the above results (Fig. 3b ). For instance, the dysregulation of platelet degranulation (crest 1: adjusted P = 1.77 × 10 −30 ; crest 2: adjusted P = 1.73 × 10 −26 ) 35 , 36 , complement cascade (crest 1: adjusted P = 3.84 × 10 −30 ; crest 2: adjusted P = 2.02 × 10 −28 ), complement and coagulation cascades (crest 1: adjusted P = 1.78 × 10 −46 ; crest 2: adjusted P = 2.02 × 10 −28 ) 37 , 38 , protein activation cascade (crest 1: adjusted P = 1.56 × 10 −17 ; crest 2: adjusted P = 1.61 × 10 −8 ) and protease binding (crest 1: adjusted P = 2.7 × 10 −6 ; crest 2: adjusted P = 0.0114) 39 have various effects on the cardiovascular system and can contribute to various CVDs. Furthermore, blood coagulation (crest 1: adjusted P = 1.48 × 10 −28 ; crest 2: adjusted P = 9.10 × 10 −17 ) and fibrinolysis (crest 1: adjusted P = 2.11 × 10 −15 ; crest 2: adjusted P = 1.64 × 10 −10 ) were also identified, which are essential processes for maintaining blood fluidity, and dysregulation in these modules can lead to thrombotic and cardiovascular events 40 , 41 . We also identified certain dysregulated metabolic modules associated with CVD. For example, aging has been linked to an incremental rise in plasma phenylalanine levels (crest 1: adjusted P = 9.214 × 10 −4 ; crest 2: adjusted P = 0.0453), which can contribute to the development of cardiac hypertrophy, fibrosis and dysfunction 26 . Branched-chain amino acids (BCAAs), including valine, leucine and isoleucine (crest 1: adjusted P : not significant (NS); crest 2: adjusted P = 0.0173), have also been implicated in CVD development 42 , 43 and T2D, highlighting their relevance in CVD pathophysiology. Furthermore, research suggests that alpha-linolenic acid (ALA) and linoleic acid metabolism (crest 1: adjusted P : NS; crest 2: adjusted P = 0.0217) may be protective against coronary heart disease 44 , 45 . Our investigation also identified lipid metabolism modules that are associated with CVD, including high-density lipoprotein (HDL) remodeling (crest 1: adjusted P = 1.073 × 10 −8 ; crest 2: adjusted P = 2.589 × 10 −9 ) and glycerophospholipid metabolism (crest 1: adjusted P : NS; crest 2: adjusted P = 0.0033), which influence various CVDs 46 , 47 , 48 .
In addition, the dysregulation of skin and muscle stability was found to be increased at crest 1 and crest 2, as evidenced by the identification of numerous modules associated with these processes (Fig. 5a,b ). This suggests that the aging of skin and muscle is markedly accelerated at crest 1 and crest 2. The extracellular matrix (ECM) provides structural stability, mechanical strength, elasticity and hydration to the tissues and cells, and the ECM of the skin is mainly composed of collagen, elastin and glycosaminoglycans (GAGs) 49 . Phosphatidylinositols are a class of phospholipids that have various roles in cytoskeleton organization 50 . Notably, the dysregulation of ECM structural constituent (crest 1: adjusted P = 3.32 × 10 −8 ; crest 2: adjusted P = 1.61 × 10 −8 ), GAG binding (crest 1: adjusted P = 1.805 × 10 −8 ; crest 2: adjusted P = 4.093 × 10 −6 ) and phosphatidylinositol binding (crest 1: adjusted P = 3.391 × 10 −6 ; crest 2: adjusted P = 7.832 × 10 −6 ) were identified 51 , 52 . We also identified cytolysis (crest 1: adjusted P = 2.973 × 10 −5 ; crest 2: adjusted P : NS), which can increase water loss 53 . The dysregulated actin binding (crest 1: adjusted P = 3.536 × 10 −8 ; crest 2: adjusted P = 3.435 × 10 −9 ), actin filament organization (crest 1: adjusted P = 8.406 × 10 −9 ; crest 2: adjusted P = 1.157 × 10 −9 ) and regulation of actin cytoskeleton (crest 1: adjusted P = 0.00090242; crest 2: adjusted P = 6.788 × 10 −4 ) were identified, which affect the structure and function of various tissues 54 , 55 , 56 , 57 , 58 . Additionally, cell adhesion is the attachment of a cell to another cell or to ECM via adhesion molecules 59 . We identified the positive regulation of cell adhesion (crest 1: adjusted P = 3.618 × 10 −5 ; crest 2: adjusted P = 8.272 × 10 −9 ) module, which can prevent or delay skin aging 60 , 61 . Threonine can affect sialic acid production, which is involved in cell adhesion 62 . We also identified the glycine, serine and threonine metabolism (crest 1: adjusted P : NS; crest 2: adjusted P = 0.00506) 62 . Additionally, scavenging of heme from plasma was identified (crest 1: adjusted P = 1.176 × 10 −11 ; crest 2: adjusted P = 1.694 × 10 −8 ), which can modulate skin aging as excess-free heme can damage cellular components 63 , 64 . Rho GTPases regulate a wide range of cellular responses, including changes to the cytoskeleton and cell adhesion (RHO GTPase cycle, crest 1: adjusted P = 9.956 × 10 −10 ; crest 2: adjusted P = 1.546 × 10 −5 ) 65 . In relation to muscle, previous studies demonstrated that muscle mass decreases by approximately 3–8% per decade after the age of 30, with an even higher decline rate after the age of 60, which consistently coincides with the observed second crest 66 . Interestingly, we identified dysregulation in the module associated with the structural constituent of muscle (crest 1: adjusted P = 0.00565; crest 2: adjusted P = 0.0162), consistent with previous findings 66 . Furthermore, we identified the pathway associated with caffeine metabolism (crest 1: adjusted P = 0.00378; crest 2: adjusted P = 0.0162), which is consistent with our observations above (Fig. 2b ) and implies that the capacity to metabolize caffeine undergoes a notable alteration not only around 60 years of age but also around the age of 40 years.
In crest 1, we identified specific modules associated with lipid and alcohol metabolism. Previous studies established that lipid metabolism declines with human aging 67 . Our analysis revealed several modules related to lipid metabolism, including plasma lipoprotein remodeling (crest 1: adjusted P = 2.269 × 10 −9 ), chylomicron assembly (crest 1: adjusted P = 9.065 × 10 −7 ) and ATP-binding cassette (ABC) transporters (adjusted P = 1.102 × 10 −4 ). Moreover, we discovered a module linked to alcohol metabolism (alcohol binding, adjusted P = 8.485 × 10 −7 ), suggesting a decline in alcohol metabolization efficiency with advancing age, particularly around the age of 40, when it significantly diminishes. In crest 2, we observed prominent modules related to immune dysfunction, encompassing acute-phase response (adjusted P = 2.851 × 10 −8 ), antimicrobial humoral response (adjusted P = 2.181 × 10 −5 ), zymogen activation (adjusted P = 4.367 × 10 −6 ), complement binding (adjusted P = 0.002568), mononuclear cell differentiation (adjusted P = 9.352 × 10 −8 ), viral process (adjusted P = 5.124 × 10 −7 ) and regulation of hemopoiesis (adjusted P = 3.522 × 10 −7 ) (Fig. 5a ). Age-related changes in the immune system, collectively known as immunosenescence, have been extensively documented 68 , 69 , 70 , and our results demonstrate a rapid decline at age 60. Furthermore, we also identified modules associated with kidney function (glomerular filtration, adjusted P = 0.00869) and carbohydrate metabolism (carbohydrate binding, adjusted P = 0.01045). Our previous findings indicated a decline in kidney function around the age of 60 years (Fig. 3c ), as did the present result of this observation. Previous studies indicated the influence of carbohydrates on aging, characterized by the progressive decline of physiological functions and increased susceptibility to diseases over time 71 , 72 .
In summary, our analysis identifies many dysregulated functional modules identified in both crest 1 and crest 2 that underlie the risk for various diseases and alterations of biological functions. Notably, we observed an overlap of dysregulated functional modules among clusters 2, 4 and 6 because they overlap at the molecular level, as depicted in Fig. 5b . This indicates that certain molecular components are shared among these clusters and the identified crests. However, it is important to note that numerous molecules are specific to each of the two approaches employed in our study. This suggests that these two approaches complement each other in identifying nonlinear changes in molecules and functions during human aging. By using both approaches, we were able to capture a more comprehensive understanding of the molecular alterations associated with aging and their potential implications for diseases.
Analyzing a longitudinal multi-omics dataset involving 108 participants, we successfully captured the dynamic and nonlinear molecular changes that occur during human aging. Our study’s strength lies in the comprehensive nature of the dataset, which includes multiple time point samples for each participant. This longitudinal design enhances the reliability and robustness of our findings compared to cross-sectional studies with only one time point sample for each participant. The first particularly intriguing finding from our analysis is that only a small fraction of molecules (6.6%) displayed linear changes throughout human aging (Fig. 1d ). This observation is consistent with previous research and underscores the limitations of relying solely on linear regression to understand the complexity of aging-related molecular changes 5 . Instead, our study revealed that a considerable number of molecules (81.0%) exhibited nonlinear patterns (Fig. 1e ). Notably, this nonlinear trend was observed across all types of omics data with remarkably high consistency (Fig. 1e,g ), highlighting the widespread functionally relevant nature of these dynamic changes. By unveiling the nonlinear molecular alterations associated with aging, our research contributes to a more comprehensive understanding of the aging process and its molecular underpinnings.
To further investigate the nonlinear changing molecules observed in our study, we employed a trajectory clustering approach to group molecules with similar temporal patterns. This analysis revealed the presence of three distinct clusters (Fig. 2c ) that exhibited clear and compelling patterns across the human lifespan. These clusters suggest that there are specific age ranges, such as around 60 years old, where distinct and extensive molecular changes occur (Fig. 2c ). Functional analysis revealed several modules that exhibited nonlinear changes during human aging. For example, we identified a module associated with oxidative stress, which is consistent with previous studies linking oxidative stress to the aging process 23 (Fig. 3a ). Our analysis indicates that this pathway increases significantly after the age of 60 years. In cluster 5, we identified a transcriptomics module associated with mRNA stabilization and autophagy (Fig. 3a ). Both of these processes have been implicated in the aging process and are involved in maintaining cellular homeostasis and removing damaged components. Furthermore, our analysis uncovered nonlinear changes in disease risk across aging. In cluster 2, we identified the phenylalanine metabolism pathway (Fig. 3b ), which has been associated with cardiac dysfunction during aging 26 . Additionally, we found that the clinical laboratory tests blood urea nitrogen and serum/plasma glucose increase significantly with age (cluster 2; Fig. 3c ), indicating a nonlinear decline in kidney function and an increased risk of T2D with age, with a critical threshold occurring approximately at the age of 60 years. In cluster 4, we identified pathways related to cardiovascular health, such as the biosynthesis of unsaturated fatty acids and caffeine metabolism (Fig. 3b ). Overall, our study provides compelling evidence for the existence of nonlinear changes in molecular profiles during human aging. By elucidating the specific functional modules and disease-related pathways that exhibit such nonlinear changes, we contribute to a better understanding of the complex molecular dynamics underlying the aging process and its implications for disease risk.
Although the trajectory clustering approach proves effective in identifying molecules that undergo nonlinear changes, it may not be as proficient in capturing substantial alterations that occur at specific time points without exhibiting a consistent pattern in other stages. We then employed a modified version of the DE-SWAN algorithm 14 to comprehensively investigate changes in multi-omics profiling throughout human aging. This approach enabled us to identify waves of dysregulated molecules and microbes across the human lifespan. Our analysis revealed two prominent crests occurring around the ages of 40 years and 60 years, which were consistent across various omics data types, suggesting their universal nature (Fig. 4a,e ). Notably, in the proteomics data, we observed crests around the ages of 40 years and 60 years, which aligns approximately with a previous study (which reported crests at ages 34 years, 60 years and 78 years) 14 . Due to the age range of our cohort being 25–75 years, we did not detect the third peak. Furthermore, the differences in proteomics data acquisition platforms (mass spectrometry versus SomaScan) 14 , 73 resulted in different identified proteins, with only a small overlap (1,305 proteins versus 302 proteins, of which only 75 were shared). This discrepancy may explain the age variation of the first crest identified in the two studies (approximately 10 years). However, despite the differences in the two proteomics datasets, the wave patterns observed in both studies were highly similar 14 (Fig. 4a ). Remarkably, by considering multiple omics data types, we consistently identified similar crests for each type, indicating the universality of these waves of change across plasma molecules and microbes from various body sites (Fig. 4e and Supplementary Fig. 3 ).
The analysis of molecular functionality in the two distinct crests revealed the presence of several modules, indicating a nonlinear increase in the risks of various diseases (Fig. 5a ). Both crest 1 and crest 2 exhibit the identification of multiple modules associated with CVD, which aligns with the aforementioned findings (Fig. 3b ). Moreover, we observed an escalated dysregulation in skin and muscle functioning in both crest 1 and crest 2. Additionally, we identified a pathway linked to caffeine metabolism, indicating a noticeable alteration in caffeine metabolization not only around the age of 60 but also around the age of 40. This shift may be due to either a metabolic shift or a change in caffeine consumption. In crest 1, we also identified specific modules associated with lipid and alcohol metabolism, whereas crest 2 demonstrated prominent modules related to immune dysfunction. Furthermore, we also detected modules associated with kidney function and carbohydrate metabolism, which is consistent with our above results. These findings reinforce our previous observations regarding a decline in kidney function around the age of 60 years (Fig. 3c ) while shedding light on the impact of dysregulated functional modules in both crest 1 and crest 2, suggesting nonlinear changes in disease risk and functional dysregulation. Notably, we identified an overlap of dysregulated functional modules among clusters 2, 4 and 6, indicating molecular-level similarities between these clusters and the identified crests (Fig. 5b ). This suggests the presence of shared molecular components among these clusters and crests. However, it is crucial to note that there are also numerous molecules specific to each of the two approaches employed in our study, indicating that these approaches complement each other in identifying nonlinear changes in molecules and functions during human aging.
The present research is subject to certain constraints. We accounted for many basic characteristics (confounders) of participants in the cohort; but because this study primarily reflects between-individual differences, there may be additional confounders due to the different age distributions of the participants. For example, we identified a notable decrease in oxygen carrier activity around age 60 (Figs. 2c and 3a ) and marked variations in alcohol and caffeine metabolism around ages 40 and 60 (Fig. 3a ). However, these findings might be shaped by participants’ lifestyle—that is, physical activity and their alcohol and caffeine intake. Regrettably, we do not have such detailed behavioral data for the entire group, necessitating validation in upcoming research. Although initial BMI and insulin sensitivity measurements were available at cohort entry, subsequent metrics during the observation span were absent, marking a study limitation.
A further constraint is our cohort’s modest size, encompassing merely 108 individuals (eight individuals between 25 years and 40 years of age), which hampers the full utilization of deep learning and may affect the robustness of the identification of nonlinear changing features in Fig. 1e . Although advanced computational techniques, including deep learning, are pivotal for probing nonlinear patterns, our sample size poses restrictions. Expanding the cohort size in subsequent research would be instrumental in harnessing the full potential of machine learning tools. Another limitation of our study is that the recruitment of participants was within the community around Stanford University, driven by rigorous sample collection procedures and the substantial expenses associated with setting up a longitudinal cohort. Although our participants exhibited a considerable degree of ethnic age and biological sex diversity (Fig. 1a and Supplementary Data ), it is important to acknowledge that our cohort may not fully represent the diversity of the broader population. The selectivity of our cohort limits the generalizability of our findings. Future studies should aim to include a more diverse cohort to enhance the external validity and applicability of the results.
In addition, the mean observation span for participants was 626 days, which is insufficient for detailed inflection point analyses. Our cohort’s age range of 25–70 years lacks individuals who lie outside of this range. The molecular nonlinearity detected might be subject to inherent variations or oscillations, a factor to consider during interpretation. Our analysis has not delved into the nuances of the dynamical systems theory, which provides a robust mathematical framework for understanding observed behaviors. Delving into this theory in future endeavors may yield enhanced clarity and interpretation of the data.
Moreover, it should be noted that, in our study, the observed nonlinear molecular changes occurred across individuals of varying ages rather than within the same individuals. This is attributed to the fact that, despite our longitudinal study, the follow-up period for our participants was relatively brief for following aging patterns (median, 1.7 years; Extended Data Fig. 1g ). Such a timeframe is inadequate for detecting nonlinear molecular changes that unfold over decades throughout the human lifespan. Addressing this limitation in future research is essential.
Lastly, our study’s molecular data are derived exclusively from blood samples, casting doubt on its direct relevance to specific tissues, such as the skin or muscles. We propose that blood gene expression variations might hint at overarching physiological alterations, potentially impacting the ECM in tissues, including skin and muscle. Notably, some blood-based biomarkers and transcripts have demonstrated correlations with tissue modifications, inflammation and other elements influencing the ECM across diverse tissues 74 , 75 .
In our future endeavors, the definitive confirmation of our findings hinges on determining if nonlinear molecular patterns align with nonlinear changes in functional capacities, disease occurrences and mortality hazards. For a holistic grasp of this, amalgamating multifaceted data from long-term cohort studies covering several decades becomes crucial. Such data should encompass molecular markers, comprehensive medical records, functional assessments and mortality data. Moreover, employing cutting-edge statistical techniques is vital to intricately decipher the ties between these nonlinear molecular paths and health-centric results.
In summary, the unique contribution of our study lies not merely in reaffirming the nonlinear nature of aging but also in the depth and breadth of the multi-omics data that we analyzed. Our study goes beyond stating that aging is nonlinear by identifying specific patterns, inflection points and potential waves in aging across multiple layers of biological data during human aging. Identifying specific clusters with distinct patterns, functional implications and disease risks enhances our understanding of the aging process. By considering the nonlinear dynamics of aging-related changes, we can gain insights into specific periods of significant changes (around age 40 and age 60) and the molecular mechanisms underlying age-related diseases, which could lead to the development of early diagnosis and prevention strategies. These comprehensive multi-omics data and the approach allow for a more nuanced understanding of the complexities involved in the aging process, which we think adds value to the existing body of research. However, further research is needed to validate and expand upon these findings, potentially incorporating larger cohorts to capture the full complexity of aging.
The participant recruitment, sample collection, data acquisition and data processing were documented in previous studies conducted by Zhou et al. 76 , Ahadi et al. 5 , Schüssler-Fiorenza Rose et al. 77 , Hornburg et al. 78 and Zhou et al. 79 .
Participant recruitment
Participants provided informed written consent for the study under research protocol 23602, which was approved by the Stanford University institutional review board. This study adheres to all relevant ethical regulations, ensuring informed consents were obtained from all participants. All participants consented to publication of potentially identifiable information. The cohort comprised 108 participants who underwent follow-up assessments. Exclusion criteria encompassed conditions such as anemia, kidney disease, a history of CVD, cancer, chronic inflammation or psychiatric illnesses as well as any prior bariatric surgery or liposuction. Each participant who met the eligibility criteria and provided informed consent underwent a one-time modified insulin suppression test to quantify insulin-mediated glucose uptake at the beginning of the enrollment 76 . The steady-state plasma glucose (SSPG) levels served as a direct indicator of each individual’s insulin sensitivity in processing a glucose load. We categorized individuals with SSPG levels below 150 mg dl −1 as insulin sensitive and those with levels of 150 mg dl −1 or higher as insulin resistant 80 , 81 . Thirty-eight participants were missing SSPG values, rendering their insulin resistance or sensitivity status undetermined. We also collected fasting plasma glucose (FPG) data for 69 participants at enrollment. Based on the FPG levels, two participants were identified as having diabetes at enrollment, with FPG levels exceeding 126 mg dl −1 ( Supplementary Data ). Additionally, we measured hemoglobin A1C (HbA1C) levels during each visit, using it as a marker for average glucose levels over the past 3 months: 6.5% or higher indicates diabetes. Accordingly, four participants developed diabetes during the study period. At the beginning of the enrollment, BMI was also measured for each participant. Participants received no compensation.
Comprehensive sample collection was conducted during the follow-up period, and multi-omics data were acquired (Fig. 1b ). For each visit, the participants self-reported as healthy or non-healthy 76 . To ensure accuracy and minimize the impact of confounding factors, only samples from individuals classified as healthy were selected for subsequent analysis.
Transcriptomics
Transcriptomic profiling was conducted on flash-frozen PBMCs. RNA isolation was performed using a QIAGEN All Prep kit. Subsequently, RNA libraries were assembled using an input of 500 ng of total RNA. In brief, ribosomal RNA (rRNA) was selectively eliminated from the total RNA pool, followed by purification and fragmentation. Reverse transcription was carried out using a random primer outfitted with an Illumina-specific adaptor to yield a cDNA library. A terminal tagging procedure was used to incorporate a second adaptor sequence. The final cDNA library underwent amplification. RNA sequencing libraries underwent sequencing on an Illumina HiSeq 2000 platform. Library quantification was performed via an Agilent Bioanalyzer and Qubit fluorometric quantification (Thermo Fisher Scientific) using a high-sensitivity dsDNA kit. After normalization, barcoded libraries were pooled at equimolar ratios into a multiplexed sequencing library. An average of 5–6 libraries were processed per HiSeq 2000 lane. Standard Illumina pipelines were employed for image analysis and base calling. Read alignment to the hg19 reference genome and personal exomes was achieved using the TopHat package, followed by transcript assembly and expression quantification via HTseq and DESeq2. In the realm of data pre-processing, genes with an average read count across all samples lower than 0.5 were excluded. Samples exhibiting an average read count lower than 0.5 across all remaining genes were likewise removed. For subsequent global variance and correlation assessments, genes with an average read count of less than 1 were eliminated.
Plasma sample tryptic peptides were fractionated using a NanoLC 425 System (SCIEX) operating at a flow rate of 5 μl min −1 under a trap-elute configuration with a 0.5 × 10 mm ChromXP column (SCIEX). The liquid chromatography gradient was programmed for a 43-min run, transitioning from 4% to 32% of mobile phase B, with an overall run time of 1 h. Mobile phase A consisted of water with 0.1% formic acid, and mobile phase B was formulated with 100% acetonitrile and 0.1% formic acid. An 8-μg aliquot of non-depleted plasma was loaded onto a 15-cm ChromXP column. Mass spectrometry analysis was executed employing SWATH acquisition on a TripleTOF 6600 system. A set of 100 variable Q1 window SWATH acquisition methods was designed in high-sensitivity tandem mass spectrometry (MS/MS) mode. Subsequent data analysis included statistical scoring of peak groups from individual runs via pyProphet 82 , followed by multi-run alignment through TRIC60, ultimately generating a finalized data matrix with a false discovery rate (FDR) of 1% at the peptide level and 10% at the protein level. Protein quantitation was based on the sum of the three most abundant peptide signals for each protein. Batch effect normalization was achieved by subtracting principal components that primarily exhibited batch-associated variation, using Perseus software v.1.4.2.40.
Untargeted metabolomics
A ternary solvent system of acetone, acetonitrile and methanol in a 1:1:1 ratio was used for metabolite extraction. The extracted metabolites were dried under a nitrogen atmosphere and reconstituted in a 1:1 methanol:water mixture before analysis. Metabolite profiles were generated using both hydrophilic interaction chromatography (HILIC) and reverse-phase liquid chromatography (RPLC) under positive and negative ion modes. Thermo Q Exactive Plus mass spectrometers were employed for HILIC and RPLC analyses, respectively, in full MS scan mode. MS/MS data were acquired using quality control (QC) samples. For the HILIC separations, a ZIC-HILIC column was used with mobile phase solutions of 10 mM ammonium acetate in 50:50 and 95:5 acetonitrile:water ratios. In the case of RPLC, a Zorbax SBaq column was used, and the mobile phase consisted of 0.06% acetic acid in water and methanol. Metabolic feature detection was performed using Progenesis QI software. Features from blanks and those lacking sufficient linearity upon dilution were excluded. Only features appearing in more than 33% of the samples were retained for subsequent analyses, and any missing values were imputed using the k -nearest neighbors approach. We employed locally estimated scatterplot smoothing (LOESS) normalization 83 to correct the metabolite-specific signal drift over time. The metid package 84 was used for metabolite annotation.
Cytokine data
A panel of 62 human cytokines, chemokines and growth factors was analyzed in EDTA-anticoagulated plasma samples using Luminex-based multiplex assays with conjugated antibodies (Affymetrix). Raw fluorescence measurements were standardized to median fluorescence intensity values and subsequently subjected to variance-stabilizing transformation to account for batch-related variations. As previously reported 76 , data points characterized by background noise, termed CHEX, that deviate beyond five standard deviations from the mean (mean ± 5 × s.d.) were excluded from the analyses.
Clinical laboratory test
The tests encompassed a comprehensive metabolic panel, a full blood count, glucose and HbA1C levels, insulin assays, high-sensitivity C-reactive protein (hsCRP), immunoglobulin M (IgM) and lipid, kidney and liver panels.
Lipid extraction and quantification procedures were executed in accordance with established protocols 78 . In summary, complex lipids were isolated from 40 μl of EDTA plasma using a solvent mixture comprising methyl tertiary-butyl ether, methanol and water, followed by a biphasic separation. Subsequent lipid analysis was conducted on the Lipidyzer platform, incorporating a differential mobility spectrometry device (SelexION Technology) and a QTRAP 5500 mass spectrometer (SCIEX).
Immediately after arrival, samples were stored at −80 °C. Stool and nasal samples were processed and sequenced in-house at the Jackson Laboratory for Genomic Medicine, whereas oral and skin samples were outsourced to uBiome for additional processing. Skin and oral samples underwent 30 min of beads-beating lysis, followed by a silica-guanidinium thiocyanate-based nucleic acid isolation protocol. The V4 region of the 16S rRNA gene was amplified using specific primers, after which the DNA was barcoded and sequenced on an Illumina NextSeq 500 platform via a 2 × 150-bp paired-end protocol. Similarly, stool and nasal samples were processed for 16S rRNA V1–V3 region amplification using a different set of primers and sequenced on an Illumina MiSeq platform. For data processing, the raw sequencing data were demultiplexed using BCL2FASTQ software and subsequently filtered for quality. Reads with a Q-score lower than 30 were excluded. The DADA2 R package was used for further sequence data processing, which included filtering out reads with ambiguous bases and errors, removing chimeras and aligning sequences against a validated 16S rRNA gene database. Relative abundance calculations for amplicon sequence variants (ASVs) were performed, and samples with inadequate sequencing depth (<1,000 reads) were excluded. Local outlier factor (LOF) was calculated for each point on a depth-richness plot, and samples with abnormal LOF were removed. In summary, rigorous procedures were followed in both the collection and processing stages, leveraging automated systems and specialized software to ensure the quality and integrity of the microbiome data across multiple body sites.
Statistics and reproducibility
For all data processing, statistical analysis and data visualization tasks, RStudio, along with R language (v.4.2.1), was employed. A comprehensive list of the packages used can be found in the Supplementary Note . The Benjamini–Hochberg method was employed to account for multiple comparisons. Spearman correlation coefficients were calculated using the R functions ‘cor’ and ‘cor.test’. Principal-component analysis (PCA) was conducted using the R function ‘princomp’. Before all the analyses, the confounders, such as BMI, sex, IRIS and ethnicity, were adjusted using the previously published method 19 . In brief, we used the intensity of each feature as the dependent variable (Y) and the confounding factors as the independent variables (X) to build a linear regression model. The residuals from this model were then used as the adjusted values for that specific feature.
All the omics data were acquired randomly. No statistical methods were used to predetermine the sample size, but our sample sizes are similar to those reported in previous publications 5 , 76 , 77 , 78 , 79 , and no data were excluded from the analyses. Additionally, the investigators were blinded to allocation during experiments and outcome assessment to the conditions of the experiments. Data distribution was assumed to be normal, but this was not formally tested.
The icons used in figures are from iconfont.cn, which can be used for non-commercial purposes under the MIT license ( https://pub.dev/packages/iconfont/license ).
Cross-sectional dataset generation
The ‘cross-sectional’ dataset was created by briefly extracting information from the longitudinal dataset. The mean value was calculated to represent each molecule’s intensity for each participant. Similarly, the age of each participant was determined by calculating the mean value of ages across all sample collection time points.
Linear changing molecule detection
We detected linear changing molecules during human aging using Spearman correlation and linear regression modeling. The confounders, such as BMI, sex, IRIS and ethnicity, were adjusted using the previously published method 19 . Our analysis revealed a high correlation between these two approaches in identifying such molecules. Based on these findings, we used the Spearman correlation approach to showcase the linear changing molecules during human aging. The permutation test was also used to get the permutated P values for each feature. In brief, each feature was subjected to sample label shuffling followed by a recalculation of the Spearman correlation. This process was reiterated 10,000 times, yielding 10,000 permuted Spearman correlations. The original Spearman correlation was then compared against these permuted values to obtain the permuted P values.
Dysregulated molecules compared to baseline during human aging
To depict the dysregulated molecules during human aging compared to the baseline, we categorized the participants into different age stages based on their ages. The baseline stage was defined as individuals aged 25–40 years. For each age stage group, we employed the Wilcoxon test to identify dysregulated molecules in comparison to the baseline, considering a significance threshold of P < 0.05. Before the statistical analysis, all the confounders were corrected. Subsequently, we visualized the resulting dysregulated molecules at different age stages using a Sankey plot. The permutation test was also used to get the permutated P values for each feature. In brief, we shuffled the sample labels and recalculated the absolute mean difference between the two groups, against which the actual absolute mean difference was benchmarked to derive the permuted P values. To identify the molecules and microbes that exhibited significant changes at any given age stage, we adjusted the P values for each feature by multiplying them by 6. This adjustment adheres to the Bonferroni correction method, ensuring a rigorous evaluation of statistical significance.
Evaluation of the age reflected by different types of omics data
To assess whether each type of omics data accurately reflects the ages of individuals in our dataset, we conducted a PCA. Subsequently, we computed the Spearman correlation coefficient between the ages of participants and the first principal component (PC1). The absolute value of this coefficient was used to evaluate the degree to which the omics data reflect the ages (Fig. 2a ). PLS regression was also used to compare the strength of the age effect to the different omics data types. In brief, the ‘pls’ function from the R package mixOmics was used to construct the regression model between omics data and ages. Then, the ‘perf’ function was used to assess the performance of all the modules with sevenfold cross-validation. The R 2 was extracted to assess the strength of the age effect on the different omics data types.
To accommodate the varying time points of biological and omics data, we employed the LOESS approach. This approach allowed us to smooth and predict the multi-omics data at specific time points (that is, every half year) 14 , 85 . In brief, for each molecule, we fitted a LOESS regression model. During the fitting process, the LOESS argument ‘span’ was optimized through cross-validation. This ensured that the LOESS model provided an accurate and non-overfitting fit to the data (Supplementary Fig. 2a,b ). Once we obtained the LOESS prediction model, we applied it to predict the intensity of each molecule at every half-year time point.
Trajectory clustering analysis
To conduct trajectory clustering analysis, we employed the fuzzy c-means clustering approach available in the R package ‘Mfuzz.’ This approach was previously described in our publication 19 . The analysis proceeded in several steps. First, the omics data were auto-scaled to ensure comparable ranges. Next, we computed the minimum centroid distances for a range of cluster numbers, specifically from 2 to 22, in step 1. These minimum centroid distances served as a cluster validity index, helping us determine the optimal cluster number. Based on predefined rules, we selected the optimal cluster number. To refine the accuracy of this selection, we merged clusters with center expression data correlations greater than 0.8 into a single cluster. This step aimed to capture similar patterns within the data. The resulting optimal cluster number was then used for the fuzzy c-means clustering. Only molecules with memberships above 0.5 were retained within each cluster for further analysis. This threshold ensured that the molecules exhibited a strong association with their assigned cluster and contributed considerably to the cluster’s characteristics.
Pathway enrichment analysis and functional module identification
Transcriptomics and proteomics pathway enrichment.
Pathway enrichment analysis was conducted using the ‘clusterProfiler’ R package 86 . The GO, KEGG and Reactome databases were used. The P values were adjusted using the Benjamini–Hochberg method, with a significance threshold set at <0.05. To minimize redundant enriched pathways and GO terms, we employed a series of analyses. First, for enriched GO terms, we used the ‘Wang’ algorithm from the R package ‘simplifyEnrichment’ to calculate the similarity between GO terms. Only connections with a similarity score greater than 0.7 were retained to construct the GO term similarity network. Subsequently, community analysis was performed using the ‘igraph’ R package to partition the network into distinct modules. The GO term with the smallest enrichment adjusted P value was chosen as the representative within each module. The same approach was applied to the enriched KEGG and Reactome pathways, with one slight modification. In this case, the ‘jaccard’ algorithm was used to calculate the similarity between pathways, and a similarity cutoff of 0.5 was employed for the Jaccard index. After removing redundant enriched pathways, we combined all the remaining GO terms and pathways. Subsequently, we calculated the similarity between these merged entities using the Jaccard index. This similarity analysis aimed to capture the overlap and relationships between the different GO terms and pathways. Using the same approach as before, we performed community analysis to identify distinct biological functional modules based on the merged GO terms and pathways.
Identification of functional modules
First, we used the ‘Wang’ algorithm for the GO database and the ‘jaccard’ algorithm for the KEGG and Reactome databases to calculate the similarity between pathways. The enriched pathways served as nodes in a similarity network, with edges representing the similarity between two nodes. Next, we employed the R package ‘igraph’ to identify modules within the network based on edge betweenness. By gradually removing edges with the highest edge betweenness scores, we constructed a hierarchical map known as a dendrogram, representing a rooted tree of the graph. The leaf nodes correspond to individual pathways, and the root node represents the entire graph 87 . We then merged pathways within each module, selecting the pathway with the smallest adjusted P value to represent the module. After this step, we merged pathways from all three databases into modules. Subsequently, we repeated the process by calculating the similarity between modules from all three databases using the ‘jaccard’ algorithm. Once again, we employed the same approach described above to identify the functional modules.
Metabolomics pathway enrichment
To perform pathway enrichment analysis for metabolomics data, we used the human KEGG pathway database. This database was obtained from KEGG using the R package ‘massDatabase’ 88 . For pathway enrichment analysis, we employed the hypergeometric distribution test from the ‘TidyMass’ project 89 . This statistical test allowed us to assess the enrichment of metabolites within each pathway. To account for multiple tests, P values were adjusted using the Benjamini–Hochberg method. We considered pathways with Benjamini–Hochberg-adjusted P values lower than 0.05 as significantly enriched.
Modified DE-SWAN
The DE-SWAN algorithm 14 was used. To begin, a unique age is selected as the center of a 20-year window. Molecule levels in individuals younger than and older than that age are compared using the Wilcoxon test to assess differential expression. P values are calculated for each molecule, indicating the significance of the observed differences. To ensure sufficient sample sizes for statistical analysis in each time window, the initial window ranges from ages 25 to 50. The left half of this window covers ages 25–40, whereas the right half spans ages 41–50. The window then moves in one-year steps; this is why Fig. 4 displays an age range of 40–65 years. To account for multiple comparisons, these P values are adjusted using Benjamini–Hochberg correction. To evaluate the robustness and relevance of the DE-SWAN results, the algorithm is tested with various parcel widths, including 15 years, 20 years, 25 years and 30 years. Additionally, different q value thresholds, such as <0.0001, <0.001, <0.01 and <0.05, are applied. By comparing the results obtained with these different parameters to results obtained by chance, we can assess the significance of the findings. To generate random results for comparison, the phenotypes of the individuals are randomly permuted, and the modified DE-SWAN algorithm is applied to the permuted dataset. This allows us to determine whether the observed results obtained with DE-SWAN are statistically significant and not merely a result of chance.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The raw data used in this study can be accessed without any restrictions on the National Institutes of Health Human Microbiome 2 project site ( https://portal.hmpdacc.org ). Both the raw and processed data are also available on the Stanford iPOP site ( http://med.stanford.edu/ipop.html ). Researchers and interested individuals can visit these websites to access the data. For further details and inquiries about the study, we recommend contacting the corresponding author, who can provide additional information and address any specific questions related to the research.
Code availability
The statistical analysis and data processing in this study were performed using R v.4.2.1, along with various base packages and additional packages. Detailed information about the specific packages used can be found in the Supplementary Note , which accompanies the manuscript. Furthermore, all the custom scripts developed for this study have been made openly accessible and can be found on the GitHub repository at https://github.com/jaspershen-lab/ipop_aging . By visiting this repository, researchers and interested individuals can access and use the custom scripts for their own analyses or to replicate the study’s findings.
Hou, Y. et al. Ageing as a risk factor for neurodegenerative disease. Nat. Rev. Neurol. 15 , 565–581 (2019).
Article PubMed Google Scholar
Chen, R. et al. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell 148 , 1293–1307 (2012).
Article CAS PubMed PubMed Central Google Scholar
Valdes, A. M., Glass, D. & Spector, T. D. Omics technologies and the study of human ageing. Nat. Rev. Genet. 14 , 601–607 (2013).
Rutledge, J., Oh, H. & Wyss-Coray, T. Measuring biological age using omics data. Nat. Rev. Genet. 23 , 715–727 (2022).
Ahadi, S. et al. Personal aging markers and ageotypes revealed by deep longitudinal profiling. Nat. Med. 26 , 83–90 (2020).
Ram, U. et al. Age-specific and sex-specific adult mortality risk in India in 2014: analysis of 0.27 million nationally surveyed deaths and demographic estimates from 597 districts. Lancet Glob. Health 3 , e767–e775 (2015).
Rodgers, J. L. et al. Cardiovascular risks associated with gender and aging. J. Cardiovasc. Dev. Dis. 6 , 19 (2019).
CAS PubMed PubMed Central Google Scholar
Poewe, W. et al. Parkinson disease. Nat. Rev. Dis. Primers 3 , 17013 (2017).
Hy, L. X. & Keller, D. M. Prevalence of AD among whites: a summary by levels of severity. Neurology 55 , 198–204 (2000).
Article CAS PubMed Google Scholar
Nussbaum, R. L. & Ellis, C. E. Alzheimer’s disease and Parkinson’s disease. N. Engl. J. Med. 348 , 1356–1364 (2003).
Xiong, Y. et al. Vimar/RAP1GDS1 promotes acceleration of brain aging after flies and mice reach middle age. Commun. Biol. 6 , 420 (2023).
Sherwood, C. C. et al. Aging of the cerebral cortex differs between humans and chimpanzees. Proc. Natl Acad. Sci. USA 108 , 13029–13034 (2011).
Márquez, E. J. et al. Sexual-dimorphism in human immune system aging. Nat. Commun. 11 , 751 (2020).
Article PubMed PubMed Central Google Scholar
Lehallier, B. et al. Undulating changes in human plasma proteome profiles across the lifespan. Nat. Med. 25 , 1843–1850 (2019).
Fehlmann, T. et al. Common diseases alter the physiological age-related blood microRNA profile. Nat. Commun. 11 , 5958 (2020).
Shavlakadze, T. et al. Age-related gene expression signature in rats demonstrate early, late, and linear transcriptional changes from multiple tissues. Cell Rep. 28 , 3263–3273 (2019).
Vershinina, O., Bacalini, M. G., Zaikin, A., Franceschi, C. & Ivanchenko, M. Disentangling age-dependent DNA methylation: deterministic, stochastic, and nonlinear. Sci Rep. 11 , 9201 (2021).
Li, J. et al. Determining a multimodal aging clock in a cohort of Chinese women. Med 4 , 825–848 (2023).
Shen, X. Multi-omics microsampling for the profiling of lifestyle-associated changes in health. Nat. Biomed. Eng . 8 , 11–29 (2024).
Takahashi, T. A. & Johnson, K. M. Menopause. Med. Clin. North Am. 99 , 521–534 (2015).
Umbayev, B. et al. Role of a small GTPase Cdc42 in aging and age-related diseases. Biogerontology 24 , 27–46 (2023).
Yi, S.-J. & Kim, K. New insights into the role of histone changes in aging. Int. J. Mol. Sci. 21 , 8241 (2020).
Liguori, I. et al. Oxidative stress, aging, and diseases. Clin. Interv. Aging 13 , 757–772 (2018).
Borbolis, F. & Syntichaki, P. Cytoplasmic mRNA turnover and ageing. Mech. Ageing Dev. 152 , 32–42 (2015).
Kaushik, S. et al. Autophagy and the hallmarks of aging. Ageing Res. Rev. 72 , 101468 (2021).
Czibik, G. et al. Dysregulated phenylalanine catabolism plays a key role in the trajectory of cardiac aging. Circulation 144 , 559–574 (2021).
Rousselle, A. et al. CXCL5 limits macrophage foam cell formation in atherosclerosis. J. Clin. Invest. 123 , 1343–1347 (2013).
Fazeli, P. K., Lee, H. & Steinhauser, M. L. Aging is a powerful risk factor for type 2 diabetes mellitus independent of body mass index. Gerontology 66 , 209–210 (2019).
Allayee, H., Roth, N. & Hodis, H. N. Polyunsaturated fatty acids and cardiovascular disease: implications for nutrigenetics. J. Nutrigenet. Nutrigenomics 2 , 140–148 (2009).
Sacks, F. M. et al. Dietary fats and cardiovascular disease: a presidential advisory from the American Heart Association. Circulation 136 , e1–e23 (2017).
Qi, W. et al. The ω-3 fatty acid α-linolenic acid extends Caenorhabditis elegans lifespan via NHR-49/PPARα and oxidation to oxylipins. Aging Cell 16 , 1125–1135 (2017).
Bird, A. W. et al. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature 419 , 411–415 (2002).
Sivanand, S. et al. Nuclear acetyl-CoA production by ACLY promotes homologous recombination. Mol. Cell 67 , 252–265 (2017).
Zhao, S. et al. Regulation of cellular metabolism by protein lysine acetylation. Science 327 , 1000–1004 (2010).
Vericel, E. et al. Platelets and aging I.—Aggregation, arachidonate metabolism and antioxidant status. Thromb. Res. 49 , 331–342 (1988).
Gu, S. X. & Dayal, S. Redox mechanisms of platelet activation in aging. Antioxidants (Basel) 11 , 995 (2022).
Oikonomopoulou, K., Ricklin, D., Ward, P. A. & Lambris, J. D. Interactions between coagulation and complement–their role in inflammation. Semin. Immunopathol. 34 , 151–165 (2012).
Wasiak, S. et al. Downregulation of the complement cascade in vitro, in mice and in patients with cardiovascular disease by the bet protein inhibitor apabetalone (RVX-208). J. Cardiovasc. Transl. 10 , 337–347 (2017).
Article Google Scholar
Slack, M. A. & Gordon, S. M. Protease activity in vascular disease. Arterioscl. Thromb. Vasc. Biol. 39 , e210–e218 (2019).
Mari, D. et al. Hemostasis and ageing. Immun. Ageing 5 , 12 (2008).
Lowe, G. & Rumley, A. The relevance of coagulation in cardiovascular disease: what do the biomarkers tell us? Thromb. Haemostasis 112 , 860–867 (2014).
Li, Y. et al. Branched chain amino acids exacerbate myocardial ischemia/reperfusion vulnerability via enhancing GCN2/ATF6/PPAR-α pathway-dependent fatty acid oxidation. Theranostics 10 , 5623–5640 (2020).
McGarrah, R. W. & White, P. J. Branched-chain amino acids in cardiovascular disease. Nat. Rev. Cardiol. 20 , 77–89 (2023).
Arsenian, M. Potential cardiovascular applications of glutamate, aspartate, and other amino acids. Clin. Cardiol. 21 , 620–624 (1998).
Grajeda-Iglesias, C. & Aviram, M. Specific amino acids affect cardiovascular diseases and atherogenesis via protection against macrophage foam cell formation: review article. Rambam Maimonides Med. J. 9 , e0022 (2018).
Chen, H. et al. Comprehensive metabolomics identified the prominent role of glycerophospholipid metabolism in coronary artery disease progression. Front. Mol. Biosci. 8 , 632950 (2021).
Giammanco, A. et al. Hyperalphalipoproteinemia and beyond: the role of HDL in cardiovascular diseases. Life (Basel) 11 , 581 (2021).
CAS PubMed Google Scholar
Zhu, Q. et al. Comprehensive metabolic profiling of inflammation indicated key roles of glycerophospholipid and arginine metabolism in coronary artery disease. Front. Immunol. 13 , 829425 (2022).
Yue, B. Biology of the extracellular matrix: an overview. J. Glaucoma 23 , S20–S23 (2014).
Zambrzycka, A. Aging decreases phosphatidylinositol-4,5-bisphosphate level but has no effect on activities of phosphoinositide kinases. Pol. J. Pharmacol. 56 , 651–654 (2004).
Lee, D. H., Oh, J.-H. & Chung, J. H. Glycosaminoglycan and proteoglycan in skin aging. J. Dermatol. Sci. 83 , 174–181 (2016).
Khan, A. U., Qu, R., Fan, T., Ouyang, J. & Dai, J. A glance on the role of actin in osteogenic and adipogenic differentiation of mesenchymal stem cells. Stem Cell Res. Ther. 11 , 283 (2020).
Lago, J. C. & Puzzi, M. B. The effect of aging in primary human dermal fibroblasts. PLoS ONE 14 , e0219165 (2019).
Pollard, T. D. Actin and actin-binding proteins. Cold Spring Harb. Perspect. Biol. 8 , a018226 (2016).
Lai, W.-F. & Wong, W.-T. Roles of the actin cytoskeleton in aging and age-associated diseases. Ageing Res. Rev. 58 , 101021 (2020).
Garcia, G., Homentcovschi, S., Kelet, N. & Higuchi-Sanabria, R. Imaging of actin cytoskeletal integrity during aging in C. elegans . Methods Mol. Biol. 2364 , 101–137 (2022).
Kim, Y. J. et al. Links of cytoskeletal integrity with disease and aging. Cells 11 , 2896 (2022).
Oosterheert, W., Klink, B. U., Belyy, A., Pospich, S. & Raunser, S. Structural basis of actin filament assembly and aging. Nature 611 , 374–379 (2022).
Bruzzone, A. et al. Dosage-dependent regulation of cell proliferation and adhesion through dual β2-adrenergic receptor/cAMP signals. FASEB J. 28 , 1342–1354 (2014).
McEver, R. P. & Luscinskas, F. W. Cell adhesion. In Hematology 7th edn (eds Hoffman, R. et al.) 127–134 (Elsevier, 2018).
Persa, O. D., Koester, J. & Niessen, C. M. Regulation of cell polarity and tissue architecture in epidermal aging and cancer. J. Invest. Dermatol. 141 , 1017–1023 (2021).
Canfield, C.-A. & Bradshaw, P. C. Amino acids in the regulation of aging and aging-related diseases. Transl. Med. Aging 3 , 70–89 (2019).
Chiabrando, D., Vinchi, F., Fiorito, V., Mercurio, S. & Tolosano, E. Heme in pathophysiology: a matter of scavenging, metabolism and trafficking across cell membranes. Front. Pharmacol. 5 , 61 (2014).
Aggarwal, S. et al. Heme scavenging reduces pulmonary endoplasmic reticulum stress, fibrosis, and emphysema. JCI Insight 3 , e120694 (2018).
Hodge, R. G. & Ridley, A. J. Regulating Rho GTPases and their regulators. Nat. Rev. Mol. Cell Biol. 17 , 496–510 (2016).
Siparsky, P. N., Kirkendall, D. T. & Garrett, W. E. Muscle changes in aging. Sports Health 6 , 36–40 (2014).
Johnson, A. A. & Stolzing, A. The role of lipid metabolism in aging, lifespan regulation, and age-related disease. Aging Cell 18 , e13048 (2019).
Paganelli, R., Scala, E., Quinti, I. & Ansotegui, I. J. Humoral immunity in aging. Aging Clin. Exp. Res. 6 , 143–150 (1994).
Article CAS Google Scholar
Goronzy, J. J. & Weyand, C. M. Understanding immunosenescence to improve responses to vaccines. Nat. Immunol. 14 , 428–436 (2013).
Cunha, L. L., Perazzio, S. F., Azzi, J., Cravedi, P. & Riella, L. V. Remodeling of the immune response with aging: immunosenescence and its potential impact on COVID-19 immune response. Front. Immunol. 11 , 1748 (2020).
Lee, D., Son, H. G., Jung, Y. & Lee, S.-J. V. The role of dietary carbohydrates in organismal aging. Cell. Mol. Life Sci. 74 , 1793–1803 (2017).
Franco-Juárez, B. et al. Effects of high dietary carbohydrate and lipid intake on the lifespan of C. elegans . Cells 10 , 2359 (2021).
Gold, L. et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS ONE 5 , e15004 (2010).
Galliera, E., Tacchini, L. & Corsi Romanelli, M. M. Matrix metalloproteinases as biomarkers of disease: updates and new insights. Clin. Chem. Lab. Med. 53 , 349–355 (2015).
Golusda, L., Kühl, A. A., Siegmund, B. & Paclik, D. Extracellular matrix components as diagnostic tools in inflammatory bowel disease. Biology (Basel) 10 , 1024 (2021).
Zhou, W. et al. Longitudinal multi-omics of host–microbe dynamics in prediabetes. Nature 569 , 663–671 (2019).
Schüssler-Fiorenza Rose, et al. A longitudinal big data approach for precision health. Nat. Med. 25 , 792–804 (2019).
Hornburg, D. et al. Dynamic lipidome alterations associated with human health, disease and ageing. Nat. Metab. 5 , 1578–1594 (2023).
Zhou, X. et al. Longitudinal profiling of the microbiome at four body sites reveals core stability and individualized dynamics during health and disease. Cell Host Microbe 32 , 506–526 (2024).
Contreras, P. H., Serrano, F. G., Salgado, A. M. & Vigil, P. Insulin sensitivity and testicular function in a cohort of adult males suspected of being insulin-resistant. Front. Med. (Lausanne) 5 , 190 (2018).
Evans, D. J., Murray, R. & Kissebah, A. H. Relationship between skeletal muscle insulin resistance, insulin-mediated glucose disposal, and insulin binding. Effects of obesity and body fat topography. J. Clin. Invest. 74 , 1515–1525 (1984).
Röst, H. L., Schmitt, U., Aebersold, R. & Malmström, L. pyOpenMS: a Python-based interface to the OpenMS mass-spectrometry algorithm library. Proteomics 14 , 74–77 (2014).
Shen, X. et al. Normalization and integration of large-scale metabolomics data using support vector regression. Metabolomics 12 , 89 (2016).
Shen, X. et al. metID: an R package for automatable compound annotation for LC−MS-based data. Bioinformatics 38 , 568–569 (2022).
Marabita, F. et al. Multiomics and digital monitoring during lifestyle changes reveal independent dimensions of human biology and health. Cell Syst. 13 , 241–255 (2022).
Wu, T. et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation (Camb.) 2 , 100141 (2021).
Newman, M. E. J. & Girvan, M. Finding and evaluating community structure in networks. Phys. Rev. E 69 , 026113 (2004).
Shen, X., Wang, C. & Snyder, M. P. massDatabase: utilities for the operation of the public compound and pathway database. Bioinformatics 38 , 4650–4651 (2022).
Shen, X. et al. TidyMass an object-oriented reproducible analysis framework for LC–MS data. Nat. Commun. 13 , 4365 (2022).
Download references
Acknowledgements
We sincerely thank all the research participants for their dedicated involvement in this study. We also thank A. Chen and L. Stainton for their valuable administrative assistance. Additionally, we are deeply grateful to A.T. Brunger’s support for this work. This work was supported by National Institutes of Health (NIH) grants U54DK102556 (M.P.S.), R01 DK110186-03 (M.P.S.), R01HG008164 (M.P.S.), NIH S10OD020141 (M.P.S.), UL1 TR001085 (M.P.S.) and P30DK116074 (M.P.S.) and by the Stanford Data Science Initiative (M.P.S.). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
These authors contributed equally: Xiaotao Shen, Chuchu Wang.
Authors and Affiliations
Department of Genetics, Stanford University School of Medicine, Stanford, CA, USA
Xiaotao Shen, Xin Zhou, Wenyu Zhou, Daniel Hornburg, Si Wu & Michael P. Snyder
Lee Kong Chian School of Medicine, Nanyang Technological University, Singapore, Singapore
Xiaotao Shen
School of Chemistry, Chemical Engineering and Biotechnology, Singapore, Singapore
Howard Hughes Medical Institute, Stanford University, Stanford, CA, USA
Chuchu Wang
Department of Molecular and Cellular Physiology, Stanford University, Stanford, CA, USA
Stanford Center for Genomics and Personalized Medicine, Stanford, CA, USA
Xin Zhou & Michael P. Snyder
You can also search for this author in PubMed Google Scholar
Contributions
X.S. and M.P.S. conceptualized and designed the study. X.Z. and W.Z. prepared the microbiome data. D.H. and W.S. prepared the lipidomics data. X.S. and C.W. conducted the data analysis. X.S. and C.W. prepared the figures. X.S., C.W. and M.P.S. contributed to the writing and revision of the manuscript, with input from other authors. M.S. and X.S. supervised the overall study.
Corresponding author
Correspondence to Michael P. Snyder .
Ethics declarations
Competing interests.
M.P.S. is a co-founder of Personalis, SensOmics, Qbio, January AI, Filtricine, Protos and NiMo and is on the scientific advisory boards of Personalis, SensOmics, Qbio, January AI, Filtricine, Protos, NiMo and Genapsys. D.H. has a financial interest in PrognomIQ and Seer. All other authors have no competing interests.
Peer review
Peer review information.
Nature Aging thanks Daniel Belsky and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended data fig. 1 demographic data of all the participants in the study..
a , The ages positively correlate with BMI. The shaded area around the regression line represents the 95% confidence interval. b , Gender with age. c , Ethnicity with age. d , Insulin response with age. e , biological sample collection for all the participants. f , Overlap of the different kinds of omics data. g , The age range for each participant in this study.
Extended Data Fig. 2 Most of the molecules change nonlinearly during human aging.
a , Differential expressional microbes in different age ranges compared to baselines (25 – 40 years old, two-sided Wilcoxon test, p -value < 0.05). b , Most of the linear changing molecules and microbiota are also included in the molecules/microbes that significantly dysregulated at least one age range.
Extended Data Fig. 3 Omics data can represent aging.
PCA score plot of metabolomics data ( a ), cytokine ( b ), and oral microbiome ( c ).
Extended Data Fig. 4 Functional analysis of molecules in different clusters.
a , The Jaccard index between clusters from different datasets. b , The overlap between clusters using different types of omics data. c , Functional module detection and identification. d , Functional analysis of nonlinear changing molecules for all clusters.
Extended Data Fig. 5 Function annotation for significantly dysregulated molecules in crest 1 and 2.
a , Transcriptomics data. b , Proteomics data. c , Metabolomics data.
Extended Data Fig. 6 Pathways enrichment results for crest 1 and 2.
a , The final functional modules identified for Crest 1 and 2. b , The pathway enrichment analysis results for transcriptomics data. c , The pathway enrichment analysis results for proteomics data. d , The pathway enrichment results for metabolomics data.
Supplementary information
Supplementary figs. 1–6, reporting summary, supplementary data analysis results of the study., rights and permissions.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ .
Reprints and permissions
About this article
Cite this article.
Shen, X., Wang, C., Zhou, X. et al. Nonlinear dynamics of multi-omics profiles during human aging. Nat Aging (2024). https://doi.org/10.1038/s43587-024-00692-2
Download citation
Received : 09 December 2023
Accepted : 22 July 2024
Published : 14 August 2024
DOI : https://doi.org/10.1038/s43587-024-00692-2
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing newsletter — what matters in science, free to your inbox daily.

Title Page Setup
A title page is required for all APA Style papers. There are both student and professional versions of the title page. Students should use the student version of the title page unless their instructor or institution has requested they use the professional version. APA provides a student title page guide (PDF, 199KB) to assist students in creating their title pages.
Student title page
The student title page includes the paper title, author names (the byline), author affiliation, course number and name for which the paper is being submitted, instructor name, assignment due date, and page number, as shown in this example.
Title page setup is covered in the seventh edition APA Style manuals in the Publication Manual Section 2.3 and the Concise Guide Section 1.6
Related handouts
- Student Title Page Guide (PDF, 263KB)
- Student Paper Setup Guide (PDF, 3MB)
Student papers do not include a running head unless requested by the instructor or institution.
Follow the guidelines described next to format each element of the student title page.
|
|
|
|
|---|---|---|
| Paper title | Place the title three to four lines down from the top of the title page. Center it and type it in bold font. Capitalize of the title. Place the main title and any subtitle on separate double-spaced lines if desired. There is no maximum length for titles; however, keep titles focused and include key terms. |
|
| Author names | Place one double-spaced blank line between the paper title and the author names. Center author names on their own line. If there are two authors, use the word “and” between authors; if there are three or more authors, place a comma between author names and use the word “and” before the final author name. | Cecily J. Sinclair and Adam Gonzaga |
| Author affiliation | For a student paper, the affiliation is the institution where the student attends school. Include both the name of any department and the name of the college, university, or other institution, separated by a comma. Center the affiliation on the next double-spaced line after the author name(s). | Department of Psychology, University of Georgia |
| Course number and name | Provide the course number as shown on instructional materials, followed by a colon and the course name. Center the course number and name on the next double-spaced line after the author affiliation. | PSY 201: Introduction to Psychology |
| Instructor name | Provide the name of the instructor for the course using the format shown on instructional materials. Center the instructor name on the next double-spaced line after the course number and name. | Dr. Rowan J. Estes |
| Assignment due date | Provide the due date for the assignment. Center the due date on the next double-spaced line after the instructor name. Use the date format commonly used in your country. | October 18, 2020 |
|
| Use the page number 1 on the title page. Use the automatic page-numbering function of your word processing program to insert page numbers in the top right corner of the page header. | 1 |
Professional title page
The professional title page includes the paper title, author names (the byline), author affiliation(s), author note, running head, and page number, as shown in the following example.
Follow the guidelines described next to format each element of the professional title page.
|
|
|
|
|---|---|---|
| Paper title | Place the title three to four lines down from the top of the title page. Center it and type it in bold font. Capitalize of the title. Place the main title and any subtitle on separate double-spaced lines if desired. There is no maximum length for titles; however, keep titles focused and include key terms. |
|
| Author names
| Place one double-spaced blank line between the paper title and the author names. Center author names on their own line. If there are two authors, use the word “and” between authors; if there are three or more authors, place a comma between author names and use the word “and” before the final author name. | Francesca Humboldt |
| When different authors have different affiliations, use superscript numerals after author names to connect the names to the appropriate affiliation(s). If all authors have the same affiliation, superscript numerals are not used (see Section 2.3 of the for more on how to set up bylines and affiliations). | Tracy Reuter , Arielle Borovsky , and Casey Lew-Williams | |
| Author affiliation
| For a professional paper, the affiliation is the institution at which the research was conducted. Include both the name of any department and the name of the college, university, or other institution, separated by a comma. Center the affiliation on the next double-spaced line after the author names; when there are multiple affiliations, center each affiliation on its own line.
| Department of Nursing, Morrigan University |
| When different authors have different affiliations, use superscript numerals before affiliations to connect the affiliations to the appropriate author(s). Do not use superscript numerals if all authors share the same affiliations (see Section 2.3 of the for more). | Department of Psychology, Princeton University | |
| Author note | Place the author note in the bottom half of the title page. Center and bold the label “Author Note.” Align the paragraphs of the author note to the left. For further information on the contents of the author note, see Section 2.7 of the . | n/a |
|
| The running head appears in all-capital letters in the page header of all pages, including the title page. Align the running head to the left margin. Do not use the label “Running head:” before the running head. | Prediction errors support children’s word learning |
|
| Use the page number 1 on the title page. Use the automatic page-numbering function of your word processing program to insert page numbers in the top right corner of the page header. | 1 |
Log in using your username and password
- Search More Search for this keyword Advanced search
- Latest content
- For authors
- Browse by collection
- BMJ Journals
You are here
- Volume 12, Issue 2
- Consumption and effects of caffeinated energy drinks in young people: an overview of systematic reviews and secondary analysis of UK data to inform policy
- Article Text
- Article info
- Citation Tools
- Rapid Responses
- Article metrics
- http://orcid.org/0000-0002-9571-3147 Claire Khouja 1 ,
- http://orcid.org/0000-0002-7016-978X Dylan Kneale 2 ,
- Ginny Brunton 3 ,
- Gary Raine 1 ,
- Claire Stansfield 2 ,
- Amanda Sowden 1 ,
- Katy Sutcliffe 2 ,
- James Thomas 2
- 1 Centre for Reviews and Dissemination , University of York , York , UK
- 2 EPPI-Centre, Social Science Research Unit , UCL Institute of Education, University College London , London , UK
- 3 Faculty of Health Sciences , Ontario Tech University , Oshawa , Ontario , Canada
- Correspondence to Claire Khouja; claire.khouja{at}york.ac.uk
Background This overview and analysis of UK datasets was commissioned by the UK government to address concerns about children’s consumption of caffeinated energy drinks and their effects on health and behaviour.
Methods We searched nine databases for systematic reviews, published between 2013 and July 2021, in English, assessing caffeinated energy drink consumption by people under 18 years old (children). Two reviewers rated or checked risk of bias using AMSTAR2, and extracted and synthesised findings. We searched the UK Data Service for country-representative datasets, reporting children’s energy-drink consumption, and conducted bivariate or latent class analyses.
Results For the overview, we included 15 systematic reviews; six reported drinking prevalence and 14 reported associations between drinking and health or behaviour. AMSTAR2 ratings were low or critically low. Worldwide, across reviews, from 13% to 67% of children had consumed energy drinks in the past year. Only two of the 74 studies in the reviews were UK-based. For the dataset analysis, we identified and included five UK cross-sectional datasets, and found that 3% to 32% of children, across UK countries, consumed energy drinks weekly, with no difference by ethnicity. Frequent drinking (5 or more days per week) was associated with low psychological, physical, educational and overall well-being. Evidence from reviews and datasets suggested that boys drank more than girls, and drinking was associated with more headaches, sleep problems, alcohol use, smoking, irritability, and school exclusion. GRADE (Grading of Recommendations, Assessment, Development and Evaluation) assessment suggests that the evidence is weak.
Conclusions Weak evidence suggests that up to a third of children in the UK consume caffeinated energy drinks weekly; and drinking 5 or more days per week is associated with some health and behaviour problems. Most of the evidence is from surveys, making it impossible to distinguish cause from effect. Randomised controlled trials are unlikely to be ethical; longitudinal studies could provide stronger evidence.
PROSPERO registrations CRD42018096292 – no deviations. CRD42018110498 – one deviation - a latent class analysis was conducted.
- nutrition & dietetics
- epidemiology
- public health
- community child health
Data availability statement
Data are available upon reasonable request. All the data in the overview are publicly available, but not necessarily without charge. Those for the dataset analysis are available from the UK Data Service.
This is an open access article distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited, appropriate credit is given, any changes made indicated, and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/ .
https://doi.org/10.1136/bmjopen-2020-047746
Statistics from Altmetric.com
Request permissions.
If you wish to reuse any or all of this article please use the link below which will take you to the Copyright Clearance Center’s RightsLink service. You will be able to get a quick price and instant permission to reuse the content in many different ways.
Strengths and limitations of this study
The main strength of this study was the novel use of a secondary data analysis to fill a gap in the evidence that was identified by the overview.
A strength of the overview was its robust methods, and that it only included reviews that used systematic methods.
A limitation of the overview was the strength of evidence of the primary research, most of which was from cross-sectional surveys.
The main limitations of the dataset analysis were that longitudinal data were not available, and the survey data could not be combined due to differences between surveys in their designs and measures reported.
Introduction
Caffeinated energy drinks (CEDs) are drinks containing caffeine, among other ingredients, that are marketed as boosting energy, reducing tiredness, and improving concentration. They include brands such as Red Bull, Monster Energy, and Rockstar. There is widespread concern about their consumption and effects in children and adolescents (under 18 years old). 1–4 Some professional organisations have suggested banning sales to children. 2 In the UK, warnings, aimed at children and pregnant women, are required on the packaging for drinks that contain over 150 mg/L of caffeine. 5 An average 250 mL energy drink contains a similar amount of caffeine to a 60 mL espresso, and the European Food Safety Authority proposes a safe level of 3 mg of caffeine per kg of body weight per day for children and adolescents. 6 Many drinks also contain other potentially active ingredients, such as guarana and taurine, and more sugar than other soft drinks, although there are sugar-free options. 7–9 Children may be more at risk of ill effects than adults. 10 11 Effects could be physical (eg, headaches), psychological (eg, anxiety) or behavioural (eg, school attendance or alcohol consumption). 12 Available systematic reviews report a wide range of findings, including positive effects on sports performance.
In 2018, the UK government ran a consultation on implementing a ban on sales to children, 13–15 and in March 2019 they published a policy paper. 16 The research reported here was commissioned by the Department of Health and Social Care (DHSC), England, in 2018, to identify and assess the evidence on the use of CEDs by children. As the deadline was short, and as initial searches identified several systematic reviews, a systematic review of systematic reviews (referred to as overview, from this point onwards) was conducted. As only two UK studies were identified within the reviews included in the overview, UK datasets were sought, and a secondary analysis of relevant data was carried out to supplement the international literature and ensure relevance to UK policy. Full reports are available. 17 18
The research questions (RQ) were:
RQ1. What is the nature and extent of CED consumption among people aged 17 years or under in the UK?
RQ2. What impact do CEDs have on young people’s physical and mental health, and behaviour?
This paper summarises the overview and dataset analysis. 17 18 For the overview, a literature search was conducted during May 2018 and updated on 2 July 2021. EPPI-Reviewer software 19 was used to manage the data. The gaps, identified by the overview and a search for primary studies, guided the search, conducted during August 2018, for UK datasets and their subsequent analysis. STATA v13 20 was used to analyse the datasets. Ethical approval was granted by UCL’s Ethics Committee. Protocols were registered on PROSPERO (CRD42018096292 and CRD42018110498).
Search strategies
For the overview, we searched nine databases, focusing on research in health, psychology, science or social science, or general research. We completed forward citation searching in Google Scholar for 13 included reviews. The databases searched and the MEDLINE search strategy are in the online supplemental file (section 1). The search terms were based on three concepts: caffeine, energy drink, and systematic review. The searches were limited to the publication year of 2013 onwards, to identify the most recent systematic reviews. For the dataset analysis, search terms were based on caffeine and energy drink. We searched the UK Data Service 21 (accessing over 6000 UK nation population datasets), with no restrictions.
Supplemental material
Inclusion criteria.
For the overview:
Systematic review published since 2013
Extractable data on children under 18 years of age
Available in English
Patterns of CED use or associations with physical, mental, social or behavioural effects.
Four reviewers (GB, CK, GR and CS) screened references based on their titles and abstracts, and then screened potential includes on their full texts. The four reviewers double-screened batches of 10 references until their decisions to include or exclude each paper were the same on at least nine of the 10 (90%), then they screened individually. Disagreements and indecisions were resolved by another of the four reviewers, where necessary.
For the dataset analysis:
Downloadable datasets, representative of the UK or a constituent country
Information on the levels and patterns of CED consumption
Data on children under 18 years of age (adults could provide the data on their behalf)
Reporting primary (frequency, amount, or occurrence of drinking/not drinking (comparator)) or secondary (sugar consumption, cardiovascular health, mental health, neurological conditions, educational outcomes, substance misuse, sports performance or sleep characteristics) measures.
After a pilot batch, for which two reviewers (GB and DK) assessed datasets independently and discussed their decisions to include or exclude, the remaining datasets were screened, independently.
Data extraction
From the systematic review reports that met the overview inclusion criteria, we extracted details on/for: systematic review methods; included studies; CED consumption; associations with physical, mental, social or behavioural effects; and risk of bias assessment. One reviewer (GB, CK, GR or CS) extracted these data, which were checked by another reviewer. For the dataset analysis, one reviewer (GB or DK) extracted dataset characteristics (sample size, etc); details on participants (age, gender, etc) and consumption (how it was measured, etc); well-being and health outcomes, including potential confounders; and information on missing data and for risk of bias assessment.
The data extracted from the systematic reviews were synthesised in a narrative format due to variation between reviews. Prevalence was synthesised by the measure used, where possible. Associations were synthesised by whether they were physical, mental, behavioural, or social/educational, and summary tables were produced. One reviewer (GB, CK, GR or CS) synthesised the data and another checked each synthesis.
Each dataset was analysed for prevalence and frequency of CED consumption, and any variations by children’s characteristics. Most of the cross-sectional analyses were bivariate (exploring interactions between two features), with binary and multinomial logistic regression used to control for confounders. A latent class analysis (LCA) was conducted, 22 for one dataset. The latent profiles were based on children’s health experiences, such as headaches, anxiety, or dizziness. The observed variables (11 indicators of child well-being) and latent variables (five classes of well-being) were identified from the data. Class membership was used as the dependent variable in multinomial logistic regressions. Descriptive associations were explored in bivariate analyses of the 11 indicators, separately. The results from individual datasets were synthesised in a narrative because meta-analysis was not deemed to be appropriate. Missing data were not imputed, as it was not possible to determine if they were missing at random. One reviewer (DK) analysed the data.
Risk of bias
AMSTAR2 23 was used to assess the risk of bias in the included systematic reviews, because some reviews included randomised controlled trials (RCTs) as well as non-RCTs. AMSTAR2 has questions on the protocol, inclusion criteria, search, selection, data extraction, risk of bias assessment, reporting, synthesis (RCTs and non-RCTs), and conflicts of interest; a question on relevance was added. The strength of the evidence was assessed using GRADE (Grading of Recommendations, Assessment, Development and Evaluation) criteria, 24 which can be used to determine whether the evidence is strong or weak, based on any risk of bias, including in study design and size, consistency of the results, relevance to the population, and potential publication bias. Overlap, where the same primary studies appear in more than one review, was assessed. 25 Overlap can lead to double counting of the results of a study, giving these more influence than those of other studies. 26 Two reviewers (CK and GR) assessed risk of bias; random samples were checked by a third reviewer (GB). Datasets were not formally assessed, but all datasets met the quality assurance criteria of the UK Data Service. 27 Data on exposure (quantity, frequency and type of drink), sample frame (characteristics of participants), and level of participation (response rate) were extracted, by one reviewer (DK), to determine their parameters. 17 In line with National Institutes of Health guidance, 28 no overall risk of bias score was produced for each dataset because overall scores can be misleading where the risk of bias on each criterion has a different impact on the reliability of the conclusions.
Patient and public involvement
We did not include young people in the research process.
The overview searches identified 1102 references, after deduplication (see figure 1 ); 126 were screened on full texts. We included 15 reviews; six reported information on prevalence, 12 29–33 and 14 reported associations. 12 29–32 34–42 The reasons for exclusion, based on assessment of the full text, are reported in the online supplemental file (section 2). Most were excluded because they did not use systematic review methods or did not report information on children.
- Download figure
- Open in new tab
- Download powerpoint
Flow diagram for the overview. CED, caffeinated energy drinks; T&A, title and abstract.
Three reviews focused on CEDs in children. 12 30 41 One 35 focused on children, with a section on CEDs alongside other drinks. The other 11 reported information on children alongside data for adults; one 29 with CEDs alongside other drinks, and two 31 32 focusing on alcohol mixed with CEDs. For summary and full characteristics, see the online supplemental file (section 3) and the full report. 18
For the dataset analysis, as there was no facility to export results, it was not possible to record the flow of datasets through screening. Five datasets met the inclusion criteria; analyses were not possible for one dataset 43 (see table 1 ). For full descriptions, see the full report. 17
- View inline
Description of the five datasets included in the secondary data analysis
There was a high risk of bias in all but three of the reviews—Visram et al , 12 and Bull et al 37 Yasuma et al 41 (details in the online supplemental file , section 4)—meaning that some relevant evidence may have been missed. Overlap between studies in the reviews was slight (corrected covered area 3.2%; see the online supplemental file , section 5). The reviews did not include any analyses of the UK datasets that we analysed. Within the reviews, there were four small randomised controlled trials, while most studies were surveys with a high risk of bias; the application of GRADE criteria, which are used to assess the overall strength of the evidence found, suggests that the evidence is weak. Exposure, sample frame and level of participation for the datasets are reported in appendix 1 of the full report. 17
UK studies in the overview
Of the 74 studies identified by the reviews that are summarised in the overview, two were UK surveys. One 44–46 was a longitudinal (two time-points) cross-sectional survey of 11- to 17-year-olds in the south-west of England. The other 47 was a survey of 13- to 18-year-olds across 22 European countries, one of which was the UK (2.6% of respondents).
Below and in tables 2–4 , the overview results are summarised by research question, followed by highlights of the dataset analysis within each topic. The full results of the overview 18 and dataset analysis 17 are available online.
Characteristics and main findings of reviews reporting prevalence of consumption
Prevalence of CED consumption across datasets by school year (approximately weekly consumption with weighted percentages and unweighted sample sizes - see notes below)
Characteristics and main findings of the reviews reporting associations with consumption
RQ1. Nature and extent of CED consumption
The overview included six reviews with data on prevalence of children’s CED consumption, and these are summarised in table 2 .
Across reviews, prevalence varied by study location, population age range, and definition of drinking (ever drunk, in the past year, regularly, with alcohol, etc) from 13% to 67% of children having a CED in the past year. 30 32 One meta-analysis 29 of four studies in the Gulf states suggested that about two thirds of children consumed CEDs (not further defined; 65.3%, 95% CI 41.6 to 102.3 (as reported in the paper)). Across reviews, weekly or monthly drinking ranged from 13% to 54% 48 of children. In one study, across Europe, UK children had the highest proportion of caffeine intake from CEDs, at 11%, 47 but this might reflect a lower intake from coffee or tea. Across reviews, 10% 49 to 46% 50 of children had tried CEDs with alcohol.
In the UK dataset analysis, self-reported prevalence was relatively consistent across UK countries (see table 3 ), although there were differences in the questions asked. About a quarter of children aged 13 to 14 years consumed one drink or more per week (Smoking and Drinking Survey of Young People (SDSYP) data). 51 Prevalence ranged from 3% to 32% of children—slightly lower than found in the overview.
Characteristics of drinkers
In the overview, more boys reported drinking CEDs than girls. 12 29–32 Prevalence by age was inconsistent: for example, within the reviews, one study 48 found that girls started drinking CEDs when they were younger; while one 52 suggested that drinking prevalence peaked at 14 to 15 years; and another 53 suggested that more older boys drank CEDs than younger boys, but more younger girls drank them than older girls. Prevalence by ethnicity was also inconsistent. Children with minority ethnicity drank more than white children, 12 32 but white children drank more than black or Hispanic children, when drinks were mixed with alcohol. 12 In the UK, drinking was associated with being male, older and lower socioeconomic status. 45
In the dataset analysis, the SDSYP reported the most detailed information on sociodemographic characteristics. As in most of the overview evidence, prevalence increased with age, so that between a quarter and a third of children aged 15 to 16 years reported consuming one or more CED per week. More boys (29.3%) than girls (18.1%), and more children living in the North of England than in the South (for example, 33.1% in the North-East vs 16.5% in the South-East), consumed at least one can a week. More children who were eligible for free school meals (29.5%), than those who were not eligible (22.6%), drank CEDs weekly. These differences were robust to the impact of potential confounders (see the online supplemental file , section 6). Unlike the evidence from the overview, which suggested differences in consumption by ethnicity, the proportion of weekly CED consumers was within 3 percentage points of the average across all ethnic groups.
Motives and context
Three reviews reported on motives or context for consumption. 12 29 32 The context was parties and socialising with friends or family 12 32 35 or exams. 29 Children’s motives included taste (particularly with alcohol), for energy, curiosity, friends drinking them, and parental approval or disapproval. Across the reviews, single studies suggested that more girls than boys drank CEDs to suppress appetite, 54 while more boys than girls drank them for performance in sport. 55 And about half of children knew that the drinks contained caffeine, 56 while those who knew that the content might be harmful drank less. 57
Motives and context were not measured in the UK datasets.
RQ2. Associations with drinking CEDs
Fourteen reviews reported associations and are summarised in table 4 . Most reviews included cross-sectional evidence (surveys) or individual case studies. Three reviews 12 40 42 reported prospective trials (four small RCTs in total), which assessed physical performance, cardiovascular response, or the effects of sleep education; one review reported prospective cohort studies.
As most of the evidence was from surveys, measured at a single time-point, cause cannot be distinguished from effect.
Physical health associations
Associations between drinking CEDs and physical symptoms were reported in all but one 40 of the 14 reviews. CEDs improved sports performance. 58 59 There was consistent evidence of associations with headaches, stomach aches and low appetite, 12 35 42 and with sleep problems. 12 30 35 42 Within the reviews, a trial of boys randomised to receive different doses of CED reported dose-dependent increases in diastolic blood pressure and decreases in heart rate. 60 Across reviews, 34 36–39 nine cases of adverse events were reported; eight children had cardiovascular events, and one had renal failure, following a single drink, moderate drinking, or excessive drinking (in a day or for weeks).
Analysis of the Health Behaviour in School Children (HBSC) 2013/14 data found that children drinking CEDs once a week or more, compared with those drinking less often, were statistically significantly more likely to report physical symptoms occurring more than once a week, such as headaches (22.2% vs 16.8%), sleep problems (13.6% vs 8.5%) and stomach problems (31.2% vs 23.1%).
Mental health associations
Associations between drinking CEDs and mental health were inconsistent. 12 29 30 32 35 40 42 One review reported that improvements in mental health and hyperactivity were found in children who were randomised to receive an intervention to lower their intake of CEDs. 61 Associations were found with stress, anxiety or depression, 12 30 35 40 42 but two reviews 12 40 also found studies that did not find an association. Some reviews included evidence of associations with self-harm or suicidal behaviour, 30 35 40 42 and with irritation and anger. 12 30 35 40 42
Secondary analyses of the HBSC 2013/14 data found that children who consumed CEDs at least once a week were statistically significantly more likely, than those who did not, to report low mood (20.3% vs 14.9%) and irritability (30.8% vs 18.0%) on a weekly basis.
Behavioural associations
Some evidence of associations between drinking CEDs and behaviour was reported. 12 30–32 35 42 Drinking CEDs was associated with alcohol, smoking and substance misuse at a single time point, 12 30 35 and at follow-up. 41 CED consumption at baseline predicted alcohol consumption at follow-up. 12 Consumption was associated with increased hyperactivity and inattention, and with sensation seeking. 12 30 35 Injuries were associated with drinking CEDs with alcohol 12 31 and without alcohol. 12 30
Analysis of the SDSYP data found that higher proportions of children who consumed one or more cans per week had tried alcohol (59.1%) and smoking (39.7%), compared with non-CED consumers (alcohol 28.9%, smoking 10.4%).
Social or educational associations
Consistent associations between drinking CEDs and social or educational outcomes were reported. 12 32 Within reviews, one UK study 45 found an association between drinking CEDs once a week or more and poor school attendance. CEDs mixed with alcohol were associated with lower grades and more absence from school. 32
Analysis of the SDSYP data found that almost half of children who had been truant or excluded reported drinking a can of CED on a weekly basis (49.5%), compared with less than a fifth of those who had not been truant or excluded (18.5%).
Well-being profiles
Using the HBSC 2013/14 dataset, we identified 11 indicators of well-being: weekly experience of irritability, sleep difficulties, nervousness, dizziness, headaches, stomach aches, and low mood; as well as low life satisfaction, feeling pressured by schoolwork some or a lot of the time, dislike of school, and low self-rated academic achievement. From these, using LCA, we identified five profiles: low psychological well-being (18.2% of children), high overall well-being (48.6%), low educational well-being (6.7% of children), low physical well-being (13.0%), and low overall well-being (13.5%). See the online supplemental file (section 6) for details.
After controlling for age, gender, rurality, smoking status, alcohol status and Family Affluence Scale (a measure of socioeconomic status; for more information see Hartley et al 62 ), the relative risk of having a low well-being profile, compared with a high well-being profile, was substantially higher for children who consumed CEDs at least 5 days a week (frequent), compared with those who rarely or never did. Relative to a high well-being profile, frequent consumers had a higher risk of low psychological well-being (RR 2.11, 95% CI 1.56 to 2.85) and low physical well-being (RR 2.52, 95% CI 1.76 to 3.61), and were over four times more likely to have low educational well-being (RR 4.81, 95% CI 3.59 to 6.44) and low overall well-being (RR 4.15, 95% CI 2.85 to 6.00). These data suggest that CED consumption is a marker of low well-being, but the analyses also showed that consumption was one of a cluster of factors (eg, smoking and drinking alcohol) in children with low well-being.
Summary of the evidence
Prevalence varied according to the measures used and the ages of children. In the overview, CED consumption prevalence was up to 67% of children in the past year and, in the dataset analyses, up to 32% of children were consuming a CED at least 1 day a week, meaning that up to a third of UK children are regularly consuming caffeine. Evidence from the overview and the dataset analyses consistently suggests that boys drink more than girls, and that drinking tends to increase with age. Some evidence from the overview suggested higher prevalence in children from ethnic minority backgrounds, but no such association was detected in the UK data analysis. This could be due to factors such as area of residence or social class affecting well-being in children from ethnic minorities, where well-being is driving the differences in prevalence of CED consumption, rather than minority background. Reviews included in the overview found that most drinking of CEDs occurred at parties, around exams, with friends, or with family, and motives included taste, energy, curiosity, appetite suppression, and sports performance, which was reported to be improved. There was some evidence that knowledge of content was low, and that children who knew that the content might be harmful drank less, suggesting that education could reduce drinking.
Evidence from the overview suggests worse sleep, and raised blood pressure, with CED consumption, compared with reduced or no consumption. Both the overview and the dataset analysis found that children who consumed CEDs reported headaches, stomach aches and sleep issues more frequently than those who did not; although most studies were cross-sectional, some in the overview were longitudinal, showing changes over time. 18 The overview identified consistent evidence of associations with self-harm, suicide behaviour, alcohol use*, smoking*, substance misuse*, hyperactivity, irritation*, anger, and school performance, attendance, and exclusion (*also found in the UK dataset analysis). This was consistent with findings reported in non-systematic reviews. 10 63 64
The UK dataset analysis suggested that children who consumed CEDs 5 or more days a week had lower psychological, physical, educational and overall well-being than non-drinkers. It remains unclear whether drinking CEDs contributes to low well-being, or low well-being leads to CED consumption, or both. Alternatively, there may be a common cause, such as social inequality.
Strengths and limitations
The overview was limited by the amount of information reported in the included systematic reviews, and by their method limitations; all had a high risk of bias. They mainly included cross-sectional surveys or case reports, which means that cause or effect cannot be determined where an association is found. However, some prospective studies, including four small RCTs, were included in the reviews and where there were common measures, the evidence from these RCTs and from most of the cross-sectional studies within the reviews was consistent. This suggests that the associations found could be reliable. A strength of our work is that the UK evidence in the overview (two studies within the reviews) was supplemented by the analysis of UK data, which was mostly consistent with the non-UK evidence. These data support the idea that there is a link between drinking CEDs and poorer health and behaviour in children, although the cause is unclear. Overlap between reviews in the overview was slight (unsurprisingly, given the different foci of the reviews). There was no overlap between the reviews and the dataset analysis, meaning that the latter added new information. The wide range of tools used to measure prevalence made it difficult to summarise the overview evidence, and meta-analysis of the individual participant UK data was not possible, meaning that the conclusions are based on weaker evidence from single sources.
Recommendations for research
Standardisation is needed in the measurement of the prevalence of drinking—defining the dosage (in drinks and/or caffeine), timing (daily, weekly, etc) and population (age, ethnicity, etc). There was little evidence on children under 12 years old, and both the overview and dataset analysis found little evidence from the UK. Longitudinal data, from the UK datasets, should be collected to understand better the impact of consumption. RCTs may not be ethical, even where benefits are predicted, such as where children who consume CEDs are randomised to interventions to reduce or stop their drinking to see if this improves their well-being.
Based on a comprehensive overview of available systematic reviews, we conclude that up to half of children, worldwide, drink CEDs weekly or monthly, and based on the dataset analysis, up to a third of UK children do so. There is weak but consistent evidence, from reviews and UK datasets, that poorer health and well-being is found in children who drink CEDs. In the absence of RCTs, which are unlikely to be ethical, longitudinal studies could provide stronger evidence.
Ethics statements
Patient consent for publication.
Not applicable.
Ethics approval
This study does not involve human participants.
Acknowledgments
Thank you to Irene Kwan for assisting with data extraction for the review.
- Committee on Nutrition and the Council on Sports Medicine and Fitness
- UK Government.
- Haskell CF ,
- Kennedy DO ,
- Wesnes KA , et al
- Keast RSJ ,
- Swinburn BA ,
- Sayompark D , et al
- Curran CP ,
- Marczinski CA
- Henderson R , et al
- Cheetham M ,
- Riby DM , et al
- UK Government
- Department of Health and Social Care (DHSC)
- Department of Health and Social Care
- Brunton G ,
- Sowden A , et al
- Raine G , et al
- Brunton J ,
- ↵ Uk data service . Available: https://ukdataservice.ac.uk/ [Accessed 3 Apr 2020 ].
- Huang L , et al
- Reeves BC ,
- Wells G , et al
- Brennan S ,
- McKenzie J ,
- Middleton P
- Antoine S-L ,
- Mathes T , et al
- McKenzie JE ,
- UK Data Service
- National Institutes of Health
- El Kashef A ,
- AlGhaferi H
- Stockwell T
- Verster JC ,
- Johnson SJ , et al
- Babayan Z , et al
- Bleich SN ,
- Vercammen KA
- Burnett K , et al
- Goldfarb M ,
- Tellier C ,
- Thanassoulis G
- Cervellin G ,
- Sanchis-Gomar F
- Richards G ,
- Imamura K ,
- Watanabe K , et al
- Nadeem IM ,
- Shanmugaraj A ,
- Sakha S , et al
- University of London, I.o.E., Centre for Longitudinal Studies,
- EFSA Panel on Dietetic Products, Nutrition and Allergies (NDA)
- Al-Hazzaa H ,
- Waly MI , et al
- Jasionowski A
- Nobile CGA , et al
- NHS Digital
- Gambon DL ,
- Boutkabout C , et al
- Hammond D ,
- McCrory C , et al
- Bryant Ludden A ,
- Musaiger AO ,
- Gallimberti L ,
- Chindamo S , et al
- Abian-Vicen J ,
- Salinero JJ , et al
- Gallo-Salazar C ,
- Abián-Vicén J , et al
- Temple JL ,
- Briatico LN
- Man Yu MW , et al
- Hartley JEK ,
- Zucconi S ,
- Volpato C ,
- Adinolfi F , et al
- Seifert SM ,
- Schaechter JL ,
- Hershorin ER , et al
- Public Health England
- Northern Ireland Statistics and Research Agency
Supplementary materials
Supplementary data.
This web only file has been produced by the BMJ Publishing Group from an electronic file supplied by the author(s) and has not been edited for content.
- Data supplement 1
- Press_Release.pdf
Twitter @katysutcliffe
Contributors GB, CK, GR and CS worked on all stages of the overview. GB, CK, DK and GR worked on the overview update. GB and DK completed all stages of the secondary data analysis. GB, KS, AS and JT supervised the work. All authors discussed the results and contributed to the final manuscript. JT is the guarantor of this work.
Funding This overview and secondary data analysis was funded by the National Institute for Health Research (NIHR) Policy Research Programme (PRP) for the Department of Health and Social Care (DHSC). It was funded through the NIHR PRP contract with the EPPI Centre at UCL (Reviews facility to support national policy development and implementation, PR-R6-0113-11003). Any views expressed in this publication are those of the author(s) and not necessarily those of the NHS, the NIHR or the DHSC.
Competing interests None declared.
Provenance and peer review Not commissioned; externally peer reviewed.
Supplemental material This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Read the full text or download the PDF:

IMAGES
COMMENTS
The research papers that proposed new methods in . ... The lifecycle for data analysis will help to manage and organize the tasks connected to big data research and analysis. Data Analytics ...
Data analysis is simply the process of converting the gathered data to meanin gf ul information. Different techniques such as modeling to reach trends, relatio nships, and therefore conclusions to ...
Yonjoo Cho is an associate professor of Instructional Systems Technology focusing on human resource development (HRD) at Indiana University. Her research interests include action learning in organizations, international HRD, and women in leadership. She serves as an associate editor of Human Resource Development Review and served as a board member of the Academy of Human Resource Development ...
This paper presents a variety of data analysis techniques described by various qualitative researchers, such as LeCompte and Schensul, Wolcott, and Miles and Huberman. It further shares several ...
Qualitative methods play a vital role in applied research because they provide deeper examinations and realizations of the human experience (Grinnell & Unrau, 2011; Padgett, 2008; Watkins, 2012; Watkins & Gioia, 2015).Compared to quantitative methods, qualitative research methods help researchers acquire more in-depth information—or the words behind the numbers—for a phenomenon of interest ...
Epistemology and methodology guide methods; these are the specific actions taken during the research process, including data collection, analysis, and reporting (Carter & Little, 2007; Chamberlain, 2015; Jackson et al., 2007). RCA can best be seen as a discrete method, a way of conducting data analysis, able to be applied within a number of ...
The SAGE Handbook of. tive Data AnalysisUwe FlickMapping the FieldData analys. s is the central step in qualitative research. Whatever the data are, it is their analysis that, in a de. isive way, forms the outcomes of the research. Sometimes, data collection is limited to recording and docu-menting naturally occurring ph.
Objectives. The objectives of IJDATS are to promote discussions, deliberations and debates on different data analysis principles, architectures, techniques, methodologies, models, as well as the appropriate strategies and applications for various decision-making environments. Two main data analysis schools of thoughts, in terms of quantitative and qualitative, can intersect, interchange, and ...
Qualitative data analysis is a slow process of moving backwards and forwards between the research question (RQ), theory, and your data from the transcribed interviews while thinking about the context of your study. Although it takes time, it is essential to be systematic and rigorous to create trustworthy findings.
The term "Data analysis" refers to the processing of ... This research contributes to the creation of a research vector on the role of data science in central banking. ... The advanced analytics methods based on machine learning techniques discussed in this paper can be applied to enhance the capabilities of an application in terms of data ...
Definition of research in data analysis: According to LeCompte and Schensul, research data analysis is a process used by researchers to reduce data to a story and interpret it to derive insights. The data analysis process helps reduce a large chunk of data into smaller fragments, which makes sense. Three essential things occur during the data ...
Then, the data analysis methods will be discussed. For doing so, the first six main categories are described briefly. Then, the statistical tools of the most commonly used methods including descriptive, explanatory, and inferential analyses are investigated in detail. Finally, we focus more on qualitative data analysis to get familiar with the ...
The paper outlines an overview about contemporary state of art and trends in the field of data analysis. Collecting, storing, merging and sorting enormous amounts of data have been a major ...
We introduce and discuss general concepts underlying this research, namely on data analysis and agile project management aspects. 2.1 Data Analysis and Visualization. Data analysis is integral across various domains, in particular helping businesses maximize their potential through data mining [].In project management, statistical techniques and artificial intelligence, particularly neural ...
Below we present a step-by-step guide for analysing data for two different types of research questions. The data analysis methods described here are based on basic content analysis as described by Elo and Kyngäs 4 and Graneheim and Lundman, 5 and the integrative review as described by Whittemore and Knafl, 6 but modified to be applicable to ...
Qualitative data analysis methods. Wow, that's a mouthful. If you're new to the world of research, qualitative data analysis can look rather intimidating. So much bulky terminology and so many abstract, fluffy concepts. ... choosing a right method for a paper is always a hard job for a student, this is a useful information, but it would be ...
Statistical methods involved in carrying out a study include planning, designing, collecting data, analysing, drawing meaningful interpretation and reporting of the research findings. The statistical analysis gives meaning to the meaningless numbers, thereby breathing life into a lifeless data. The results and inferences are precise only if ...
Data Analysis Techniques in Research: While various groups, institutions, and professionals may have diverse approaches to data analysis, a universal definition captures its essence. Data analysis involves refining, transforming, and interpreting raw data to derive actionable insights that guide informed decision-making for businesses. ...
Data Analysis. Definition: Data analysis refers to the process of inspecting, cleaning, transforming, and modeling data with the goal of discovering useful information, drawing conclusions, and supporting decision-making. It involves applying various statistical and computational techniques to interpret and derive insights from large datasets.
INTRODUCTION. In an earlier paper, 1 we presented an introduction to using qualitative research methods in pharmacy practice. In this article, we review some principles of the collection, analysis, and management of qualitative data to help pharmacists interested in doing research in their practice to continue their learning in this area.
Feature papers represent the most advanced research with significant potential for high impact in the field. A Feature Paper should be a substantial original Article that involves several techniques or approaches, provides an outlook for future research directions and describes possible research applications. ... Data analysis techniques ...
Below we present a step-by-step guide for analysing data for two different types of research ques-tions. The data analysis methods described here are based on basic content analysis as described by Elo and Kyngas4. Anna Jervaeus, Alfred Nobels alle 23, 141 83 Huddinge, Sweden. Email: [email protected].
Quantitative data analysis is a systematic process of both collecting and evaluating measurable. and verifiable data. It contains a statistical mechanism of assessing or analyzing quantitative ...
For all data processing, statistical analysis and data visualization tasks, RStudio, along with R language (v.4.2.1), was employed. A comprehensive list of the packages used can be found in the ...
For a professional paper, the affiliation is the institution at which the research was conducted. Include both the name of any department and the name of the college, university, or other institution, separated by a comma. Center the affiliation on the next double-spaced line after the author names; when there are multiple affiliations, center ...
We assess whether V s (10-13), V p (), and bulk density ρ b data are consistent with liquid water-saturated pores in the mid-crust (11.5 ± 3.1 to 20 ± 5km) within 50 km of the InSight lander.The mid-crust is one of four robust seismically detectable kilometer-scale layers beneath InSight (10-13) and may be global ().V p and layer thickness have been challenging to obtain for other ...
This paper, therefore, presents a new step-by-step research methodology for using publicly available secondary data to mitigate the risks associated with using secondary qualitative data analysis. We set a clear distinction between overall research methodology and the data analysis method.
Background This overview and analysis of UK datasets was commissioned by the UK government to address concerns about children's consumption of caffeinated energy drinks and their effects on health and behaviour. Methods We searched nine databases for systematic reviews, published between 2013 and July 2021, in English, assessing caffeinated energy drink consumption by people under 18 years ...
Twenty-two (76%) 12,15,17-26,28-34,36-38 of the 29 primary studies documented the methods of data analysis undertaken in each respective study. The most common method was Rasch analysis, ... there is merit in exploring the difference in between PAPERS and COSMIN as further research, taking into consideration the pragmatic and ...